Project
averaged GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
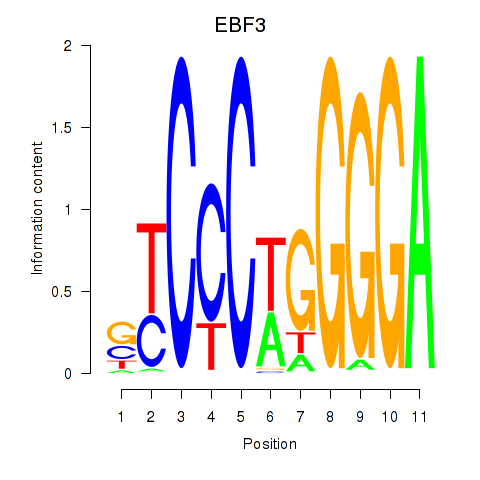
Results for EBF3
Z-value: 1.17
Transcription factors associated with EBF3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EBF3
|
ENSG00000108001.9 | EBF transcription factor 3 |
Activity profile of EBF3 motif
Sorted Z-values of EBF3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_153517473 | 13.64 |
ENST00000368715.1
|
S100A4
|
S100 calcium binding protein A4 |
| chr1_-_113247543 | 12.36 |
ENST00000414971.1
ENST00000534717.1 |
RHOC
|
ras homolog family member C |
| chr7_-_100881041 | 11.28 |
ENST00000412417.1
ENST00000414035.1 |
CLDN15
|
claudin 15 |
| chr8_+_22436248 | 11.08 |
ENST00000308354.7
|
PDLIM2
|
PDZ and LIM domain 2 (mystique) |
| chr7_+_75932863 | 10.96 |
ENST00000429938.1
|
HSPB1
|
heat shock 27kDa protein 1 |
| chr22_-_37640277 | 10.56 |
ENST00000401529.3
ENST00000249071.6 |
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chr1_-_154943212 | 10.09 |
ENST00000368445.5
ENST00000448116.2 ENST00000368449.4 |
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1 |
| chr7_-_6523688 | 9.67 |
ENST00000490996.1
|
KDELR2
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2 |
| chr1_-_154943002 | 9.53 |
ENST00000606391.1
|
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1 |
| chr11_-_67120974 | 9.44 |
ENST00000539074.1
ENST00000312419.3 |
POLD4
|
polymerase (DNA-directed), delta 4, accessory subunit |
| chr6_-_7911042 | 9.44 |
ENST00000379757.4
|
TXNDC5
|
thioredoxin domain containing 5 (endoplasmic reticulum) |
| chr16_+_3068393 | 9.03 |
ENST00000573001.1
|
TNFRSF12A
|
tumor necrosis factor receptor superfamily, member 12A |
| chr19_-_40331345 | 8.66 |
ENST00000597224.1
|
FBL
|
fibrillarin |
| chr11_+_75273101 | 8.23 |
ENST00000533603.1
ENST00000358171.3 ENST00000526242.1 |
SERPINH1
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1, (collagen binding protein 1) |
| chr19_+_50180317 | 7.90 |
ENST00000534465.1
|
PRMT1
|
protein arginine methyltransferase 1 |
| chr11_+_75273246 | 7.77 |
ENST00000526397.1
ENST00000529643.1 ENST00000525492.1 ENST00000530284.1 ENST00000532356.1 ENST00000524558.1 |
SERPINH1
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1, (collagen binding protein 1) |
| chr1_+_32757668 | 7.67 |
ENST00000373548.3
|
HDAC1
|
histone deacetylase 1 |
| chr8_+_22436635 | 7.53 |
ENST00000452226.1
ENST00000397760.4 ENST00000339162.7 ENST00000397761.2 |
PDLIM2
|
PDZ and LIM domain 2 (mystique) |
| chr7_-_137686791 | 7.41 |
ENST00000452463.1
ENST00000330387.6 ENST00000456390.1 |
CREB3L2
|
cAMP responsive element binding protein 3-like 2 |
| chr3_-_142608001 | 7.24 |
ENST00000295992.3
|
PCOLCE2
|
procollagen C-endopeptidase enhancer 2 |
| chr11_+_114128522 | 7.21 |
ENST00000535401.1
|
NNMT
|
nicotinamide N-methyltransferase |
| chr5_+_154393260 | 6.56 |
ENST00000435029.4
|
KIF4B
|
kinesin family member 4B |
| chrX_+_69509927 | 6.45 |
ENST00000374403.3
|
KIF4A
|
kinesin family member 4A |
| chr19_+_50180409 | 6.15 |
ENST00000391851.4
|
PRMT1
|
protein arginine methyltransferase 1 |
| chr19_+_13049413 | 6.14 |
ENST00000316448.5
ENST00000588454.1 |
CALR
|
calreticulin |
| chr3_+_184018352 | 6.14 |
ENST00000435761.1
ENST00000439383.1 |
PSMD2
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 2 |
| chr22_+_24309089 | 5.94 |
ENST00000215770.5
|
DDTL
|
D-dopachrome tautomerase-like |
| chr19_+_42381337 | 5.94 |
ENST00000597454.1
ENST00000444740.2 |
CD79A
|
CD79a molecule, immunoglobulin-associated alpha |
| chr19_+_50180507 | 5.90 |
ENST00000454376.2
ENST00000524771.1 |
PRMT1
|
protein arginine methyltransferase 1 |
| chr19_+_49403562 | 5.89 |
ENST00000407032.1
ENST00000452087.1 ENST00000411700.1 |
NUCB1
|
nucleobindin 1 |
| chr12_-_54694807 | 5.88 |
ENST00000435572.2
|
NFE2
|
nuclear factor, erythroid 2 |
| chr12_-_106641728 | 5.84 |
ENST00000378026.4
|
CKAP4
|
cytoskeleton-associated protein 4 |
| chr2_-_219151487 | 5.64 |
ENST00000444881.1
|
TMBIM1
|
transmembrane BAX inhibitor motif containing 1 |
| chr22_-_37640456 | 5.58 |
ENST00000405484.1
ENST00000441619.1 ENST00000406508.1 |
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chr3_-_145878954 | 5.40 |
ENST00000282903.5
ENST00000360060.3 |
PLOD2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 |
| chr5_-_180669197 | 5.37 |
ENST00000502905.1
|
GNB2L1
|
guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 |
| chr22_-_19466732 | 5.35 |
ENST00000263202.10
ENST00000360834.4 |
UFD1L
|
ubiquitin fusion degradation 1 like (yeast) |
| chr22_-_19466683 | 5.31 |
ENST00000399523.1
ENST00000421968.2 ENST00000447868.1 |
UFD1L
|
ubiquitin fusion degradation 1 like (yeast) |
| chr14_+_101193164 | 5.28 |
ENST00000341267.4
|
DLK1
|
delta-like 1 homolog (Drosophila) |
| chr17_-_40075197 | 5.24 |
ENST00000590770.1
ENST00000590151.1 |
ACLY
|
ATP citrate lyase |
| chr12_-_54694758 | 5.11 |
ENST00000553070.1
|
NFE2
|
nuclear factor, erythroid 2 |
| chr3_+_38179969 | 5.07 |
ENST00000396334.3
ENST00000417037.2 ENST00000424893.1 ENST00000495303.1 ENST00000443433.2 ENST00000421516.1 |
MYD88
|
myeloid differentiation primary response 88 |
| chr11_-_46142948 | 5.07 |
ENST00000257821.4
|
PHF21A
|
PHD finger protein 21A |
| chr3_-_149293990 | 5.05 |
ENST00000472417.1
|
WWTR1
|
WW domain containing transcription regulator 1 |
| chr15_+_74218787 | 5.05 |
ENST00000261921.7
|
LOXL1
|
lysyl oxidase-like 1 |
| chr19_-_45826125 | 5.04 |
ENST00000221476.3
|
CKM
|
creatine kinase, muscle |
| chr14_+_101193246 | 5.01 |
ENST00000331224.6
|
DLK1
|
delta-like 1 homolog (Drosophila) |
| chr17_+_39975455 | 4.96 |
ENST00000455106.1
|
FKBP10
|
FK506 binding protein 10, 65 kDa |
| chr17_-_40075219 | 4.91 |
ENST00000537919.1
ENST00000352035.2 ENST00000353196.1 ENST00000393896.2 |
ACLY
|
ATP citrate lyase |
| chr7_+_100464760 | 4.89 |
ENST00000200457.4
|
TRIP6
|
thyroid hormone receptor interactor 6 |
| chr11_+_20385327 | 4.87 |
ENST00000451739.2
ENST00000532505.1 |
HTATIP2
|
HIV-1 Tat interactive protein 2, 30kDa |
| chr5_-_180669236 | 4.85 |
ENST00000507756.1
|
GNB2L1
|
guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 |
| chr19_+_33865218 | 4.84 |
ENST00000585933.2
|
CEBPG
|
CCAAT/enhancer binding protein (C/EBP), gamma |
| chr11_-_67169253 | 4.79 |
ENST00000527663.1
ENST00000312989.7 |
PPP1CA
|
protein phosphatase 1, catalytic subunit, alpha isozyme |
| chr14_-_106725723 | 4.76 |
ENST00000390609.2
|
IGHV3-23
|
immunoglobulin heavy variable 3-23 |
| chr7_+_77167376 | 4.76 |
ENST00000435495.2
|
PTPN12
|
protein tyrosine phosphatase, non-receptor type 12 |
| chr1_+_26869597 | 4.76 |
ENST00000530003.1
|
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1 |
| chr8_+_145582217 | 4.64 |
ENST00000530047.1
ENST00000527078.1 |
SLC52A2
|
solute carrier family 52 (riboflavin transporter), member 2 |
| chr22_-_24316648 | 4.61 |
ENST00000403754.3
ENST00000430101.2 ENST00000398344.4 |
DDT
|
D-dopachrome tautomerase |
| chr3_-_67705006 | 4.61 |
ENST00000492795.1
ENST00000493112.1 ENST00000307227.5 |
SUCLG2
|
succinate-CoA ligase, GDP-forming, beta subunit |
| chr19_-_39108552 | 4.56 |
ENST00000591517.1
|
MAP4K1
|
mitogen-activated protein kinase kinase kinase kinase 1 |
| chr16_+_69958887 | 4.55 |
ENST00000568684.1
|
WWP2
|
WW domain containing E3 ubiquitin protein ligase 2 |
| chr14_-_106586656 | 4.54 |
ENST00000390602.2
|
IGHV3-13
|
immunoglobulin heavy variable 3-13 |
| chr19_-_39108643 | 4.50 |
ENST00000396857.2
|
MAP4K1
|
mitogen-activated protein kinase kinase kinase kinase 1 |
| chrX_-_152989798 | 4.48 |
ENST00000441714.1
ENST00000442093.1 ENST00000429550.1 ENST00000345046.6 |
BCAP31
|
B-cell receptor-associated protein 31 |
| chr3_-_142607740 | 4.47 |
ENST00000485766.1
|
PCOLCE2
|
procollagen C-endopeptidase enhancer 2 |
| chr7_-_6523755 | 4.45 |
ENST00000436575.1
ENST00000258739.4 |
DAGLB
KDELR2
|
diacylglycerol lipase, beta KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2 |
| chr7_-_132766818 | 4.38 |
ENST00000262570.5
|
CHCHD3
|
coiled-coil-helix-coiled-coil-helix domain containing 3 |
| chr6_-_45983581 | 4.38 |
ENST00000339561.6
|
CLIC5
|
chloride intracellular channel 5 |
| chr3_-_189840223 | 4.37 |
ENST00000427335.2
|
LEPREL1
|
leprecan-like 1 |
| chr1_-_206785789 | 4.34 |
ENST00000437518.1
ENST00000367114.3 |
EIF2D
|
eukaryotic translation initiation factor 2D |
| chr14_-_24664776 | 4.34 |
ENST00000530468.1
ENST00000528010.1 ENST00000396854.4 ENST00000524835.1 ENST00000261789.4 ENST00000525592.1 |
TM9SF1
|
transmembrane 9 superfamily member 1 |
| chr17_+_39975544 | 4.33 |
ENST00000544340.1
|
FKBP10
|
FK506 binding protein 10, 65 kDa |
| chr7_-_99698338 | 4.24 |
ENST00000354230.3
ENST00000425308.1 |
MCM7
|
minichromosome maintenance complex component 7 |
| chr8_+_145582231 | 4.24 |
ENST00000526338.1
ENST00000402965.1 ENST00000534725.1 ENST00000532887.1 ENST00000329994.2 |
SLC52A2
|
solute carrier family 52 (riboflavin transporter), member 2 |
| chr10_-_102089729 | 4.23 |
ENST00000465680.2
|
PKD2L1
|
polycystic kidney disease 2-like 1 |
| chr3_-_52273098 | 4.23 |
ENST00000499914.2
ENST00000305533.5 ENST00000597542.1 |
TWF2
TLR9
|
twinfilin actin-binding protein 2 toll-like receptor 9 |
| chr2_-_188378368 | 4.21 |
ENST00000392365.1
ENST00000435414.1 |
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr7_-_44180673 | 4.18 |
ENST00000457314.1
ENST00000447951.1 ENST00000431007.1 |
MYL7
|
myosin, light chain 7, regulatory |
| chr8_+_145582633 | 4.17 |
ENST00000540505.1
|
SLC52A2
|
solute carrier family 52 (riboflavin transporter), member 2 |
| chr16_+_33605231 | 4.16 |
ENST00000570121.2
|
IGHV3OR16-12
|
immunoglobulin heavy variable 3/OR16-12 (non-functional) |
| chr11_+_60223225 | 4.16 |
ENST00000524807.1
ENST00000345732.4 |
MS4A1
|
membrane-spanning 4-domains, subfamily A, member 1 |
| chr15_-_20193370 | 4.16 |
ENST00000558565.2
|
IGHV3OR15-7
|
immunoglobulin heavy variable 3/OR15-7 (pseudogene) |
| chr11_+_60223312 | 4.15 |
ENST00000532491.1
ENST00000532073.1 ENST00000534668.1 ENST00000528313.1 ENST00000533306.1 |
MS4A1
|
membrane-spanning 4-domains, subfamily A, member 1 |
| chr7_-_132766800 | 4.15 |
ENST00000542753.1
ENST00000448878.1 |
CHCHD3
|
coiled-coil-helix-coiled-coil-helix domain containing 3 |
| chr11_-_64545941 | 4.12 |
ENST00000377387.1
|
SF1
|
splicing factor 1 |
| chrX_+_155110956 | 4.05 |
ENST00000286448.6
ENST00000262640.6 ENST00000460621.1 |
VAMP7
|
vesicle-associated membrane protein 7 |
| chr14_+_105952648 | 3.99 |
ENST00000330233.7
|
CRIP1
|
cysteine-rich protein 1 (intestinal) |
| chr7_-_44180884 | 3.94 |
ENST00000458240.1
ENST00000223364.3 |
MYL7
|
myosin, light chain 7, regulatory |
| chr14_-_24664540 | 3.91 |
ENST00000530563.1
ENST00000528895.1 ENST00000528669.1 ENST00000532632.1 |
TM9SF1
|
transmembrane 9 superfamily member 1 |
| chrX_+_48644962 | 3.87 |
ENST00000376670.3
ENST00000376665.3 |
GATA1
|
GATA binding protein 1 (globin transcription factor 1) |
| chr14_-_107131560 | 3.86 |
ENST00000390632.2
|
IGHV3-66
|
immunoglobulin heavy variable 3-66 |
| chr11_+_20385231 | 3.83 |
ENST00000530266.1
ENST00000421577.2 ENST00000443524.2 ENST00000419348.2 |
HTATIP2
|
HIV-1 Tat interactive protein 2, 30kDa |
| chr6_+_43737939 | 3.77 |
ENST00000372067.3
|
VEGFA
|
vascular endothelial growth factor A |
| chr6_-_10419871 | 3.74 |
ENST00000319516.4
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) |
| chr7_+_77167343 | 3.73 |
ENST00000433369.2
ENST00000415482.2 |
PTPN12
|
protein tyrosine phosphatase, non-receptor type 12 |
| chr9_-_139581848 | 3.72 |
ENST00000538402.1
ENST00000371694.3 |
AGPAT2
|
1-acylglycerol-3-phosphate O-acyltransferase 2 |
| chr19_-_39108568 | 3.70 |
ENST00000586296.1
|
MAP4K1
|
mitogen-activated protein kinase kinase kinase kinase 1 |
| chr20_-_48732472 | 3.65 |
ENST00000340309.3
ENST00000415862.2 ENST00000371677.3 ENST00000420027.2 |
UBE2V1
|
ubiquitin-conjugating enzyme E2 variant 1 |
| chr1_+_11072696 | 3.64 |
ENST00000240185.3
ENST00000476201.1 |
TARDBP
|
TAR DNA binding protein |
| chr7_-_100240328 | 3.63 |
ENST00000462107.1
|
TFR2
|
transferrin receptor 2 |
| chr11_-_65381643 | 3.55 |
ENST00000309100.3
ENST00000529839.1 ENST00000526293.1 |
MAP3K11
|
mitogen-activated protein kinase kinase kinase 11 |
| chr11_+_1860200 | 3.55 |
ENST00000381911.1
|
TNNI2
|
troponin I type 2 (skeletal, fast) |
| chr10_+_73724123 | 3.53 |
ENST00000373115.4
|
CHST3
|
carbohydrate (chondroitin 6) sulfotransferase 3 |
| chr14_+_105953246 | 3.49 |
ENST00000392531.3
|
CRIP1
|
cysteine-rich protein 1 (intestinal) |
| chr11_+_1942580 | 3.48 |
ENST00000381558.1
|
TNNT3
|
troponin T type 3 (skeletal, fast) |
| chr11_-_64546202 | 3.47 |
ENST00000377390.3
ENST00000227503.9 ENST00000377394.3 ENST00000422298.2 ENST00000334944.5 |
SF1
|
splicing factor 1 |
| chr9_+_135854091 | 3.46 |
ENST00000450530.1
ENST00000534944.1 |
GFI1B
|
growth factor independent 1B transcription repressor |
| chr6_+_31555045 | 3.45 |
ENST00000396101.3
ENST00000490742.1 |
LST1
|
leukocyte specific transcript 1 |
| chr3_-_49170522 | 3.44 |
ENST00000418109.1
|
LAMB2
|
laminin, beta 2 (laminin S) |
| chr16_+_67233007 | 3.41 |
ENST00000360833.1
ENST00000393997.2 |
ELMO3
|
engulfment and cell motility 3 |
| chr11_+_128562372 | 3.37 |
ENST00000344954.6
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr16_-_33647696 | 3.36 |
ENST00000558425.1
ENST00000569103.2 |
RP11-812E19.9
|
Uncharacterized protein |
| chr1_+_153330322 | 3.36 |
ENST00000368738.3
|
S100A9
|
S100 calcium binding protein A9 |
| chr11_-_33744487 | 3.34 |
ENST00000426650.2
|
CD59
|
CD59 molecule, complement regulatory protein |
| chr2_-_219134822 | 3.32 |
ENST00000444053.1
ENST00000248450.4 |
AAMP
|
angio-associated, migratory cell protein |
| chr19_+_46010674 | 3.28 |
ENST00000245932.6
ENST00000592139.1 ENST00000590603.1 |
VASP
|
vasodilator-stimulated phosphoprotein |
| chr1_-_222885770 | 3.26 |
ENST00000355727.2
ENST00000340020.6 |
AIDA
|
axin interactor, dorsalization associated |
| chr5_-_133340326 | 3.26 |
ENST00000425992.1
ENST00000395044.3 ENST00000395047.2 |
VDAC1
|
voltage-dependent anion channel 1 |
| chr4_+_37892682 | 3.25 |
ENST00000508802.1
ENST00000261439.4 ENST00000402522.1 |
TBC1D1
|
TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1 |
| chr7_-_100881109 | 3.23 |
ENST00000308344.5
|
CLDN15
|
claudin 15 |
| chr11_+_2466218 | 3.22 |
ENST00000155840.5
|
KCNQ1
|
potassium voltage-gated channel, KQT-like subfamily, member 1 |
| chr17_-_80023401 | 3.20 |
ENST00000354321.7
ENST00000306796.5 |
DUS1L
|
dihydrouridine synthase 1-like (S. cerevisiae) |
| chr3_-_16524357 | 3.19 |
ENST00000432519.1
|
RFTN1
|
raftlin, lipid raft linker 1 |
| chr19_+_41107249 | 3.19 |
ENST00000396819.3
|
LTBP4
|
latent transforming growth factor beta binding protein 4 |
| chr14_+_105953204 | 3.18 |
ENST00000409393.2
|
CRIP1
|
cysteine-rich protein 1 (intestinal) |
| chr14_-_106994333 | 3.17 |
ENST00000390624.2
|
IGHV3-48
|
immunoglobulin heavy variable 3-48 |
| chr9_-_111775772 | 3.15 |
ENST00000325580.6
ENST00000374593.4 ENST00000374595.4 ENST00000325551.4 |
CTNNAL1
|
catenin (cadherin-associated protein), alpha-like 1 |
| chr11_+_1860682 | 3.14 |
ENST00000381906.1
|
TNNI2
|
troponin I type 2 (skeletal, fast) |
| chr19_+_42381173 | 3.12 |
ENST00000221972.3
|
CD79A
|
CD79a molecule, immunoglobulin-associated alpha |
| chr18_-_23670546 | 3.11 |
ENST00000542743.1
ENST00000545952.1 ENST00000539849.1 ENST00000415083.2 |
SS18
|
synovial sarcoma translocation, chromosome 18 |
| chrX_-_74376108 | 3.10 |
ENST00000339447.4
ENST00000373394.3 ENST00000529949.1 ENST00000534524.1 ENST00000253577.3 |
ABCB7
|
ATP-binding cassette, sub-family B (MDR/TAP), member 7 |
| chr3_-_49170405 | 3.08 |
ENST00000305544.4
ENST00000494831.1 |
LAMB2
|
laminin, beta 2 (laminin S) |
| chrX_-_48827976 | 3.07 |
ENST00000218176.3
|
KCND1
|
potassium voltage-gated channel, Shal-related subfamily, member 1 |
| chr8_+_22446763 | 3.03 |
ENST00000450780.2
ENST00000430850.2 ENST00000447849.1 |
AC037459.4
|
Uncharacterized protein |
| chr9_-_139581875 | 3.01 |
ENST00000371696.2
|
AGPAT2
|
1-acylglycerol-3-phosphate O-acyltransferase 2 |
| chr22_-_43411106 | 3.00 |
ENST00000453643.1
ENST00000263246.3 ENST00000337959.4 |
PACSIN2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr14_-_106816253 | 2.96 |
ENST00000390615.2
|
IGHV3-33
|
immunoglobulin heavy variable 3-33 |
| chr4_-_140223614 | 2.95 |
ENST00000394223.1
|
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr11_-_6640585 | 2.93 |
ENST00000533371.1
ENST00000528657.1 ENST00000436873.2 ENST00000299427.6 |
TPP1
|
tripeptidyl peptidase I |
| chr12_-_51663728 | 2.91 |
ENST00000603864.1
ENST00000605426.1 |
SMAGP
|
small cell adhesion glycoprotein |
| chr1_+_161719552 | 2.90 |
ENST00000367943.4
|
DUSP12
|
dual specificity phosphatase 12 |
| chr2_+_27301435 | 2.89 |
ENST00000380320.4
|
EMILIN1
|
elastin microfibril interfacer 1 |
| chr2_-_174830430 | 2.86 |
ENST00000310015.6
ENST00000455789.2 |
SP3
|
Sp3 transcription factor |
| chr2_+_46769798 | 2.85 |
ENST00000238738.4
|
RHOQ
|
ras homolog family member Q |
| chr14_-_106552755 | 2.84 |
ENST00000390600.2
|
IGHV3-9
|
immunoglobulin heavy variable 3-9 |
| chr2_+_119981384 | 2.81 |
ENST00000393108.2
ENST00000354888.5 ENST00000450943.2 ENST00000393110.2 ENST00000393106.2 ENST00000409811.1 ENST00000393107.2 |
STEAP3
|
STEAP family member 3, metalloreductase |
| chr18_-_48723690 | 2.78 |
ENST00000406189.3
|
MEX3C
|
mex-3 RNA binding family member C |
| chr17_-_62009702 | 2.76 |
ENST00000006750.3
|
CD79B
|
CD79b molecule, immunoglobulin-associated beta |
| chr14_-_106518922 | 2.76 |
ENST00000390598.2
|
IGHV3-7
|
immunoglobulin heavy variable 3-7 |
| chr5_-_146833222 | 2.75 |
ENST00000534907.1
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr1_+_2487800 | 2.65 |
ENST00000355716.4
|
TNFRSF14
|
tumor necrosis factor receptor superfamily, member 14 |
| chr11_+_1860832 | 2.64 |
ENST00000252898.7
|
TNNI2
|
troponin I type 2 (skeletal, fast) |
| chr14_-_106791536 | 2.63 |
ENST00000390613.2
|
IGHV3-30
|
immunoglobulin heavy variable 3-30 |
| chr17_-_62009621 | 2.62 |
ENST00000349817.2
ENST00000392795.3 |
CD79B
|
CD79b molecule, immunoglobulin-associated beta |
| chr11_-_72432950 | 2.61 |
ENST00000426523.1
ENST00000429686.1 |
ARAP1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr16_-_28506840 | 2.59 |
ENST00000569430.1
|
CLN3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr5_-_142783694 | 2.58 |
ENST00000394466.2
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr5_-_180668605 | 2.56 |
ENST00000504128.1
|
GNB2L1
|
guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 |
| chr1_+_153747746 | 2.53 |
ENST00000368661.3
|
SLC27A3
|
solute carrier family 27 (fatty acid transporter), member 3 |
| chr16_+_67233412 | 2.53 |
ENST00000477898.1
|
ELMO3
|
engulfment and cell motility 3 |
| chrX_-_10645773 | 2.52 |
ENST00000453318.2
|
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr16_-_89007491 | 2.48 |
ENST00000327483.5
ENST00000564416.1 |
CBFA2T3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3 |
| chr19_+_984313 | 2.47 |
ENST00000251289.5
ENST00000587001.2 ENST00000607440.1 |
WDR18
|
WD repeat domain 18 |
| chr19_-_6591113 | 2.44 |
ENST00000423145.3
ENST00000245903.3 |
CD70
|
CD70 molecule |
| chr20_+_62496596 | 2.44 |
ENST00000369927.4
ENST00000346249.4 ENST00000348257.5 ENST00000352482.4 ENST00000351424.4 ENST00000217121.5 ENST00000358548.4 |
TPD52L2
|
tumor protein D52-like 2 |
| chr14_-_23540826 | 2.40 |
ENST00000357481.2
|
ACIN1
|
apoptotic chromatin condensation inducer 1 |
| chr3_-_88108192 | 2.32 |
ENST00000309534.6
|
CGGBP1
|
CGG triplet repeat binding protein 1 |
| chr17_+_73663402 | 2.31 |
ENST00000355423.3
|
SAP30BP
|
SAP30 binding protein |
| chr17_-_79881408 | 2.30 |
ENST00000392366.3
|
MAFG
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog G |
| chr10_+_99332198 | 2.27 |
ENST00000307518.5
ENST00000298808.5 ENST00000370655.1 |
ANKRD2
|
ankyrin repeat domain 2 (stretch responsive muscle) |
| chr7_-_105752651 | 2.24 |
ENST00000470347.1
ENST00000455385.2 |
SYPL1
|
synaptophysin-like 1 |
| chr10_+_114709999 | 2.23 |
ENST00000355995.4
ENST00000545257.1 ENST00000543371.1 ENST00000536810.1 ENST00000355717.4 ENST00000538897.1 ENST00000534894.1 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr2_-_27632390 | 2.21 |
ENST00000350803.4
ENST00000344034.4 |
PPM1G
|
protein phosphatase, Mg2+/Mn2+ dependent, 1G |
| chr7_-_6098770 | 2.18 |
ENST00000536084.1
ENST00000446699.1 ENST00000199389.6 |
EIF2AK1
|
eukaryotic translation initiation factor 2-alpha kinase 1 |
| chr6_+_43738444 | 2.16 |
ENST00000324450.6
ENST00000417285.2 ENST00000413642.3 ENST00000372055.4 ENST00000482630.2 ENST00000425836.2 ENST00000372064.4 ENST00000372077.4 ENST00000519767.1 |
VEGFA
|
vascular endothelial growth factor A |
| chr5_+_34915444 | 2.14 |
ENST00000336767.5
|
BRIX1
|
BRX1, biogenesis of ribosomes, homolog (S. cerevisiae) |
| chr17_+_75315534 | 2.14 |
ENST00000590294.1
ENST00000329047.8 |
SEPT9
|
septin 9 |
| chr5_-_138725594 | 2.14 |
ENST00000302125.8
|
MZB1
|
marginal zone B and B1 cell-specific protein |
| chr6_+_7107999 | 2.13 |
ENST00000491191.1
ENST00000379938.2 ENST00000471433.1 |
RREB1
|
ras responsive element binding protein 1 |
| chr13_+_29233218 | 2.12 |
ENST00000380842.4
|
POMP
|
proteasome maturation protein |
| chr7_+_48128194 | 2.11 |
ENST00000416681.1
ENST00000331803.4 ENST00000432131.1 |
UPP1
|
uridine phosphorylase 1 |
| chr14_+_24590560 | 2.10 |
ENST00000558325.1
|
RP11-468E2.6
|
RP11-468E2.6 |
| chr12_-_7261772 | 2.10 |
ENST00000545280.1
ENST00000543933.1 ENST00000545337.1 ENST00000544702.1 ENST00000266542.4 |
C1RL
|
complement component 1, r subcomponent-like |
| chr5_-_138725560 | 2.09 |
ENST00000412103.2
ENST00000457570.2 |
MZB1
|
marginal zone B and B1 cell-specific protein |
| chr9_+_134103496 | 2.08 |
ENST00000498010.1
ENST00000476004.1 ENST00000528406.1 |
NUP214
|
nucleoporin 214kDa |
| chr14_-_65569244 | 2.08 |
ENST00000557277.1
ENST00000556892.1 |
MAX
|
MYC associated factor X |
| chr1_-_156828810 | 2.07 |
ENST00000368195.3
|
INSRR
|
insulin receptor-related receptor |
| chr10_-_102090243 | 2.06 |
ENST00000338519.3
ENST00000353274.3 ENST00000318222.3 |
PKD2L1
|
polycystic kidney disease 2-like 1 |
| chr2_+_89986318 | 2.05 |
ENST00000491977.1
|
IGKV2D-29
|
immunoglobulin kappa variable 2D-29 |
| chr1_+_32084794 | 2.05 |
ENST00000373705.1
|
HCRTR1
|
hypocretin (orexin) receptor 1 |
| chr7_+_48128316 | 2.05 |
ENST00000341253.4
|
UPP1
|
uridine phosphorylase 1 |
| chrX_-_153637612 | 2.04 |
ENST00000369807.1
ENST00000369808.3 |
DNASE1L1
|
deoxyribonuclease I-like 1 |
| chr1_+_110198944 | 2.03 |
ENST00000369833.1
|
GSTM4
|
glutathione S-transferase mu 4 |
| chr9_-_130635741 | 2.03 |
ENST00000223836.10
|
AK1
|
adenylate kinase 1 |
| chr2_-_90538397 | 2.00 |
ENST00000443397.3
|
RP11-685N3.1
|
Uncharacterized protein |
| chr16_+_33020496 | 2.00 |
ENST00000565407.2
|
IGHV3OR16-8
|
immunoglobulin heavy variable 3/OR16-8 (non-functional) |
| chrX_+_105937068 | 2.00 |
ENST00000324342.3
|
RNF128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr1_-_1310530 | 1.97 |
ENST00000338370.3
ENST00000321751.5 ENST00000378853.3 |
AURKAIP1
|
aurora kinase A interacting protein 1 |
| chr10_+_99332529 | 1.96 |
ENST00000455090.1
|
ANKRD2
|
ankyrin repeat domain 2 (stretch responsive muscle) |
Network of associatons between targets according to the STRING database.
First level regulatory network of EBF3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 20.0 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 3.3 | 13.1 | GO:0032218 | riboflavin transport(GO:0032218) |
| 3.1 | 9.3 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 2.7 | 11.0 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 2.2 | 2.2 | GO:0046984 | regulation of hemoglobin biosynthetic process(GO:0046984) |
| 2.2 | 8.7 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 2.1 | 10.7 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 2.1 | 12.4 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 2.0 | 22.1 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 2.0 | 15.7 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 1.8 | 5.4 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 1.6 | 4.8 | GO:0060319 | regulation of primitive erythrocyte differentiation(GO:0010725) eosinophil fate commitment(GO:0035854) primitive erythrocyte differentiation(GO:0060319) |
| 1.5 | 7.7 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 1.5 | 7.6 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 1.4 | 5.6 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) |
| 1.4 | 2.8 | GO:0003415 | chondrocyte hypertrophy(GO:0003415) |
| 1.4 | 4.1 | GO:1903595 | positive regulation of histamine secretion by mast cell(GO:1903595) |
| 1.3 | 6.5 | GO:0072313 | metanephric glomerular epithelium development(GO:0072244) metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 1.3 | 14.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 1.2 | 3.6 | GO:2000909 | regulation of cholesterol import(GO:0060620) negative regulation of cholesterol import(GO:0060621) regulation of sterol import(GO:2000909) negative regulation of sterol import(GO:2000910) |
| 1.2 | 4.8 | GO:0036496 | regulation of translational initiation by eIF2 alpha dephosphorylation(GO:0036496) |
| 1.1 | 9.0 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 1.1 | 3.4 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 1.1 | 4.3 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 1.1 | 4.2 | GO:1901895 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) negative regulation of calcium-transporting ATPase activity(GO:1901895) |
| 1.0 | 5.2 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 1.0 | 4.2 | GO:0006218 | uridine catabolic process(GO:0006218) uridine metabolic process(GO:0046108) |
| 1.0 | 6.1 | GO:0002501 | peptide antigen assembly with MHC protein complex(GO:0002501) |
| 1.0 | 5.1 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 1.0 | 14.6 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.9 | 3.7 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.9 | 4.6 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.9 | 4.5 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.8 | 4.2 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 0.8 | 3.3 | GO:0001971 | negative regulation of activation of membrane attack complex(GO:0001971) |
| 0.8 | 3.1 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.8 | 3.8 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.7 | 3.6 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.7 | 8.5 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.7 | 4.9 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.7 | 4.8 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.7 | 5.9 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.6 | 2.6 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.6 | 10.2 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.6 | 5.0 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.6 | 2.4 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.6 | 3.6 | GO:0098707 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.6 | 1.8 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.6 | 7.2 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.6 | 1.2 | GO:0060769 | positive regulation of epithelial cell proliferation involved in prostate gland development(GO:0060769) |
| 0.6 | 13.0 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.6 | 2.8 | GO:0033216 | ferric iron import(GO:0033216) ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.5 | 15.8 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.5 | 4.3 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.5 | 3.2 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.5 | 3.2 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.5 | 4.2 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.5 | 19.6 | GO:0007176 | regulation of epidermal growth factor-activated receptor activity(GO:0007176) |
| 0.5 | 4.4 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.5 | 2.9 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.5 | 1.0 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.5 | 2.9 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.5 | 4.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.5 | 5.0 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.5 | 5.5 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.4 | 6.7 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.4 | 1.8 | GO:0010909 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.4 | 1.3 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.4 | 1.3 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.4 | 1.7 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.4 | 0.9 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.4 | 1.2 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.4 | 3.2 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.4 | 0.8 | GO:1903423 | positive regulation of synaptic vesicle endocytosis(GO:1900244) positive regulation of synaptic vesicle recycling(GO:1903423) |
| 0.4 | 3.5 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.4 | 3.1 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.4 | 9.5 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.4 | 2.6 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.4 | 5.2 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.4 | 3.6 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.4 | 2.5 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.3 | 2.4 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.3 | 1.4 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.3 | 3.0 | GO:0018916 | nitrobenzene metabolic process(GO:0018916) |
| 0.3 | 1.0 | GO:0002818 | intracellular defense response(GO:0002818) |
| 0.3 | 1.0 | GO:2000157 | regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.3 | 1.0 | GO:2000977 | regulation of mitotic cell cycle, embryonic(GO:0009794) midbrain-hindbrain boundary morphogenesis(GO:0021555) trochlear nerve development(GO:0021558) mitotic cell cycle, embryonic(GO:0045448) regulation of timing of neuron differentiation(GO:0060164) regulation of forebrain neuron differentiation(GO:2000977) |
| 0.3 | 3.0 | GO:0030238 | male sex determination(GO:0030238) |
| 0.3 | 1.0 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 0.3 | 1.6 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.3 | 0.3 | GO:0090027 | negative regulation of monocyte chemotaxis(GO:0090027) |
| 0.3 | 8.1 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.3 | 3.1 | GO:0015886 | heme transport(GO:0015886) |
| 0.3 | 9.4 | GO:0032201 | telomere maintenance via semi-conservative replication(GO:0032201) |
| 0.3 | 3.0 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.3 | 3.3 | GO:0048262 | determination of dorsal/ventral asymmetry(GO:0048262) |
| 0.3 | 20.1 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.3 | 1.1 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.3 | 0.5 | GO:0055073 | cadmium ion homeostasis(GO:0055073) |
| 0.3 | 0.5 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.2 | 2.0 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.2 | 1.0 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.2 | 1.7 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.2 | 9.4 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.2 | 0.7 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.2 | 1.4 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.2 | 11.0 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) |
| 0.2 | 0.9 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.2 | 21.0 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.2 | 1.7 | GO:0002326 | B cell lineage commitment(GO:0002326) immunoglobulin V(D)J recombination(GO:0033152) |
| 0.2 | 14.4 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.2 | 3.2 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.2 | 1.4 | GO:0006642 | triglyceride mobilization(GO:0006642) plasma membrane long-chain fatty acid transport(GO:0015911) |
| 0.2 | 2.5 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.2 | 1.0 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.2 | 3.4 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.2 | 3.7 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.2 | 0.6 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.2 | 0.5 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.2 | 8.2 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.2 | 1.8 | GO:0021957 | corticospinal tract morphogenesis(GO:0021957) |
| 0.2 | 2.9 | GO:0090005 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.2 | 2.4 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.2 | 0.5 | GO:1904954 | Spemann organizer formation(GO:0060061) canonical Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation(GO:1904954) |
| 0.2 | 0.5 | GO:0039526 | suppression by virus of host apoptotic process(GO:0019050) modulation by virus of host molecular function(GO:0039506) suppression by virus of host molecular function(GO:0039507) suppression by virus of host catalytic activity(GO:0039513) modulation by virus of host catalytic activity(GO:0039516) modulation by virus of host apoptotic process(GO:0039526) suppression by virus of host cysteine-type endopeptidase activity involved in apoptotic process(GO:0039650) negative regulation by symbiont of host catalytic activity(GO:0052053) negative regulation by symbiont of host molecular function(GO:0052056) modulation by symbiont of host catalytic activity(GO:0052148) |
| 0.2 | 1.8 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.2 | 1.6 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.2 | 1.6 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.2 | 1.1 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.2 | 3.1 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.2 | 0.9 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.2 | 1.2 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 4.6 | GO:0042438 | melanin metabolic process(GO:0006582) melanin biosynthetic process(GO:0042438) |
| 0.1 | 2.9 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 1.0 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.1 | 2.6 | GO:0045187 | regulation of circadian sleep/wake cycle(GO:0042749) regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.1 | 3.5 | GO:0051569 | regulation of histone H3-K4 methylation(GO:0051569) |
| 0.1 | 1.7 | GO:1902659 | regulation of glucose mediated signaling pathway(GO:1902659) |
| 0.1 | 11.5 | GO:0033275 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.1 | 1.5 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.1 | 0.4 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.1 | 10.2 | GO:0042100 | B cell proliferation(GO:0042100) |
| 0.1 | 2.1 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 0.3 | GO:0044550 | secondary metabolite biosynthetic process(GO:0044550) |
| 0.1 | 0.4 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.1 | 1.1 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.1 | 3.3 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.1 | 1.8 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 1.2 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.1 | 1.0 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.1 | 0.3 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.1 | 1.5 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.1 | 1.3 | GO:0034384 | high-density lipoprotein particle clearance(GO:0034384) |
| 0.1 | 5.0 | GO:0035904 | aorta development(GO:0035904) |
| 0.1 | 0.8 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 0.4 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.1 | 2.1 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.1 | 1.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 2.6 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.1 | 1.1 | GO:0043401 | steroid hormone mediated signaling pathway(GO:0043401) |
| 0.1 | 0.4 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.1 | 1.1 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.1 | 7.5 | GO:0002479 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent(GO:0002479) |
| 0.1 | 1.2 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.1 | 0.3 | GO:0010816 | neuropeptide catabolic process(GO:0010813) substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.1 | 1.4 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 11.8 | GO:0001837 | epithelial to mesenchymal transition(GO:0001837) |
| 0.1 | 0.3 | GO:0002027 | regulation of heart rate(GO:0002027) |
| 0.1 | 1.8 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.1 | 1.5 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 | 2.4 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.1 | 9.8 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.1 | 8.0 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.1 | 0.5 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.1 | 0.6 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.1 | 1.0 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.1 | 1.0 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 1.3 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 2.9 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.1 | 0.2 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.1 | 2.1 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 1.5 | GO:0030100 | regulation of endocytosis(GO:0030100) |
| 0.1 | 1.2 | GO:0042246 | tissue regeneration(GO:0042246) |
| 0.0 | 3.5 | GO:0048678 | response to axon injury(GO:0048678) |
| 0.0 | 2.8 | GO:0018196 | peptidyl-asparagine modification(GO:0018196) protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 2.2 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 2.3 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.5 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 2.0 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 | 0.2 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.0 | 0.6 | GO:0005984 | disaccharide metabolic process(GO:0005984) |
| 0.0 | 0.3 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 2.9 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 3.4 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 2.0 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) |
| 0.0 | 0.8 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.2 | GO:0044546 | NLRP3 inflammasome complex assembly(GO:0044546) |
| 0.0 | 3.3 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 0.2 | GO:0099624 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) atrial cardiac muscle cell membrane repolarization(GO:0099624) |
| 0.0 | 1.3 | GO:0042733 | embryonic digit morphogenesis(GO:0042733) |
| 0.0 | 2.4 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 1.5 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.3 | GO:0034447 | very-low-density lipoprotein particle clearance(GO:0034447) |
| 0.0 | 0.8 | GO:0048147 | negative regulation of fibroblast proliferation(GO:0048147) |
| 0.0 | 1.7 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.1 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.4 | GO:0050858 | negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) |
| 0.0 | 0.4 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.0 | 0.5 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 2.1 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.3 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.0 | 0.4 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 2.4 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.9 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 4.4 | GO:0051170 | nuclear import(GO:0051170) |
| 0.0 | 0.3 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.1 | GO:0019359 | nicotinamide nucleotide biosynthetic process(GO:0019359) pyridine nucleotide biosynthetic process(GO:0019363) |
| 0.0 | 0.9 | GO:0048704 | embryonic skeletal system morphogenesis(GO:0048704) |
| 0.0 | 0.9 | GO:0050672 | negative regulation of mononuclear cell proliferation(GO:0032945) negative regulation of lymphocyte proliferation(GO:0050672) negative regulation of leukocyte proliferation(GO:0070664) |
| 0.0 | 1.4 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 3.9 | GO:0006364 | rRNA processing(GO:0006364) |
| 0.0 | 0.5 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.7 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 2.3 | GO:0006909 | phagocytosis(GO:0006909) |
| 0.0 | 1.1 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 | 0.2 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 19.6 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 2.9 | 8.7 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 2.4 | 9.4 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 2.2 | 6.5 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 2.1 | 14.4 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 2.0 | 5.9 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 1.2 | 8.5 | GO:0061617 | MICOS complex(GO:0061617) |
| 1.1 | 20.0 | GO:0034709 | methylosome(GO:0034709) |
| 1.0 | 4.9 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.8 | 6.1 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.8 | 4.6 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.7 | 4.5 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.7 | 3.6 | GO:0035370 | UBC13-UEV1A complex(GO:0035370) |
| 0.7 | 3.6 | GO:0005726 | perichromatin fibrils(GO:0005726) interchromatin granule(GO:0035061) |
| 0.7 | 4.2 | GO:0032009 | early phagosome(GO:0032009) |
| 0.7 | 2.1 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.5 | 12.8 | GO:0005861 | troponin complex(GO:0005861) |
| 0.5 | 4.8 | GO:0042587 | glycogen granule(GO:0042587) PTW/PP1 phosphatase complex(GO:0072357) |
| 0.5 | 3.6 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.4 | 20.1 | GO:0019814 | immunoglobulin complex(GO:0019814) |
| 0.4 | 1.4 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.3 | 12.0 | GO:0032420 | stereocilium(GO:0032420) |
| 0.3 | 1.6 | GO:0060091 | kinocilium(GO:0060091) |
| 0.3 | 4.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.3 | 2.9 | GO:0032039 | integrator complex(GO:0032039) |
| 0.3 | 6.1 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) cytosolic proteasome complex(GO:0031597) |
| 0.3 | 7.7 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.3 | 6.7 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.3 | 1.8 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.3 | 3.6 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.3 | 5.8 | GO:0042599 | lamellar body(GO:0042599) |
| 0.3 | 20.7 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.3 | 2.4 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.3 | 15.5 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.3 | 14.1 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.3 | 1.5 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.2 | 1.7 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) calcium channel complex(GO:0034704) |
| 0.2 | 8.2 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.2 | 4.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.2 | 0.9 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.2 | 3.3 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.2 | 19.1 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.2 | 8.4 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.2 | 11.8 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.2 | 1.1 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.1 | 1.5 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 16.0 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 1.0 | GO:0044194 | cytolytic granule(GO:0044194) phagocytic vesicle lumen(GO:0097013) |
| 0.1 | 3.3 | GO:0046930 | pore complex(GO:0046930) |
| 0.1 | 18.6 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 7.3 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 1.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 2.1 | GO:0031105 | septin complex(GO:0031105) |
| 0.1 | 1.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 0.5 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 8.1 | GO:0031672 | A band(GO:0031672) |
| 0.1 | 11.4 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.1 | 3.3 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 13.4 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 1.8 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 4.6 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.1 | 1.3 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 0.3 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.1 | 2.9 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 1.8 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 2.9 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 5.1 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 3.1 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.1 | 5.9 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 5.9 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 3.1 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.1 | 1.3 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 5.2 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 0.7 | GO:0001741 | XY body(GO:0001741) |
| 0.1 | 4.3 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 2.8 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 1.0 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 5.0 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 1.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 3.5 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 5.1 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 3.6 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 9.1 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.5 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.3 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.9 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 1.5 | GO:0012505 | endomembrane system(GO:0012505) |
| 0.0 | 2.0 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 1.4 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 3.4 | GO:0034774 | secretory granule lumen(GO:0034774) |
| 0.0 | 2.7 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 5.2 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.9 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.0 | 0.4 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.5 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 0.1 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.8 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 2.0 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 1.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 3.8 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 8.3 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 0.4 | GO:0005719 | nuclear euchromatin(GO:0005719) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.7 | 11.0 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 3.5 | 14.1 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 3.3 | 20.0 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 3.3 | 13.1 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 2.8 | 19.6 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 2.0 | 10.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 1.8 | 7.2 | GO:0030760 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 1.8 | 5.4 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 1.4 | 4.2 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 1.4 | 4.2 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 1.2 | 4.6 | GO:0033981 | D-dopachrome decarboxylase activity(GO:0033981) |
| 1.2 | 4.6 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 1.1 | 3.4 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 1.0 | 4.2 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 1.0 | 5.0 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 1.0 | 5.9 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 1.0 | 2.9 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 1.0 | 2.9 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.9 | 3.6 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.9 | 3.6 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.9 | 3.6 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.9 | 3.5 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.9 | 3.5 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.8 | 6.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.8 | 13.6 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.7 | 2.2 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.7 | 12.7 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.7 | 2.8 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.7 | 19.4 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.7 | 9.5 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.7 | 9.3 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.7 | 2.6 | GO:0016499 | orexin receptor activity(GO:0016499) |
| 0.6 | 2.4 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 0.6 | 1.7 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.6 | 3.4 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.5 | 7.7 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.5 | 8.2 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.5 | 3.2 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.5 | 2.6 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.5 | 2.9 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.5 | 8.4 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.5 | 1.8 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.4 | 9.4 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.4 | 6.3 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.4 | 13.0 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.4 | 3.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.4 | 20.1 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.4 | 3.3 | GO:0015288 | porin activity(GO:0015288) |
| 0.4 | 11.2 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.3 | 2.8 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.3 | 1.0 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.3 | 9.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.3 | 9.3 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.3 | 3.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.3 | 4.9 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.3 | 6.7 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.3 | 1.2 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.3 | 29.0 | GO:0005518 | collagen binding(GO:0005518) |
| 0.3 | 1.2 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.3 | 5.2 | GO:0019864 | IgG binding(GO:0019864) |
| 0.3 | 1.7 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.3 | 3.9 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.3 | 1.4 | GO:0070697 | activin receptor binding(GO:0070697) |
| 0.3 | 1.8 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.3 | 4.9 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.2 | 1.7 | GO:0019863 | IgE binding(GO:0019863) |
| 0.2 | 1.7 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.2 | 1.7 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.2 | 6.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.2 | 1.4 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.2 | 1.8 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.2 | 1.1 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.2 | 1.3 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.2 | 1.2 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.2 | 3.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.2 | 5.9 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.2 | 5.9 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.2 | 2.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.2 | 0.5 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.2 | 3.0 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.2 | 0.5 | GO:0051139 | metal ion:proton antiporter activity(GO:0051139) |
| 0.2 | 1.4 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.2 | 2.0 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.2 | 1.5 | GO:0043426 | MRF binding(GO:0043426) |
| 0.2 | 1.0 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.2 | 7.6 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.2 | 4.5 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.2 | 0.8 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.2 | 6.6 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 1.5 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.1 | 3.0 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.1 | 0.9 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.1 | 20.9 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 3.2 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.1 | 4.2 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.1 | 2.0 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.1 | 5.1 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.1 | 1.6 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 4.8 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 1.3 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.1 | 1.0 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.1 | 1.7 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.1 | 2.0 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.1 | 3.0 | GO:0001848 | complement binding(GO:0001848) |
| 0.1 | 2.2 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 4.8 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 4.6 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 0.5 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.1 | 0.3 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.1 | 2.6 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 3.3 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 9.1 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.1 | 1.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.4 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.1 | 1.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 1.5 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 1.8 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.1 | 7.8 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.1 | 1.7 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.1 | 1.6 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.1 | 0.4 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 5.3 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 14.6 | GO:0019887 | protein kinase regulator activity(GO:0019887) |
| 0.1 | 7.5 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.1 | 2.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 4.3 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 4.8 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 3.4 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.1 | 0.2 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.1 | 0.8 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 1.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 0.4 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 2.9 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.2 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.1 | 0.7 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 0.3 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.1 | 3.1 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 5.9 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 1.1 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 1.0 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.7 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.3 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 4.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.5 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 1.4 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 2.5 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.2 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 2.3 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 2.0 | GO:0004536 | deoxyribonuclease activity(GO:0004536) |
| 0.0 | 9.2 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.1 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.0 | 1.8 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 1.0 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.1 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 2.6 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 1.9 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.8 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.8 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 1.4 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 4.3 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.1 | GO:0004473 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) oxaloacetate decarboxylase activity(GO:0008948) |
| 0.0 | 1.0 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.2 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 2.3 | GO:0061659 | ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.0 | 10.6 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 2.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.3 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 19.6 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.5 | 7.7 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.4 | 12.4 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.4 | 13.6 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.4 | 5.9 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.4 | 19.4 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.3 | 16.7 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.3 | 4.8 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.3 | 3.6 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.3 | 12.8 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.3 | 6.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.3 | 8.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.2 | 2.9 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.2 | 14.3 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.2 | 7.4 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 9.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 8.2 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 10.0 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 5.7 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 22.1 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 4.6 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 1.3 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 4.4 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 4.3 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.1 | 2.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 25.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 1.0 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 6.7 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 1.5 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 2.8 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 3.8 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 1.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 1.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 1.0 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 1.7 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 9.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 2.5 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 2.4 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 2.0 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.6 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.0 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 1.2 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.2 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 0.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.1 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.9 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.5 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 1.2 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 3.5 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.1 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 19.8 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.8 | 9.4 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.7 | 9.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.6 | 37.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.5 | 13.0 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.5 | 20.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.4 | 6.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.4 | 15.8 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.4 | 10.1 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.3 | 6.7 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.3 | 5.9 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.3 | 13.4 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.3 | 4.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.3 | 12.7 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.3 | 9.4 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.3 | 7.7 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.3 | 4.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.2 | 1.5 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.2 | 5.5 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.2 | 17.1 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.2 | 4.6 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.2 | 3.6 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.2 | 4.8 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 3.9 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.1 | 4.0 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 1.4 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 4.8 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 15.9 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 2.0 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 4.8 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.1 | 3.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 8.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 0.7 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 2.5 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 2.8 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.1 | 1.8 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 2.2 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 3.3 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.1 | 10.1 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.1 | 2.8 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.1 | 2.1 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.1 | 1.3 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.1 | 1.7 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.1 | 1.5 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 1.0 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 2.0 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 5.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 1.8 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.1 | 3.3 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 0.5 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.1 | 2.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 2.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.5 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 2.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.8 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 2.3 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.0 | 0.5 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 4.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.4 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 2.2 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.7 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.7 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 0.4 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.4 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 1.4 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 1.0 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 1.0 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.0 | 2.7 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 1.5 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.9 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 1.5 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.2 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |