Project
averaged GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
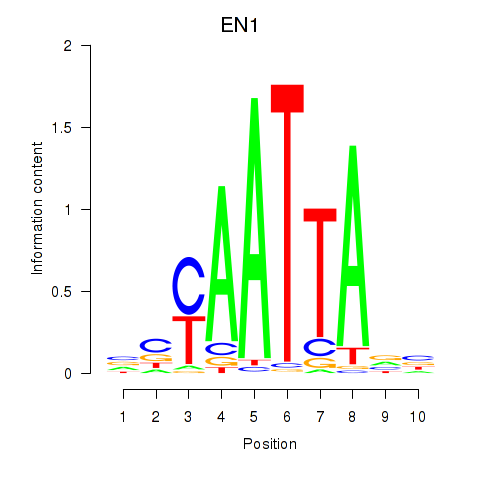
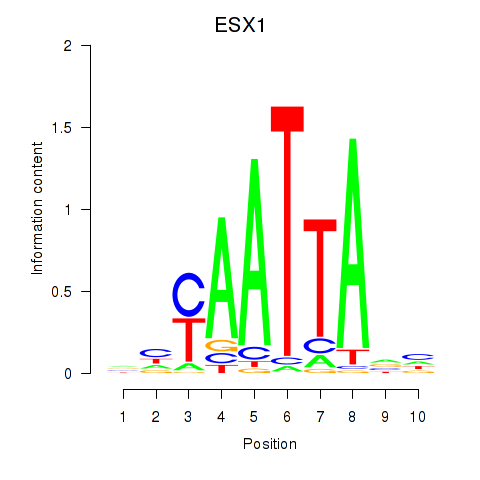
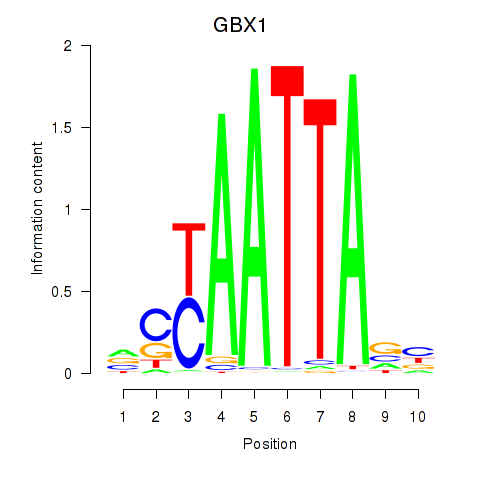
Results for EN1_ESX1_GBX1
Z-value: 1.15
Transcription factors associated with EN1_ESX1_GBX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EN1
|
ENSG00000163064.6 | engrailed homeobox 1 |
|
ESX1
|
ENSG00000123576.5 | ESX homeobox 1 |
|
GBX1
|
ENSG00000164900.4 | gastrulation brain homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GBX1 | hg19_v2_chr7_-_150864635_150864785 | -0.19 | 4.5e-03 | Click! |
| EN1 | hg19_v2_chr2_-_119605253_119605264 | 0.18 | 6.3e-03 | Click! |
Activity profile of EN1_ESX1_GBX1 motif
Sorted Z-values of EN1_ESX1_GBX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_111794446 | 32.90 |
ENST00000527950.1
|
CRYAB
|
crystallin, alpha B |
| chrX_-_13835147 | 28.51 |
ENST00000493677.1
ENST00000355135.2 |
GPM6B
|
glycoprotein M6B |
| chr3_+_111718036 | 27.74 |
ENST00000455401.2
|
TAGLN3
|
transgelin 3 |
| chr3_+_111718173 | 25.42 |
ENST00000494932.1
|
TAGLN3
|
transgelin 3 |
| chr3_+_111717600 | 25.31 |
ENST00000273368.4
|
TAGLN3
|
transgelin 3 |
| chr3_+_111717511 | 24.32 |
ENST00000478951.1
ENST00000393917.2 |
TAGLN3
|
transgelin 3 |
| chr3_+_115342349 | 22.62 |
ENST00000393780.3
|
GAP43
|
growth associated protein 43 |
| chr11_-_117747434 | 21.52 |
ENST00000529335.2
ENST00000530956.1 ENST00000260282.4 |
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr11_-_117747607 | 21.49 |
ENST00000540359.1
ENST00000539526.1 |
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr12_-_6233828 | 21.46 |
ENST00000572068.1
ENST00000261405.5 |
VWF
|
von Willebrand factor |
| chr11_-_117748138 | 21.40 |
ENST00000527717.1
|
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr7_+_100136811 | 18.86 |
ENST00000300176.4
ENST00000262935.4 |
AGFG2
|
ArfGAP with FG repeats 2 |
| chr16_+_58283814 | 17.84 |
ENST00000443128.2
ENST00000219299.4 |
CCDC113
|
coiled-coil domain containing 113 |
| chr12_-_16759711 | 17.29 |
ENST00000447609.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr19_-_7968427 | 17.14 |
ENST00000539278.1
|
AC010336.1
|
Uncharacterized protein |
| chr5_+_66300446 | 15.28 |
ENST00000261569.7
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr10_-_93392811 | 14.56 |
ENST00000238994.5
|
PPP1R3C
|
protein phosphatase 1, regulatory subunit 3C |
| chr1_-_118472216 | 13.82 |
ENST00000369443.5
|
GDAP2
|
ganglioside induced differentiation associated protein 2 |
| chr12_+_81110684 | 13.59 |
ENST00000228644.3
|
MYF5
|
myogenic factor 5 |
| chr1_+_160160283 | 13.48 |
ENST00000368079.3
|
CASQ1
|
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr2_-_224467093 | 12.76 |
ENST00000305409.2
|
SCG2
|
secretogranin II |
| chr12_+_7014064 | 12.44 |
ENST00000443597.2
|
LRRC23
|
leucine rich repeat containing 23 |
| chr13_-_67802549 | 12.13 |
ENST00000328454.5
ENST00000377865.2 |
PCDH9
|
protocadherin 9 |
| chr4_-_134070250 | 12.06 |
ENST00000505289.1
ENST00000509715.1 |
RP11-9G1.3
|
RP11-9G1.3 |
| chr6_+_153552455 | 11.92 |
ENST00000392385.2
|
AL590867.1
|
Uncharacterized protein; cDNA FLJ59044, highly similar to LINE-1 reverse transcriptase homolog |
| chr1_-_151762943 | 11.76 |
ENST00000368825.3
ENST00000368823.1 ENST00000458431.2 ENST00000368827.6 ENST00000368824.3 |
TDRKH
|
tudor and KH domain containing |
| chr8_+_105235572 | 11.72 |
ENST00000523362.1
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr1_+_160160346 | 11.53 |
ENST00000368078.3
|
CASQ1
|
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr12_+_7013897 | 11.52 |
ENST00000007969.8
ENST00000323702.5 |
LRRC23
|
leucine rich repeat containing 23 |
| chr12_+_26348246 | 11.34 |
ENST00000422622.2
|
SSPN
|
sarcospan |
| chr14_+_104182061 | 10.33 |
ENST00000216602.6
|
ZFYVE21
|
zinc finger, FYVE domain containing 21 |
| chr14_+_104182105 | 10.27 |
ENST00000311141.2
|
ZFYVE21
|
zinc finger, FYVE domain containing 21 |
| chr11_+_94706973 | 10.04 |
ENST00000536741.1
|
KDM4D
|
lysine (K)-specific demethylase 4D |
| chr16_-_29910853 | 9.96 |
ENST00000308713.5
|
SEZ6L2
|
seizure related 6 homolog (mouse)-like 2 |
| chr4_-_8873531 | 9.74 |
ENST00000400677.3
|
HMX1
|
H6 family homeobox 1 |
| chr14_-_104181771 | 9.71 |
ENST00000554913.1
ENST00000554974.1 ENST00000553361.1 ENST00000555055.1 ENST00000555964.1 ENST00000556682.1 ENST00000445556.1 ENST00000553332.1 ENST00000352127.7 |
XRCC3
|
X-ray repair complementing defective repair in Chinese hamster cells 3 |
| chr11_-_128894053 | 9.66 |
ENST00000392657.3
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr1_+_44115814 | 9.43 |
ENST00000372396.3
|
KDM4A
|
lysine (K)-specific demethylase 4A |
| chr3_-_112127981 | 9.42 |
ENST00000486726.2
|
RP11-231E6.1
|
RP11-231E6.1 |
| chr12_-_91546926 | 9.40 |
ENST00000550758.1
|
DCN
|
decorin |
| chr7_-_137028534 | 9.13 |
ENST00000348225.2
|
PTN
|
pleiotrophin |
| chr12_+_53773944 | 9.07 |
ENST00000551969.1
ENST00000327443.4 |
SP1
|
Sp1 transcription factor |
| chr11_-_129062093 | 9.05 |
ENST00000310343.9
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr10_-_50970322 | 9.02 |
ENST00000374103.4
|
OGDHL
|
oxoglutarate dehydrogenase-like |
| chr12_-_114841703 | 9.01 |
ENST00000526441.1
|
TBX5
|
T-box 5 |
| chr14_+_29236269 | 8.87 |
ENST00000313071.4
|
FOXG1
|
forkhead box G1 |
| chr1_+_28261533 | 8.84 |
ENST00000411604.1
ENST00000373888.4 |
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr3_+_156544057 | 8.65 |
ENST00000498839.1
ENST00000470811.1 ENST00000356539.4 ENST00000483177.1 ENST00000477399.1 ENST00000491763.1 |
LEKR1
|
leucine, glutamate and lysine rich 1 |
| chr8_+_9413410 | 8.61 |
ENST00000520408.1
ENST00000310430.6 ENST00000522110.1 |
TNKS
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase |
| chr13_-_88323218 | 8.53 |
ENST00000436290.2
ENST00000453832.2 ENST00000606590.1 |
MIR4500HG
|
MIR4500 host gene (non-protein coding) |
| chr4_-_46911248 | 8.45 |
ENST00000355591.3
ENST00000505102.1 |
COX7B2
|
cytochrome c oxidase subunit VIIb2 |
| chr19_+_4007644 | 8.42 |
ENST00000262971.2
|
PIAS4
|
protein inhibitor of activated STAT, 4 |
| chr4_-_46911223 | 8.39 |
ENST00000396533.1
|
COX7B2
|
cytochrome c oxidase subunit VIIb2 |
| chr16_-_31085514 | 8.10 |
ENST00000300849.4
|
ZNF668
|
zinc finger protein 668 |
| chr7_-_137028498 | 7.88 |
ENST00000393083.2
|
PTN
|
pleiotrophin |
| chr1_+_202317815 | 7.81 |
ENST00000608999.1
ENST00000336894.4 ENST00000480184.1 |
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr8_-_87755878 | 7.78 |
ENST00000320005.5
|
CNGB3
|
cyclic nucleotide gated channel beta 3 |
| chr4_-_57547870 | 7.54 |
ENST00000381260.3
ENST00000420433.1 ENST00000554144.1 ENST00000557328.1 |
HOPX
|
HOP homeobox |
| chr12_+_7014126 | 7.52 |
ENST00000415834.1
ENST00000436789.1 |
LRRC23
|
leucine rich repeat containing 23 |
| chr7_-_73038822 | 7.36 |
ENST00000414749.2
ENST00000429400.2 ENST00000434326.1 |
MLXIPL
|
MLX interacting protein-like |
| chr20_-_18447667 | 7.27 |
ENST00000262547.5
ENST00000329494.5 ENST00000357236.4 |
DZANK1
|
double zinc ribbon and ankyrin repeat domains 1 |
| chr5_-_42811986 | 7.17 |
ENST00000511224.1
ENST00000507920.1 ENST00000510965.1 |
SEPP1
|
selenoprotein P, plasma, 1 |
| chr5_-_42812143 | 7.11 |
ENST00000514985.1
|
SEPP1
|
selenoprotein P, plasma, 1 |
| chr2_+_171034646 | 6.98 |
ENST00000409044.3
ENST00000408978.4 |
MYO3B
|
myosin IIIB |
| chr14_-_74551096 | 6.98 |
ENST00000350259.4
|
ALDH6A1
|
aldehyde dehydrogenase 6 family, member A1 |
| chr10_-_50970382 | 6.96 |
ENST00000419399.1
ENST00000432695.1 |
OGDHL
|
oxoglutarate dehydrogenase-like |
| chrX_+_13671225 | 6.76 |
ENST00000545566.1
ENST00000544987.1 ENST00000314720.4 |
TCEANC
|
transcription elongation factor A (SII) N-terminal and central domain containing |
| chr8_-_18744528 | 6.64 |
ENST00000523619.1
|
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chr16_-_55866997 | 6.63 |
ENST00000360526.3
ENST00000361503.4 |
CES1
|
carboxylesterase 1 |
| chrX_+_77166172 | 6.58 |
ENST00000343533.5
ENST00000350425.4 ENST00000341514.6 |
ATP7A
|
ATPase, Cu++ transporting, alpha polypeptide |
| chr4_-_57547454 | 6.57 |
ENST00000556376.2
|
HOPX
|
HOP homeobox |
| chr6_+_72926145 | 6.54 |
ENST00000425662.2
ENST00000453976.2 |
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr4_+_71587669 | 6.51 |
ENST00000381006.3
ENST00000226328.4 |
RUFY3
|
RUN and FYVE domain containing 3 |
| chr7_-_73038867 | 6.40 |
ENST00000313375.3
ENST00000354613.1 ENST00000395189.1 ENST00000453275.1 |
MLXIPL
|
MLX interacting protein-like |
| chr8_+_94241867 | 6.40 |
ENST00000598428.1
|
AC016885.1
|
Uncharacterized protein |
| chr15_+_58702742 | 6.37 |
ENST00000356113.6
ENST00000414170.3 |
LIPC
|
lipase, hepatic |
| chr17_-_41466555 | 6.34 |
ENST00000586231.1
|
LINC00910
|
long intergenic non-protein coding RNA 910 |
| chr9_-_5304432 | 6.26 |
ENST00000416837.1
ENST00000308420.3 |
RLN2
|
relaxin 2 |
| chr13_-_28545276 | 6.16 |
ENST00000381020.7
|
CDX2
|
caudal type homeobox 2 |
| chr4_-_89205879 | 6.16 |
ENST00000608933.1
ENST00000315194.4 ENST00000514204.1 |
PPM1K
|
protein phosphatase, Mg2+/Mn2+ dependent, 1K |
| chr17_-_9929581 | 6.12 |
ENST00000437099.2
ENST00000396115.2 |
GAS7
|
growth arrest-specific 7 |
| chr19_-_51522955 | 6.01 |
ENST00000358789.3
|
KLK10
|
kallikrein-related peptidase 10 |
| chr14_-_74551172 | 5.99 |
ENST00000553458.1
|
ALDH6A1
|
aldehyde dehydrogenase 6 family, member A1 |
| chr3_+_152552685 | 5.97 |
ENST00000305097.3
|
P2RY1
|
purinergic receptor P2Y, G-protein coupled, 1 |
| chr3_-_160823158 | 5.89 |
ENST00000392779.2
ENST00000392780.1 ENST00000494173.1 |
B3GALNT1
|
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chr17_-_43045439 | 5.88 |
ENST00000253407.3
|
C1QL1
|
complement component 1, q subcomponent-like 1 |
| chr6_-_49681235 | 5.83 |
ENST00000339139.4
|
CRISP2
|
cysteine-rich secretory protein 2 |
| chr5_-_24645078 | 5.83 |
ENST00000264463.4
|
CDH10
|
cadherin 10, type 2 (T2-cadherin) |
| chr14_-_82000140 | 5.76 |
ENST00000555824.1
ENST00000557372.1 ENST00000336735.4 |
SEL1L
|
sel-1 suppressor of lin-12-like (C. elegans) |
| chr1_-_152386732 | 5.69 |
ENST00000271835.3
|
CRNN
|
cornulin |
| chr8_+_50824233 | 5.65 |
ENST00000522124.1
|
SNTG1
|
syntrophin, gamma 1 |
| chr6_-_100912785 | 5.56 |
ENST00000369208.3
|
SIM1
|
single-minded family bHLH transcription factor 1 |
| chr6_-_167040731 | 5.49 |
ENST00000265678.4
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
| chr13_-_36788718 | 5.45 |
ENST00000317764.6
ENST00000379881.3 |
SOHLH2
|
spermatogenesis and oogenesis specific basic helix-loop-helix 2 |
| chr19_-_58951496 | 5.36 |
ENST00000254166.3
|
ZNF132
|
zinc finger protein 132 |
| chr6_-_62996066 | 5.34 |
ENST00000281156.4
|
KHDRBS2
|
KH domain containing, RNA binding, signal transduction associated 2 |
| chr6_+_72922505 | 5.33 |
ENST00000401910.3
|
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr11_+_24518723 | 5.24 |
ENST00000336930.6
ENST00000529015.1 ENST00000533227.1 |
LUZP2
|
leucine zipper protein 2 |
| chr16_-_2264779 | 5.24 |
ENST00000333503.7
|
PGP
|
phosphoglycolate phosphatase |
| chr1_-_8877692 | 5.20 |
ENST00000400908.2
|
RERE
|
arginine-glutamic acid dipeptide (RE) repeats |
| chr3_-_160823040 | 5.20 |
ENST00000484127.1
ENST00000492353.1 ENST00000473142.1 ENST00000468268.1 ENST00000460353.1 ENST00000320474.4 ENST00000392781.2 |
B3GALNT1
|
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chr6_+_72922590 | 5.12 |
ENST00000523963.1
|
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr9_-_5339873 | 5.11 |
ENST00000223862.1
ENST00000223858.4 |
RLN1
|
relaxin 1 |
| chr6_+_39760129 | 5.08 |
ENST00000274867.4
|
DAAM2
|
dishevelled associated activator of morphogenesis 2 |
| chr17_+_42264556 | 5.04 |
ENST00000319511.6
ENST00000589785.1 ENST00000592825.1 ENST00000589184.1 |
TMUB2
|
transmembrane and ubiquitin-like domain containing 2 |
| chr5_-_160279207 | 5.02 |
ENST00000327245.5
|
ATP10B
|
ATPase, class V, type 10B |
| chr7_-_14029515 | 5.00 |
ENST00000430479.1
ENST00000405218.2 ENST00000343495.5 |
ETV1
|
ets variant 1 |
| chr2_+_196313239 | 4.96 |
ENST00000413290.1
|
AC064834.1
|
AC064834.1 |
| chr4_+_106631966 | 4.93 |
ENST00000360505.5
ENST00000510865.1 ENST00000509336.1 |
GSTCD
|
glutathione S-transferase, C-terminal domain containing |
| chr6_+_28317685 | 4.93 |
ENST00000252211.2
ENST00000341464.5 ENST00000377255.3 |
ZKSCAN3
|
zinc finger with KRAB and SCAN domains 3 |
| chr11_-_108093329 | 4.93 |
ENST00000278612.8
|
NPAT
|
nuclear protein, ataxia-telangiectasia locus |
| chr7_+_99425633 | 4.92 |
ENST00000354829.2
ENST00000421837.2 ENST00000417625.1 ENST00000342499.4 ENST00000444905.1 ENST00000415413.1 ENST00000312017.5 ENST00000222382.5 |
CYP3A43
|
cytochrome P450, family 3, subfamily A, polypeptide 43 |
| chr12_+_106751436 | 4.86 |
ENST00000228347.4
|
POLR3B
|
polymerase (RNA) III (DNA directed) polypeptide B |
| chr14_+_24583836 | 4.83 |
ENST00000559115.1
ENST00000558215.1 ENST00000557810.1 ENST00000561375.1 ENST00000446197.3 ENST00000559796.1 ENST00000560713.1 ENST00000560901.1 ENST00000559382.1 |
DCAF11
|
DDB1 and CUL4 associated factor 11 |
| chr22_+_29138013 | 4.80 |
ENST00000216027.3
ENST00000398941.2 |
HSCB
|
HscB mitochondrial iron-sulfur cluster co-chaperone |
| chr14_+_32798547 | 4.75 |
ENST00000557354.1
ENST00000557102.1 ENST00000557272.1 |
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr6_+_31783291 | 4.71 |
ENST00000375651.5
ENST00000608703.1 ENST00000458062.2 |
HSPA1A
|
heat shock 70kDa protein 1A |
| chr4_+_3388057 | 4.71 |
ENST00000538395.1
|
RGS12
|
regulator of G-protein signaling 12 |
| chr12_+_26348429 | 4.70 |
ENST00000242729.2
|
SSPN
|
sarcospan |
| chr19_-_39421377 | 4.63 |
ENST00000430193.3
ENST00000600042.1 ENST00000221431.6 |
SARS2
|
seryl-tRNA synthetase 2, mitochondrial |
| chr6_-_32095968 | 4.62 |
ENST00000375203.3
ENST00000375201.4 |
ATF6B
|
activating transcription factor 6 beta |
| chr14_+_74815116 | 4.62 |
ENST00000256362.4
|
VRTN
|
vertebrae development associated |
| chr22_+_41777927 | 4.60 |
ENST00000266304.4
|
TEF
|
thyrotrophic embryonic factor |
| chr5_+_31193847 | 4.60 |
ENST00000514738.1
ENST00000265071.2 |
CDH6
|
cadherin 6, type 2, K-cadherin (fetal kidney) |
| chr8_-_22089845 | 4.56 |
ENST00000454243.2
|
PHYHIP
|
phytanoyl-CoA 2-hydroxylase interacting protein |
| chr8_-_86290333 | 4.52 |
ENST00000521846.1
ENST00000523022.1 ENST00000524324.1 ENST00000519991.1 ENST00000520663.1 ENST00000517590.1 ENST00000522579.1 ENST00000522814.1 ENST00000522662.1 ENST00000523858.1 ENST00000519129.1 |
CA1
|
carbonic anhydrase I |
| chr12_-_52828147 | 4.48 |
ENST00000252245.5
|
KRT75
|
keratin 75 |
| chr2_+_166152283 | 4.44 |
ENST00000375427.2
|
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit |
| chrX_-_130423200 | 4.40 |
ENST00000361420.3
|
IGSF1
|
immunoglobulin superfamily, member 1 |
| chr10_+_5005598 | 4.37 |
ENST00000442997.1
|
AKR1C1
|
aldo-keto reductase family 1, member C1 |
| chr1_+_196788887 | 4.36 |
ENST00000320493.5
ENST00000367424.4 ENST00000367421.3 |
CFHR1
CFHR2
|
complement factor H-related 1 complement factor H-related 2 |
| chr7_+_74072288 | 4.36 |
ENST00000443166.1
|
GTF2I
|
general transcription factor IIi |
| chr12_-_16761007 | 4.33 |
ENST00000354662.1
ENST00000441439.2 |
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr8_-_22089533 | 4.29 |
ENST00000321613.3
|
PHYHIP
|
phytanoyl-CoA 2-hydroxylase interacting protein |
| chr19_+_50016610 | 4.26 |
ENST00000596975.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr11_-_101000445 | 4.25 |
ENST00000534013.1
|
PGR
|
progesterone receptor |
| chr10_+_74451883 | 4.24 |
ENST00000373053.3
ENST00000357157.6 |
MCU
|
mitochondrial calcium uniporter |
| chr22_-_18923655 | 4.21 |
ENST00000438924.1
ENST00000457083.1 ENST00000420436.1 ENST00000334029.2 ENST00000357068.6 |
PRODH
|
proline dehydrogenase (oxidase) 1 |
| chr10_+_96522361 | 4.18 |
ENST00000371321.3
|
CYP2C19
|
cytochrome P450, family 2, subfamily C, polypeptide 19 |
| chr8_-_145159083 | 4.17 |
ENST00000398712.2
|
SHARPIN
|
SHANK-associated RH domain interactor |
| chr3_+_167453493 | 4.13 |
ENST00000295777.5
ENST00000472747.2 |
SERPINI1
|
serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
| chr19_-_19302931 | 4.12 |
ENST00000444486.3
ENST00000514819.3 ENST00000585679.1 ENST00000162023.5 |
MEF2BNB-MEF2B
MEF2BNB
MEF2B
|
MEF2BNB-MEF2B readthrough MEF2B neighbor myocyte enhancer factor 2B |
| chr17_-_64225508 | 4.11 |
ENST00000205948.6
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr3_+_155860751 | 4.07 |
ENST00000471742.1
|
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr12_-_11548496 | 4.06 |
ENST00000389362.4
ENST00000565533.1 ENST00000546254.1 |
PRB2
PRB1
|
proline-rich protein BstNI subfamily 2 proline-rich protein BstNI subfamily 1 |
| chr22_-_32766972 | 4.01 |
ENST00000382084.4
ENST00000382086.2 |
RFPL3S
|
RFPL3 antisense |
| chr10_-_99447024 | 3.92 |
ENST00000370626.3
|
AVPI1
|
arginine vasopressin-induced 1 |
| chr14_-_24584138 | 3.88 |
ENST00000558280.1
ENST00000561028.1 |
NRL
|
neural retina leucine zipper |
| chr7_+_49813255 | 3.86 |
ENST00000340652.4
|
VWC2
|
von Willebrand factor C domain containing 2 |
| chr10_-_49813090 | 3.84 |
ENST00000249601.4
|
ARHGAP22
|
Rho GTPase activating protein 22 |
| chr19_-_41870026 | 3.84 |
ENST00000243578.3
|
B9D2
|
B9 protein domain 2 |
| chr17_-_1532106 | 3.83 |
ENST00000301335.5
ENST00000382147.4 |
SLC43A2
|
solute carrier family 43 (amino acid system L transporter), member 2 |
| chr12_+_52056548 | 3.77 |
ENST00000545061.1
ENST00000355133.3 |
SCN8A
|
sodium channel, voltage gated, type VIII, alpha subunit |
| chr2_-_208989225 | 3.74 |
ENST00000264376.4
|
CRYGD
|
crystallin, gamma D |
| chr1_+_196912902 | 3.69 |
ENST00000476712.2
ENST00000367415.5 |
CFHR2
|
complement factor H-related 2 |
| chr17_-_73663168 | 3.66 |
ENST00000578201.1
ENST00000423245.2 |
RECQL5
|
RecQ protein-like 5 |
| chr6_-_22297730 | 3.64 |
ENST00000306482.1
|
PRL
|
prolactin |
| chr6_-_91296602 | 3.64 |
ENST00000369325.3
ENST00000369327.3 |
MAP3K7
|
mitogen-activated protein kinase kinase kinase 7 |
| chr11_+_94706804 | 3.60 |
ENST00000335080.5
|
KDM4D
|
lysine (K)-specific demethylase 4D |
| chrX_-_139587225 | 3.58 |
ENST00000370536.2
|
SOX3
|
SRY (sex determining region Y)-box 3 |
| chr6_+_160542870 | 3.54 |
ENST00000324965.4
ENST00000457470.2 |
SLC22A1
|
solute carrier family 22 (organic cation transporter), member 1 |
| chr9_-_95166841 | 3.52 |
ENST00000262551.4
|
OGN
|
osteoglycin |
| chr1_+_209878182 | 3.52 |
ENST00000367027.3
|
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1 |
| chr11_+_92085262 | 3.47 |
ENST00000298047.6
ENST00000409404.2 ENST00000541502.1 |
FAT3
|
FAT atypical cadherin 3 |
| chr19_+_34287751 | 3.46 |
ENST00000590771.1
ENST00000589786.1 ENST00000284006.6 ENST00000588881.1 |
KCTD15
|
potassium channel tetramerization domain containing 15 |
| chr15_+_62853562 | 3.45 |
ENST00000561311.1
|
TLN2
|
talin 2 |
| chr2_+_68962014 | 3.45 |
ENST00000467265.1
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr10_-_71169031 | 3.44 |
ENST00000373307.1
|
TACR2
|
tachykinin receptor 2 |
| chr2_-_208031943 | 3.43 |
ENST00000421199.1
ENST00000457962.1 |
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr12_+_122688090 | 3.43 |
ENST00000324189.4
ENST00000546192.1 |
B3GNT4
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 4 |
| chr17_-_18430160 | 3.42 |
ENST00000392176.3
|
FAM106A
|
family with sequence similarity 106, member A |
| chr9_-_116840728 | 3.40 |
ENST00000265132.3
|
AMBP
|
alpha-1-microglobulin/bikunin precursor |
| chr17_+_40610862 | 3.40 |
ENST00000393829.2
ENST00000546249.1 ENST00000537728.1 ENST00000264649.6 ENST00000585525.1 ENST00000343619.4 ENST00000544137.1 ENST00000589727.1 ENST00000587824.1 |
ATP6V0A1
|
ATPase, H+ transporting, lysosomal V0 subunit a1 |
| chr8_-_42698433 | 3.38 |
ENST00000345117.2
ENST00000254250.3 |
THAP1
|
THAP domain containing, apoptosis associated protein 1 |
| chrX_-_71458802 | 3.37 |
ENST00000373657.1
ENST00000334463.3 |
ERCC6L
|
excision repair cross-complementing rodent repair deficiency, complementation group 6-like |
| chr8_+_119294456 | 3.36 |
ENST00000366457.2
|
AC023590.1
|
Uncharacterized protein |
| chr4_-_74486347 | 3.34 |
ENST00000342081.3
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr7_+_100183927 | 3.34 |
ENST00000241071.6
ENST00000360609.2 |
FBXO24
|
F-box protein 24 |
| chr19_+_18942761 | 3.30 |
ENST00000599848.1
|
UPF1
|
UPF1 regulator of nonsense transcripts homolog (yeast) |
| chr19_-_48018203 | 3.29 |
ENST00000595227.1
ENST00000593761.1 ENST00000263354.3 |
NAPA
|
N-ethylmaleimide-sensitive factor attachment protein, alpha |
| chr22_-_32767017 | 3.28 |
ENST00000400234.1
|
RFPL3S
|
RFPL3 antisense |
| chr2_+_10183651 | 3.27 |
ENST00000305883.1
|
KLF11
|
Kruppel-like factor 11 |
| chr4_-_87028478 | 3.27 |
ENST00000515400.1
ENST00000395157.3 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr5_-_20575959 | 3.25 |
ENST00000507958.1
|
CDH18
|
cadherin 18, type 2 |
| chr17_-_73663245 | 3.25 |
ENST00000584999.1
ENST00000317905.5 ENST00000420326.2 ENST00000340830.5 |
RECQL5
|
RecQ protein-like 5 |
| chr4_+_88754113 | 3.25 |
ENST00000560249.1
ENST00000540395.1 ENST00000511670.1 ENST00000361056.3 |
MEPE
|
matrix extracellular phosphoglycoprotein |
| chr14_+_32798462 | 3.24 |
ENST00000280979.4
|
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr3_-_123339343 | 3.21 |
ENST00000578202.1
|
MYLK
|
myosin light chain kinase |
| chr7_-_100183742 | 3.21 |
ENST00000310300.6
|
LRCH4
|
leucine-rich repeats and calponin homology (CH) domain containing 4 |
| chr3_-_178984759 | 3.12 |
ENST00000349697.2
ENST00000497599.1 |
KCNMB3
|
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr12_-_23737534 | 3.11 |
ENST00000396007.2
|
SOX5
|
SRY (sex determining region Y)-box 5 |
| chr9_-_95166884 | 3.10 |
ENST00000375561.5
|
OGN
|
osteoglycin |
| chr1_-_190446759 | 3.06 |
ENST00000367462.3
|
BRINP3
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr1_-_10532531 | 3.03 |
ENST00000377036.2
ENST00000377038.3 |
DFFA
|
DNA fragmentation factor, 45kDa, alpha polypeptide |
| chr18_+_46065393 | 3.00 |
ENST00000256413.3
|
CTIF
|
CBP80/20-dependent translation initiation factor |
| chr11_+_119076745 | 3.00 |
ENST00000264033.4
|
CBL
|
Cbl proto-oncogene, E3 ubiquitin protein ligase |
| chr19_+_50016411 | 2.99 |
ENST00000426395.3
ENST00000600273.1 ENST00000599988.1 |
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr14_-_36789783 | 2.98 |
ENST00000605579.1
ENST00000604336.1 ENST00000359527.7 ENST00000603139.1 ENST00000318473.7 |
MBIP
|
MAP3K12 binding inhibitory protein 1 |
| chr7_-_2883928 | 2.94 |
ENST00000275364.3
|
GNA12
|
guanine nucleotide binding protein (G protein) alpha 12 |
| chrX_-_18690210 | 2.94 |
ENST00000379984.3
|
RS1
|
retinoschisin 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of EN1_ESX1_GBX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.7 | 17.0 | GO:1904395 | retinal rod cell differentiation(GO:0060221) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) negative regulation of neuromuscular junction development(GO:1904397) |
| 3.6 | 21.6 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 3.2 | 13.0 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 3.2 | 9.7 | GO:0071140 | resolution of recombination intermediates(GO:0071139) resolution of mitotic recombination intermediates(GO:0071140) |
| 3.0 | 9.0 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 2.9 | 23.1 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 2.9 | 8.6 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 2.9 | 28.5 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 2.5 | 25.0 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 2.3 | 6.9 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 2.3 | 22.6 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 2.2 | 6.6 | GO:0071284 | cellular response to lead ion(GO:0071284) |
| 2.0 | 8.0 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 2.0 | 13.8 | GO:0090324 | negative regulation of oxidative phosphorylation(GO:0090324) |
| 1.7 | 5.2 | GO:0000294 | nuclear-transcribed mRNA catabolic process, endonucleolytic cleavage-dependent decay(GO:0000294) |
| 1.7 | 8.4 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 1.6 | 28.7 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 1.5 | 4.6 | GO:0097056 | seryl-tRNA aminoacylation(GO:0006434) selenocysteinyl-tRNA(Sec) biosynthetic process(GO:0097056) |
| 1.5 | 6.2 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 1.5 | 9.1 | GO:1900239 | phenotypic switching(GO:0036166) regulation of phenotypic switching(GO:1900239) |
| 1.5 | 11.8 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 1.5 | 26.3 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 1.4 | 1.4 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 1.4 | 13.6 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 1.3 | 5.2 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 1.3 | 5.2 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 1.3 | 3.8 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 1.2 | 6.0 | GO:0060406 | positive regulation of penile erection(GO:0060406) |
| 1.2 | 4.7 | GO:0070426 | positive regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070426) positive regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070434) |
| 1.1 | 3.4 | GO:0014057 | positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
| 1.1 | 3.4 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 1.1 | 25.6 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 1.1 | 11.0 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 1.1 | 4.3 | GO:0061743 | motor learning(GO:0061743) |
| 1.1 | 4.3 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 1.1 | 2.1 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 1.1 | 4.2 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 1.0 | 7.3 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 1.0 | 3.0 | GO:1903625 | negative regulation of DNA catabolic process(GO:1903625) |
| 1.0 | 3.0 | GO:0007231 | osmosensory signaling pathway(GO:0007231) |
| 1.0 | 3.9 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 1.0 | 2.9 | GO:0071030 | nuclear mRNA surveillance of spliceosomal pre-mRNA splicing(GO:0071030) nuclear retention of unspliced pre-mRNA at the site of transcription(GO:0071048) |
| 0.9 | 6.6 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.9 | 6.4 | GO:1901374 | acetate ester transport(GO:1901374) |
| 0.9 | 4.5 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.9 | 2.7 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.9 | 1.8 | GO:0007356 | thorax and anterior abdomen determination(GO:0007356) |
| 0.9 | 4.4 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.9 | 0.9 | GO:0086092 | regulation of the force of heart contraction by cardiac conduction(GO:0086092) |
| 0.9 | 5.2 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.9 | 6.9 | GO:0097039 | CD40 signaling pathway(GO:0023035) protein linear polyubiquitination(GO:0097039) |
| 0.9 | 12.8 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.8 | 0.8 | GO:0060369 | positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.8 | 17.0 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.8 | 2.4 | GO:0002590 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) regulation of T cell antigen processing and presentation(GO:0002625) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) response to iron ion starvation(GO:1990641) |
| 0.8 | 4.8 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.8 | 60.3 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.8 | 3.9 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.8 | 9.4 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.8 | 3.8 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.7 | 2.2 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.7 | 2.2 | GO:0035995 | skeletal muscle myosin thick filament assembly(GO:0030241) detection of muscle stretch(GO:0035995) |
| 0.7 | 0.7 | GO:2000538 | regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.7 | 5.7 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.7 | 4.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.7 | 3.5 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.7 | 6.6 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.7 | 2.6 | GO:1903721 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.7 | 2.0 | GO:0015888 | thiamine transport(GO:0015888) thiamine transmembrane transport(GO:0071934) |
| 0.7 | 3.3 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.6 | 1.9 | GO:0051795 | positive regulation of catagen(GO:0051795) activation of meiosis(GO:0090427) |
| 0.6 | 1.9 | GO:0046104 | thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.6 | 2.6 | GO:0035992 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.6 | 0.6 | GO:0048520 | positive regulation of behavior(GO:0048520) |
| 0.6 | 2.4 | GO:0006710 | androgen catabolic process(GO:0006710) |
| 0.6 | 3.6 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.6 | 4.2 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.6 | 1.2 | GO:0045726 | regulation of integrin biosynthetic process(GO:0045113) positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.6 | 1.7 | GO:0021798 | neuroblast differentiation(GO:0014016) forebrain dorsal/ventral pattern formation(GO:0021798) regulation of neural retina development(GO:0061074) regulation of retina development in camera-type eye(GO:1902866) |
| 0.6 | 12.7 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.6 | 8.0 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.6 | 2.8 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) negative regulation of membrane depolarization(GO:1904180) |
| 0.5 | 4.9 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
| 0.5 | 1.1 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.5 | 1.6 | GO:0045229 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.5 | 2.1 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.5 | 3.1 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.5 | 2.6 | GO:1900005 | positive regulation of serine-type endopeptidase activity(GO:1900005) negative regulation of sperm motility(GO:1901318) positive regulation of serine-type peptidase activity(GO:1902573) |
| 0.5 | 2.0 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.5 | 1.5 | GO:0044771 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.5 | 1.5 | GO:0021538 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.5 | 6.6 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.5 | 1.4 | GO:0033025 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.4 | 3.0 | GO:0090649 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.4 | 6.4 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.4 | 1.7 | GO:0042320 | regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) |
| 0.4 | 3.3 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.4 | 2.0 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.4 | 9.5 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.4 | 4.3 | GO:0000050 | urea cycle(GO:0000050) |
| 0.4 | 4.6 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.4 | 1.1 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.4 | 7.5 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.4 | 4.1 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.4 | 1.5 | GO:0003335 | corneocyte development(GO:0003335) |
| 0.4 | 5.8 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.4 | 19.2 | GO:0006458 | 'de novo' protein folding(GO:0006458) |
| 0.4 | 1.1 | GO:0018874 | benzoate metabolic process(GO:0018874) butyrate metabolic process(GO:0019605) |
| 0.4 | 7.8 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.3 | 6.6 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.3 | 2.1 | GO:1902514 | regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.3 | 4.4 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.3 | 2.7 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.3 | 4.6 | GO:0032196 | transposition(GO:0032196) |
| 0.3 | 1.6 | GO:0060301 | vestibulocochlear nerve structural organization(GO:0021649) positive regulation of cytokine activity(GO:0060301) ganglion morphogenesis(GO:0061552) endothelial tip cell fate specification(GO:0097102) VEGF-activated neuropilin signaling pathway involved in axon guidance(GO:1902378) dorsal root ganglion morphogenesis(GO:1904835) otic placode development(GO:1905040) |
| 0.3 | 2.2 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.3 | 0.9 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.3 | 3.4 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.3 | 1.5 | GO:0050955 | thermoception(GO:0050955) |
| 0.3 | 1.1 | GO:0051342 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.3 | 3.4 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.3 | 8.5 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.3 | 2.5 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.3 | 2.8 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.3 | 1.4 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.3 | 1.3 | GO:0030070 | insulin processing(GO:0030070) |
| 0.3 | 1.9 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.3 | 2.1 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.3 | 1.3 | GO:0042421 | norepinephrine biosynthetic process(GO:0042421) |
| 0.3 | 0.8 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.2 | 2.7 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.2 | 3.5 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.2 | 0.2 | GO:1900109 | regulation of histone H3-K9 dimethylation(GO:1900109) |
| 0.2 | 6.5 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.2 | 1.8 | GO:0048842 | positive regulation of axon extension involved in axon guidance(GO:0048842) |
| 0.2 | 2.5 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.2 | 0.9 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.2 | 1.1 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.2 | 1.3 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.2 | 5.7 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.2 | 1.3 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.2 | 1.5 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.2 | 1.7 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.2 | 0.4 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.2 | 1.4 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.2 | 31.5 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.2 | 1.9 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.2 | 0.6 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.2 | 12.1 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.2 | 2.0 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.2 | 2.7 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.2 | 3.6 | GO:0007530 | sex determination(GO:0007530) |
| 0.2 | 3.5 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.2 | 0.5 | GO:2000870 | regulation of progesterone secretion(GO:2000870) |
| 0.2 | 6.9 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.2 | 5.1 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.2 | 3.2 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.2 | 3.7 | GO:0007565 | female pregnancy(GO:0007565) |
| 0.2 | 0.7 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.2 | 4.0 | GO:0045332 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.2 | 2.8 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.2 | 0.5 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.2 | 1.1 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 0.2 | 2.2 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.2 | 0.6 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.1 | 3.4 | GO:0001935 | endothelial cell proliferation(GO:0001935) |
| 0.1 | 2.5 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 | 2.2 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.1 | 1.7 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 0.4 | GO:2000523 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.1 | 2.6 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.1 | 0.8 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) |
| 0.1 | 1.5 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.1 | 15.8 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.1 | 0.4 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.1 | 3.9 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.1 | 0.6 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.1 | 0.7 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.1 | 0.5 | GO:0060850 | regulation of transcription involved in cell fate commitment(GO:0060850) |
| 0.1 | 1.8 | GO:0006706 | steroid catabolic process(GO:0006706) |
| 0.1 | 2.3 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 1.7 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.1 | 1.1 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.1 | 2.2 | GO:2000398 | regulation of T cell differentiation in thymus(GO:0033081) regulation of thymocyte aggregation(GO:2000398) |
| 0.1 | 2.6 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.1 | 0.7 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.1 | 6.4 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 0.3 | GO:0032641 | negative regulation of tolerance induction(GO:0002644) lymphotoxin A production(GO:0032641) interleukin-4 biosynthetic process(GO:0042097) lymphotoxin A biosynthetic process(GO:0042109) regulation of interleukin-4 biosynthetic process(GO:0045402) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.1 | 2.4 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 1.6 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.1 | 0.3 | GO:2001188 | regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001188) positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.1 | 3.8 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.1 | 2.8 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.1 | 1.4 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.1 | 2.9 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.1 | 1.2 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.1 | 0.7 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 6.8 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.1 | 0.3 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.1 | 4.5 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 16.2 | GO:0060271 | cilium morphogenesis(GO:0060271) |
| 0.1 | 0.6 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.1 | 2.9 | GO:0010762 | regulation of fibroblast migration(GO:0010762) |
| 0.1 | 1.6 | GO:0022604 | regulation of cell morphogenesis(GO:0022604) |
| 0.1 | 0.5 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 1.4 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.1 | 1.9 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.1 | 0.2 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.1 | 5.5 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.1 | 4.1 | GO:0007422 | peripheral nervous system development(GO:0007422) |
| 0.1 | 2.2 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 0.7 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.1 | 2.3 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.1 | 0.4 | GO:2001023 | regulation of response to drug(GO:2001023) |
| 0.1 | 7.7 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 0.1 | 2.8 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.1 | 6.9 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.1 | 1.9 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 2.6 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.1 | 20.5 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.1 | 1.5 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.1 | 0.4 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 1.8 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 0.4 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.1 | 2.4 | GO:0002763 | positive regulation of myeloid leukocyte differentiation(GO:0002763) |
| 0.1 | 4.1 | GO:0070268 | cornification(GO:0070268) |
| 0.1 | 1.0 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 0.9 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.1 | 2.9 | GO:0072164 | ureteric bud development(GO:0001657) mesonephric epithelium development(GO:0072163) mesonephric tubule development(GO:0072164) |
| 0.1 | 3.9 | GO:0031214 | biomineral tissue development(GO:0031214) |
| 0.1 | 0.4 | GO:0060644 | mammary gland epithelial cell differentiation(GO:0060644) |
| 0.1 | 3.3 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.0 | 1.5 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.0 | 2.1 | GO:0035196 | production of miRNAs involved in gene silencing by miRNA(GO:0035196) |
| 0.0 | 11.6 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 8.4 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 2.4 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 9.2 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 0.7 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.2 | GO:1900133 | renal response to blood flow involved in circulatory renin-angiotensin regulation of systemic arterial blood pressure(GO:0001999) renin secretion into blood stream(GO:0002001) regulation of renin secretion into blood stream(GO:1900133) |
| 0.0 | 1.4 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.7 | GO:0034390 | smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) |
| 0.0 | 33.2 | GO:0007417 | central nervous system development(GO:0007417) |
| 0.0 | 1.0 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 2.2 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.0 | 0.3 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.3 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 3.9 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 0.3 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) |
| 0.0 | 1.2 | GO:0060338 | regulation of type I interferon-mediated signaling pathway(GO:0060338) |
| 0.0 | 0.8 | GO:0046461 | neutral lipid catabolic process(GO:0046461) acylglycerol catabolic process(GO:0046464) |
| 0.0 | 4.8 | GO:0007411 | axon guidance(GO:0007411) |
| 0.0 | 1.2 | GO:0042246 | tissue regeneration(GO:0042246) |
| 0.0 | 1.9 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 2.5 | GO:0006024 | aminoglycan biosynthetic process(GO:0006023) glycosaminoglycan biosynthetic process(GO:0006024) |
| 0.0 | 0.6 | GO:0034142 | toll-like receptor 4 signaling pathway(GO:0034142) |
| 0.0 | 2.4 | GO:0060348 | bone development(GO:0060348) |
| 0.0 | 1.1 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.0 | 1.5 | GO:0019915 | lipid storage(GO:0019915) |
| 0.0 | 0.3 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.3 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 6.0 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 0.2 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 3.8 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 2.5 | GO:0007548 | sex differentiation(GO:0007548) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:0042148 | meiotic DNA recombinase assembly(GO:0000707) strand invasion(GO:0042148) |
| 0.0 | 1.5 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 1.2 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.2 | GO:0043010 | camera-type eye development(GO:0043010) |
| 0.0 | 1.0 | GO:0045766 | positive regulation of angiogenesis(GO:0045766) |
| 0.0 | 0.4 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 1.6 | GO:0042593 | carbohydrate homeostasis(GO:0033500) glucose homeostasis(GO:0042593) |
| 0.0 | 0.2 | GO:0071514 | genetic imprinting(GO:0071514) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 22.6 | GO:0032584 | growth cone membrane(GO:0032584) |
| 3.1 | 25.0 | GO:0014802 | terminal cisterna(GO:0014802) |
| 2.4 | 11.8 | GO:0071546 | pi-body(GO:0071546) |
| 2.3 | 16.0 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 2.1 | 32.9 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 1.7 | 5.2 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 1.6 | 9.8 | GO:0033063 | DNA recombinase mediator complex(GO:0033061) Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 1.4 | 6.9 | GO:0071797 | LUBAC complex(GO:0071797) |
| 1.3 | 8.0 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 1.1 | 5.7 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 1.1 | 4.5 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 1.1 | 15.5 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 1.1 | 4.2 | GO:1990246 | uniplex complex(GO:1990246) |
| 1.0 | 5.2 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 1.0 | 16.8 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 1.0 | 2.9 | GO:0071020 | post-spliceosomal complex(GO:0071020) |
| 0.9 | 3.6 | GO:0070081 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.9 | 23.1 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.8 | 3.3 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.8 | 7.0 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.7 | 17.8 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.7 | 12.8 | GO:0031045 | dense core granule(GO:0031045) |
| 0.6 | 16.0 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.6 | 2.4 | GO:1990357 | terminal web(GO:1990357) |
| 0.6 | 1.8 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.6 | 28.7 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.6 | 1.8 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.6 | 4.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.5 | 6.6 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.5 | 8.6 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.5 | 8.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.5 | 2.8 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.5 | 2.7 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.4 | 5.9 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.4 | 3.2 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.4 | 1.5 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.4 | 3.4 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.4 | 5.6 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.4 | 95.7 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.4 | 3.9 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.3 | 1.4 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.3 | 1.4 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.3 | 3.0 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.3 | 7.7 | GO:0071437 | invadopodium(GO:0071437) |
| 0.3 | 3.8 | GO:0036038 | MKS complex(GO:0036038) |
| 0.3 | 2.4 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.3 | 2.9 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.3 | 2.0 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.3 | 1.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.3 | 1.7 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.2 | 3.1 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.2 | 10.1 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.2 | 7.5 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.2 | 22.7 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.2 | 15.2 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.2 | 1.3 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.2 | 3.0 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.2 | 4.6 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.2 | 3.3 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.2 | 0.4 | GO:0034657 | GID complex(GO:0034657) |
| 0.2 | 2.4 | GO:0030914 | SAGA complex(GO:0000124) STAGA complex(GO:0030914) |
| 0.2 | 6.4 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.2 | 2.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 4.8 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 7.6 | GO:0031672 | A band(GO:0031672) |
| 0.1 | 1.6 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 4.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 2.9 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 1.6 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 5.2 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 5.8 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 31.7 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 0.9 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.1 | 8.1 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.1 | 10.6 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 2.7 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.1 | 0.7 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 0.6 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.1 | 6.3 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 5.8 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 4.0 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 2.2 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 10.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 1.5 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.1 | 3.6 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.1 | 1.5 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 3.0 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 5.9 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 2.8 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.1 | 11.4 | GO:1902495 | transmembrane transporter complex(GO:1902495) |
| 0.1 | 2.2 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 9.0 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 2.1 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 2.0 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 2.7 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 9.7 | GO:0045121 | membrane raft(GO:0045121) |
| 0.0 | 1.7 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 4.9 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.0 | 4.1 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 1.7 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 1.2 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 0.3 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.9 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 2.3 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 6.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.3 | GO:0097060 | synaptic membrane(GO:0097060) |
| 0.0 | 1.3 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 2.5 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.5 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 2.1 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.8 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 2.1 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.7 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 1.3 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.3 | 17.0 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 3.7 | 11.1 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 3.4 | 20.6 | GO:0000285 | 1-phosphatidylinositol-3-phosphate 5-kinase activity(GO:0000285) |
| 3.2 | 16.0 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 2.3 | 22.6 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 2.2 | 6.6 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 1.8 | 7.3 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 1.7 | 5.2 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 1.7 | 5.2 | GO:0008267 | poly-glutamine tract binding(GO:0008267) |
| 1.7 | 8.4 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 1.6 | 6.4 | GO:0005277 | acetylcholine transmembrane transporter activity(GO:0005277) secondary active organic cation transmembrane transporter activity(GO:0008513) acetate ester transmembrane transporter activity(GO:1901375) |
| 1.5 | 4.6 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 1.5 | 6.0 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 1.3 | 6.6 | GO:0004771 | sterol esterase activity(GO:0004771) methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 1.2 | 37.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 1.2 | 9.7 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 1.2 | 23.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 1.2 | 3.6 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 1.2 | 3.6 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 1.2 | 6.0 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 1.2 | 3.5 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 1.2 | 4.6 | GO:0004803 | transposase activity(GO:0004803) |
| 1.2 | 3.5 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 1.2 | 5.8 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 1.2 | 6.9 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 1.1 | 3.4 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 1.1 | 7.8 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 1.1 | 61.0 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 1.1 | 6.6 | GO:0047086 | phenanthrene 9,10-monooxygenase activity(GO:0018636) ketosteroid monooxygenase activity(GO:0047086) trans-1,2-dihydrobenzene-1,2-diol dehydrogenase activity(GO:0047115) |
| 1.1 | 11.0 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 1.1 | 4.2 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 1.0 | 3.0 | GO:0030366 | molybdopterin synthase activity(GO:0030366) |
| 1.0 | 2.9 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.9 | 2.7 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.9 | 2.7 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) procollagen galactosyltransferase activity(GO:0050211) |
| 0.9 | 24.5 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.8 | 4.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.8 | 3.8 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.7 | 2.2 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.7 | 3.4 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.7 | 3.3 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.7 | 2.0 | GO:0015403 | thiamine transmembrane transporter activity(GO:0015234) thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.6 | 1.9 | GO:0004797 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.6 | 1.8 | GO:0004088 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.6 | 1.8 | GO:0001034 | RNA polymerase III transcription factor activity, sequence-specific DNA binding(GO:0001034) |
| 0.6 | 16.6 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.6 | 4.5 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.6 | 13.0 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.6 | 4.4 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.5 | 3.8 | GO:0030345 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.5 | 3.3 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.5 | 1.6 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.5 | 1.6 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.5 | 8.0 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.5 | 1.5 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.5 | 2.4 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.5 | 2.3 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.4 | 4.7 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.4 | 2.6 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.4 | 8.2 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.4 | 1.5 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.4 | 3.7 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.4 | 6.6 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.4 | 1.1 | GO:0001181 | transcription factor activity, core RNA polymerase I binding(GO:0001181) |
| 0.4 | 89.7 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.4 | 1.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.3 | 3.8 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.3 | 4.2 | GO:0008392 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.3 | 2.4 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.3 | 9.1 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.3 | 7.0 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.3 | 3.0 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.3 | 7.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.3 | 11.5 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.3 | 4.9 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.3 | 2.9 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.3 | 2.9 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.3 | 4.6 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.2 | 21.9 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.2 | 0.7 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.2 | 7.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.2 | 14.0 | GO:0070888 | E-box binding(GO:0070888) |
| 0.2 | 1.6 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.2 | 4.0 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.2 | 0.9 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.2 | 1.3 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.2 | 1.7 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.2 | 5.3 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.2 | 2.8 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.2 | 4.6 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.2 | 1.0 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.2 | 1.2 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.2 | 1.6 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.2 | 1.5 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.2 | 1.3 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.2 | 0.4 | GO:0008670 | 2,4-dienoyl-CoA reductase (NADPH) activity(GO:0008670) |
| 0.2 | 1.4 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.2 | 4.1 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.2 | 48.0 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.2 | 4.0 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 1.8 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.1 | 3.9 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 2.6 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.1 | 2.4 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 2.1 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.1 | 3.6 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 23.3 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 2.1 | GO:0048185 | activin binding(GO:0048185) |
| 0.1 | 20.3 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 1.4 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.1 | 1.5 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.1 | 0.6 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.1 | 3.0 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 3.4 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 1.7 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 12.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 1.7 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 0.6 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 0.1 | 2.6 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 1.1 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.1 | 2.9 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.1 | 3.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 3.1 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 1.5 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 12.8 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 2.6 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 6.4 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 0.7 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.1 | 3.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 1.9 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 4.4 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 2.2 | GO:0071617 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) lysophosphatidic acid acyltransferase activity(GO:0042171) lysophospholipid acyltransferase activity(GO:0071617) |
| 0.1 | 2.6 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 3.1 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 2.7 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 2.5 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 1.3 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.1 | 2.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.7 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 0.3 | GO:0023023 | MHC protein complex binding(GO:0023023) MHC class II protein complex binding(GO:0023026) |
| 0.1 | 10.9 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.1 | 0.8 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.1 | 3.5 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.1 | 1.8 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.1 | 0.3 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 1.0 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) ErbB-3 class receptor binding(GO:0043125) |
| 0.1 | 2.5 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 0.7 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 2.6 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 1.9 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.1 | 0.4 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 0.1 | 2.7 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 2.4 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.1 | 2.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 1.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 4.8 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.1 | 0.9 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 5.2 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.1 | 45.6 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.1 | 2.5 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 0.7 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 1.5 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.1 | 0.4 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.1 | 5.3 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 1.4 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 3.9 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.1 | 0.5 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 3.3 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 1.4 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 1.1 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 1.1 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 1.4 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 18.6 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 1.6 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.2 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 1.3 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.2 | GO:0004084 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 3.0 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.2 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.3 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.0 | 2.4 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.4 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.7 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 2.8 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 1.3 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 1.7 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.7 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.7 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 1.1 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 2.2 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.5 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 2.5 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 3.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.9 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.3 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.2 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 9.5 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.5 | 16.0 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.4 | 22.7 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.4 | 21.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.3 | 8.0 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.3 | 6.6 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.3 | 6.6 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.2 | 2.9 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.2 | 6.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.2 | 4.3 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.2 | 11.8 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.2 | 11.4 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.2 | 6.3 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.2 | 7.3 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.2 | 8.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 8.1 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.1 | 5.6 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.1 | 20.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 1.6 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 2.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 1.5 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.1 | 18.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 2.4 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 2.6 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 1.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 3.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 2.9 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 17.2 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 2.4 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 1.6 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 2.8 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 1.3 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 1.5 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.2 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 3.1 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.7 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 1.1 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.3 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.6 | PID ARF6 PATHWAY | Arf6 signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 21.5 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.9 | 17.0 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.7 | 6.6 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.5 | 5.9 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.5 | 13.2 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.4 | 13.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.4 | 9.4 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.4 | 17.7 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.4 | 6.6 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.3 | 6.6 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.3 | 6.0 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.3 | 4.5 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.3 | 5.8 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.2 | 8.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 9.0 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.2 | 4.2 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.2 | 2.5 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.2 | 2.4 | REACTOME OPSINS | Genes involved in Opsins |
| 0.2 | 3.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.2 | 4.7 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.2 | 6.4 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.2 | 5.9 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.2 | 3.4 | REACTOME KERATAN SULFATE KERATIN METABOLISM | Genes involved in Keratan sulfate/keratin metabolism |
| 0.2 | 1.8 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.2 | 3.6 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.2 | 11.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.2 | 4.4 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.2 | 1.8 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.2 | 3.0 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.2 | 3.0 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 2.1 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.1 | 2.9 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 3.6 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 5.4 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.1 | 0.5 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.1 | 4.9 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 3.8 | REACTOME PERK REGULATED GENE EXPRESSION | Genes involved in PERK regulated gene expression |
| 0.1 | 1.1 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.1 | 3.0 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.1 | 1.6 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 1.9 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 1.6 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 1.5 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 3.8 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 4.1 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 2.9 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 2.9 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.1 | 1.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 2.5 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 3.6 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 2.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 2.2 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 1.9 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 1.7 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.1 | 13.2 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 2.6 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.1 | 1.2 | REACTOME ACTIVATION OF KAINATE RECEPTORS UPON GLUTAMATE BINDING | Genes involved in Activation of Kainate Receptors upon glutamate binding |
| 0.1 | 0.7 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 0.8 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 1.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 1.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 5.5 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 1.5 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 1.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 1.9 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.7 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 1.1 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 0.8 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 2.1 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.3 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 2.2 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 6.4 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 0.7 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 3.7 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 2.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.1 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 1.7 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.8 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.6 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.5 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.0 | 2.0 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |