Project
averaged GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
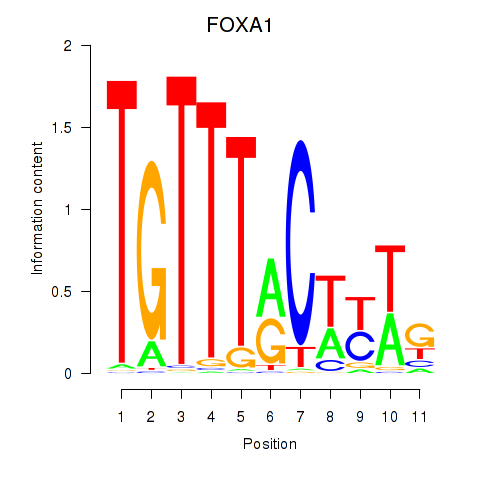
Results for FOXA1
Z-value: 1.42
Transcription factors associated with FOXA1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXA1
|
ENSG00000129514.4 | forkhead box A1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXA1 | hg19_v2_chr14_-_38064198_38064239 | 0.43 | 5.2e-11 | Click! |
Activity profile of FOXA1 motif
Sorted Z-values of FOXA1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_39684550 | 34.90 |
ENST00000455635.1
ENST00000361566.3 |
KRT19
|
keratin 19 |
| chr17_-_26695013 | 23.97 |
ENST00000555059.2
|
CTB-96E2.2
|
Homeobox protein SEBOX |
| chr19_+_18496957 | 23.00 |
ENST00000252809.3
|
GDF15
|
growth differentiation factor 15 |
| chr17_-_26694979 | 22.84 |
ENST00000438614.1
|
VTN
|
vitronectin |
| chr12_-_71551652 | 18.16 |
ENST00000546561.1
|
TSPAN8
|
tetraspanin 8 |
| chr12_-_71551868 | 16.82 |
ENST00000247829.3
|
TSPAN8
|
tetraspanin 8 |
| chr12_-_53343602 | 15.50 |
ENST00000546897.1
ENST00000552551.1 |
KRT8
|
keratin 8 |
| chr4_-_174256276 | 12.86 |
ENST00000296503.5
|
HMGB2
|
high mobility group box 2 |
| chr7_+_73242490 | 12.52 |
ENST00000431918.1
|
CLDN4
|
claudin 4 |
| chr7_+_116165754 | 12.47 |
ENST00000405348.1
|
CAV1
|
caveolin 1, caveolae protein, 22kDa |
| chr15_+_96869165 | 12.44 |
ENST00000421109.2
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr3_+_142315225 | 11.75 |
ENST00000457734.2
ENST00000483373.1 ENST00000475296.1 ENST00000495744.1 ENST00000476044.1 ENST00000461644.1 |
PLS1
|
plastin 1 |
| chr4_-_72649763 | 11.46 |
ENST00000513476.1
|
GC
|
group-specific component (vitamin D binding protein) |
| chr8_+_126442563 | 10.33 |
ENST00000311922.3
|
TRIB1
|
tribbles pseudokinase 1 |
| chr19_-_48894104 | 10.28 |
ENST00000597017.1
|
KDELR1
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 1 |
| chr3_-_185538849 | 10.22 |
ENST00000421047.2
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr3_+_52245458 | 9.88 |
ENST00000459884.1
|
ALAS1
|
aminolevulinate, delta-, synthase 1 |
| chr22_+_21128167 | 9.79 |
ENST00000215727.5
|
SERPIND1
|
serpin peptidase inhibitor, clade D (heparin cofactor), member 1 |
| chrX_-_20236970 | 9.53 |
ENST00000379548.4
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
| chr3_+_186330712 | 9.48 |
ENST00000411641.2
ENST00000273784.5 |
AHSG
|
alpha-2-HS-glycoprotein |
| chr21_-_43735446 | 9.47 |
ENST00000398431.2
|
TFF3
|
trefoil factor 3 (intestinal) |
| chr12_-_89746173 | 9.40 |
ENST00000308385.6
|
DUSP6
|
dual specificity phosphatase 6 |
| chr11_-_108464465 | 9.32 |
ENST00000525344.1
|
EXPH5
|
exophilin 5 |
| chr5_+_74011328 | 9.31 |
ENST00000513336.1
|
HEXB
|
hexosaminidase B (beta polypeptide) |
| chr4_-_155533787 | 9.19 |
ENST00000407946.1
ENST00000405164.1 ENST00000336098.3 ENST00000393846.2 ENST00000404648.3 ENST00000443553.1 |
FGG
|
fibrinogen gamma chain |
| chr14_-_36988882 | 8.95 |
ENST00000498187.2
|
NKX2-1
|
NK2 homeobox 1 |
| chr14_-_38064198 | 8.88 |
ENST00000250448.2
|
FOXA1
|
forkhead box A1 |
| chr12_+_96588143 | 8.02 |
ENST00000228741.3
ENST00000547249.1 |
ELK3
|
ELK3, ETS-domain protein (SRF accessory protein 2) |
| chr5_+_179921430 | 7.98 |
ENST00000393356.1
|
CNOT6
|
CCR4-NOT transcription complex, subunit 6 |
| chr11_+_844406 | 7.72 |
ENST00000397404.1
|
TSPAN4
|
tetraspanin 4 |
| chr17_-_19651668 | 7.60 |
ENST00000494157.2
ENST00000225740.6 |
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr19_+_39214797 | 7.56 |
ENST00000440400.1
|
ACTN4
|
actinin, alpha 4 |
| chr10_+_124320195 | 7.50 |
ENST00000359586.6
|
DMBT1
|
deleted in malignant brain tumors 1 |
| chr10_+_124320156 | 7.48 |
ENST00000338354.3
ENST00000344338.3 ENST00000330163.4 ENST00000368909.3 ENST00000368955.3 ENST00000368956.2 |
DMBT1
|
deleted in malignant brain tumors 1 |
| chr5_+_95998746 | 7.46 |
ENST00000508608.1
|
CAST
|
calpastatin |
| chr12_-_56123444 | 7.31 |
ENST00000546457.1
ENST00000549117.1 |
CD63
|
CD63 molecule |
| chr7_+_129906660 | 7.18 |
ENST00000222481.4
|
CPA2
|
carboxypeptidase A2 (pancreatic) |
| chr11_-_108464321 | 7.14 |
ENST00000265843.4
|
EXPH5
|
exophilin 5 |
| chr3_-_145878954 | 7.00 |
ENST00000282903.5
ENST00000360060.3 |
PLOD2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 |
| chr12_+_10658489 | 6.94 |
ENST00000538173.1
|
EIF2S3L
|
Putative eukaryotic translation initiation factor 2 subunit 3-like protein |
| chr12_-_56122761 | 6.94 |
ENST00000552164.1
ENST00000420846.3 ENST00000257857.4 |
CD63
|
CD63 molecule |
| chr7_-_99698338 | 6.83 |
ENST00000354230.3
ENST00000425308.1 |
MCM7
|
minichromosome maintenance complex component 7 |
| chr12_-_120765565 | 6.76 |
ENST00000423423.3
ENST00000308366.4 |
PLA2G1B
|
phospholipase A2, group IB (pancreas) |
| chr5_+_102201509 | 6.74 |
ENST00000348126.2
ENST00000379787.4 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr10_+_104535994 | 6.74 |
ENST00000369889.4
|
WBP1L
|
WW domain binding protein 1-like |
| chr19_+_16178317 | 6.73 |
ENST00000344824.6
ENST00000538887.1 |
TPM4
|
tropomyosin 4 |
| chr11_+_62186498 | 6.62 |
ENST00000278282.2
|
SCGB1A1
|
secretoglobin, family 1A, member 1 (uteroglobin) |
| chr3_+_52811596 | 6.53 |
ENST00000542827.1
ENST00000273283.2 |
ITIH1
|
inter-alpha-trypsin inhibitor heavy chain 1 |
| chr7_+_134576151 | 6.49 |
ENST00000393118.2
|
CALD1
|
caldesmon 1 |
| chr5_+_179921344 | 6.45 |
ENST00000261951.4
|
CNOT6
|
CCR4-NOT transcription complex, subunit 6 |
| chr7_+_134576317 | 6.29 |
ENST00000424922.1
ENST00000495522.1 |
CALD1
|
caldesmon 1 |
| chr2_-_152146385 | 6.27 |
ENST00000414946.1
ENST00000243346.5 |
NMI
|
N-myc (and STAT) interactor |
| chr2_-_21266935 | 6.26 |
ENST00000233242.1
|
APOB
|
apolipoprotein B |
| chr5_+_32531893 | 6.03 |
ENST00000512913.1
|
SUB1
|
SUB1 homolog (S. cerevisiae) |
| chr10_+_123923105 | 6.03 |
ENST00000368999.1
|
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr21_-_38445011 | 5.97 |
ENST00000464265.1
ENST00000399102.1 |
PIGP
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr7_+_80231466 | 5.91 |
ENST00000309881.7
ENST00000534394.1 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr3_+_148545586 | 5.83 |
ENST00000282957.4
ENST00000468341.1 |
CPB1
|
carboxypeptidase B1 (tissue) |
| chr6_+_7107999 | 5.65 |
ENST00000491191.1
ENST00000379938.2 ENST00000471433.1 |
RREB1
|
ras responsive element binding protein 1 |
| chr5_-_41213607 | 5.59 |
ENST00000337836.5
ENST00000433294.1 |
C6
|
complement component 6 |
| chr15_-_55541227 | 5.56 |
ENST00000566877.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr11_-_102401469 | 5.46 |
ENST00000260227.4
|
MMP7
|
matrix metallopeptidase 7 (matrilysin, uterine) |
| chr4_-_111119804 | 5.39 |
ENST00000394607.3
ENST00000302274.3 |
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr10_+_51549498 | 5.22 |
ENST00000358559.2
ENST00000298239.6 |
MSMB
|
microseminoprotein, beta- |
| chr17_-_17485731 | 5.21 |
ENST00000395783.1
|
PEMT
|
phosphatidylethanolamine N-methyltransferase |
| chr1_-_57431679 | 5.12 |
ENST00000371237.4
ENST00000535057.1 ENST00000543257.1 |
C8B
|
complement component 8, beta polypeptide |
| chr1_-_94079648 | 5.09 |
ENST00000370247.3
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr17_+_79953310 | 5.01 |
ENST00000582355.2
|
ASPSCR1
|
alveolar soft part sarcoma chromosome region, candidate 1 |
| chr4_-_70080449 | 4.96 |
ENST00000446444.1
|
UGT2B11
|
UDP glucuronosyltransferase 2 family, polypeptide B11 |
| chr5_+_102201430 | 4.93 |
ENST00000438793.3
ENST00000346918.2 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr17_-_46035187 | 4.85 |
ENST00000300557.2
|
PRR15L
|
proline rich 15-like |
| chr12_-_91572278 | 4.68 |
ENST00000425043.1
ENST00000420120.2 ENST00000441303.2 ENST00000456569.2 |
DCN
|
decorin |
| chr14_-_94789663 | 4.64 |
ENST00000557225.1
ENST00000341584.3 |
SERPINA6
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 6 |
| chr10_-_81320151 | 4.63 |
ENST00000372325.2
ENST00000372327.5 ENST00000417041.1 |
SFTPA2
|
surfactant protein A2 |
| chr12_-_102872317 | 4.56 |
ENST00000424202.2
|
IGF1
|
insulin-like growth factor 1 (somatomedin C) |
| chr7_+_112063192 | 4.49 |
ENST00000005558.4
|
IFRD1
|
interferon-related developmental regulator 1 |
| chr13_+_76334795 | 4.48 |
ENST00000526202.1
ENST00000465261.2 |
LMO7
|
LIM domain 7 |
| chr6_+_33589161 | 4.36 |
ENST00000605930.1
|
ITPR3
|
inositol 1,4,5-trisphosphate receptor, type 3 |
| chr1_-_43855444 | 4.34 |
ENST00000372455.4
|
MED8
|
mediator complex subunit 8 |
| chr6_+_143929307 | 4.33 |
ENST00000427704.2
ENST00000305766.6 |
PHACTR2
|
phosphatase and actin regulator 2 |
| chr7_+_77469439 | 4.32 |
ENST00000450574.1
ENST00000416283.2 ENST00000248550.7 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr13_+_76334567 | 4.27 |
ENST00000321797.8
|
LMO7
|
LIM domain 7 |
| chr20_+_306177 | 4.22 |
ENST00000544632.1
|
SOX12
|
SRY (sex determining region Y)-box 12 |
| chr17_+_8924837 | 4.09 |
ENST00000173229.2
|
NTN1
|
netrin 1 |
| chr6_-_87804815 | 4.07 |
ENST00000369582.2
|
CGA
|
glycoprotein hormones, alpha polypeptide |
| chr2_-_165424973 | 4.06 |
ENST00000543549.1
|
GRB14
|
growth factor receptor-bound protein 14 |
| chr17_+_72426891 | 4.04 |
ENST00000392627.1
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr2_-_172017343 | 3.92 |
ENST00000431350.2
ENST00000360843.3 |
TLK1
|
tousled-like kinase 1 |
| chr5_-_137674000 | 3.75 |
ENST00000510119.1
ENST00000513970.1 |
CDC25C
|
cell division cycle 25C |
| chr13_-_31038370 | 3.72 |
ENST00000399489.1
ENST00000339872.4 |
HMGB1
|
high mobility group box 1 |
| chr17_+_72427477 | 3.64 |
ENST00000342648.5
ENST00000481232.1 |
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr4_-_186456652 | 3.63 |
ENST00000284767.5
ENST00000284770.5 |
PDLIM3
|
PDZ and LIM domain 3 |
| chr14_+_64970662 | 3.59 |
ENST00000556965.1
ENST00000554015.1 |
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chr1_-_95391315 | 3.57 |
ENST00000545882.1
ENST00000415017.1 |
CNN3
|
calponin 3, acidic |
| chr4_-_186456766 | 3.49 |
ENST00000284771.6
|
PDLIM3
|
PDZ and LIM domain 3 |
| chrX_+_135614293 | 3.42 |
ENST00000370634.3
|
VGLL1
|
vestigial like 1 (Drosophila) |
| chr18_+_66465302 | 3.36 |
ENST00000360242.5
ENST00000358653.5 |
CCDC102B
|
coiled-coil domain containing 102B |
| chr12_+_21525818 | 3.29 |
ENST00000240652.3
ENST00000542023.1 ENST00000537593.1 |
IAPP
|
islet amyloid polypeptide |
| chr1_-_246729544 | 3.29 |
ENST00000544618.1
ENST00000366514.4 |
TFB2M
|
transcription factor B2, mitochondrial |
| chr14_-_37051798 | 3.25 |
ENST00000258829.5
|
NKX2-8
|
NK2 homeobox 8 |
| chr10_-_90751038 | 3.16 |
ENST00000458159.1
ENST00000415557.1 ENST00000458208.1 |
ACTA2
|
actin, alpha 2, smooth muscle, aorta |
| chr19_-_15443318 | 3.15 |
ENST00000360016.5
|
BRD4
|
bromodomain containing 4 |
| chr1_+_104159999 | 3.13 |
ENST00000414303.2
ENST00000423678.1 |
AMY2A
|
amylase, alpha 2A (pancreatic) |
| chr1_+_198126093 | 3.07 |
ENST00000367385.4
ENST00000442588.1 ENST00000538004.1 |
NEK7
|
NIMA-related kinase 7 |
| chr3_-_113897899 | 3.06 |
ENST00000383673.2
ENST00000295881.7 |
DRD3
|
dopamine receptor D3 |
| chr6_+_135502466 | 2.98 |
ENST00000367814.4
|
MYB
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr12_+_121416489 | 2.96 |
ENST00000541395.1
ENST00000544413.1 |
HNF1A
|
HNF1 homeobox A |
| chr9_-_89562104 | 2.88 |
ENST00000298743.7
|
GAS1
|
growth arrest-specific 1 |
| chr20_+_306221 | 2.87 |
ENST00000342665.2
|
SOX12
|
SRY (sex determining region Y)-box 12 |
| chr6_-_41715128 | 2.77 |
ENST00000356667.4
ENST00000373025.3 ENST00000425343.2 |
PGC
|
progastricsin (pepsinogen C) |
| chr2_-_192711968 | 2.76 |
ENST00000304141.4
|
SDPR
|
serum deprivation response |
| chr3_+_137728842 | 2.75 |
ENST00000183605.5
|
CLDN18
|
claudin 18 |
| chr10_-_32217717 | 2.73 |
ENST00000396144.4
ENST00000375245.4 ENST00000344936.2 ENST00000375250.5 |
ARHGAP12
|
Rho GTPase activating protein 12 |
| chr11_+_120973375 | 2.69 |
ENST00000264037.2
|
TECTA
|
tectorin alpha |
| chr6_-_43596899 | 2.63 |
ENST00000307126.5
ENST00000452781.1 |
GTPBP2
|
GTP binding protein 2 |
| chr4_+_55095264 | 2.60 |
ENST00000257290.5
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr6_+_26251835 | 2.58 |
ENST00000356350.2
|
HIST1H2BH
|
histone cluster 1, H2bh |
| chr13_+_78109804 | 2.58 |
ENST00000535157.1
|
SCEL
|
sciellin |
| chr13_+_78109884 | 2.52 |
ENST00000377246.3
ENST00000349847.3 |
SCEL
|
sciellin |
| chr2_+_47630255 | 2.48 |
ENST00000406134.1
|
MSH2
|
mutS homolog 2 |
| chr20_+_31823792 | 2.47 |
ENST00000375413.4
ENST00000354297.4 ENST00000375422.2 |
BPIFA1
|
BPI fold containing family A, member 1 |
| chr3_+_137717571 | 2.45 |
ENST00000343735.4
|
CLDN18
|
claudin 18 |
| chr1_+_199996733 | 2.42 |
ENST00000236914.3
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr14_+_64971292 | 2.40 |
ENST00000358738.3
ENST00000394712.2 |
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chr2_+_152266392 | 2.39 |
ENST00000444746.2
ENST00000453091.2 ENST00000428287.2 ENST00000433166.2 ENST00000420714.3 ENST00000243326.5 ENST00000414861.2 |
RIF1
|
RAP1 interacting factor homolog (yeast) |
| chr1_+_74701062 | 2.35 |
ENST00000326637.3
|
TNNI3K
|
TNNI3 interacting kinase |
| chr1_-_216596738 | 2.33 |
ENST00000307340.3
ENST00000366943.2 ENST00000366942.3 |
USH2A
|
Usher syndrome 2A (autosomal recessive, mild) |
| chr2_+_169757750 | 2.31 |
ENST00000375363.3
ENST00000429379.2 ENST00000421979.1 |
G6PC2
|
glucose-6-phosphatase, catalytic, 2 |
| chr20_-_7921090 | 2.27 |
ENST00000378789.3
|
HAO1
|
hydroxyacid oxidase (glycolate oxidase) 1 |
| chr7_+_28452130 | 2.26 |
ENST00000357727.2
|
CREB5
|
cAMP responsive element binding protein 5 |
| chr2_+_47630108 | 2.21 |
ENST00000233146.2
ENST00000454849.1 ENST00000543555.1 |
MSH2
|
mutS homolog 2 |
| chr7_+_134430212 | 2.17 |
ENST00000436461.2
|
CALD1
|
caldesmon 1 |
| chr6_+_135502408 | 2.16 |
ENST00000341911.5
ENST00000442647.2 ENST00000316528.8 |
MYB
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr1_-_111743285 | 2.15 |
ENST00000357640.4
|
DENND2D
|
DENN/MADD domain containing 2D |
| chr10_-_81708854 | 2.13 |
ENST00000372292.3
|
SFTPD
|
surfactant protein D |
| chr10_+_118350468 | 2.12 |
ENST00000358834.4
ENST00000528052.1 ENST00000442761.1 |
PNLIPRP1
|
pancreatic lipase-related protein 1 |
| chr8_+_11666649 | 2.12 |
ENST00000528643.1
ENST00000525777.1 |
FDFT1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr15_+_75335604 | 2.10 |
ENST00000563393.1
|
PPCDC
|
phosphopantothenoylcysteine decarboxylase |
| chr2_-_172017393 | 1.95 |
ENST00000442919.2
|
TLK1
|
tousled-like kinase 1 |
| chrX_+_9431324 | 1.94 |
ENST00000407597.2
ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X
|
transducin (beta)-like 1X-linked |
| chr5_+_147774275 | 1.86 |
ENST00000513826.1
|
FBXO38
|
F-box protein 38 |
| chr2_+_58655461 | 1.85 |
ENST00000429095.1
ENST00000429664.1 ENST00000452840.1 |
AC007092.1
|
long intergenic non-protein coding RNA 1122 |
| chr12_-_48398104 | 1.85 |
ENST00000337299.6
ENST00000380518.3 |
COL2A1
|
collagen, type II, alpha 1 |
| chr5_-_135701164 | 1.82 |
ENST00000355180.3
ENST00000426057.2 ENST00000513104.1 |
TRPC7
|
transient receptor potential cation channel, subfamily C, member 7 |
| chr2_-_89160770 | 1.76 |
ENST00000390240.2
|
IGKJ3
|
immunoglobulin kappa joining 3 |
| chr3_+_69985734 | 1.71 |
ENST00000314557.6
ENST00000394351.3 |
MITF
|
microphthalmia-associated transcription factor |
| chr11_+_71249071 | 1.69 |
ENST00000398534.3
|
KRTAP5-8
|
keratin associated protein 5-8 |
| chr7_+_116654935 | 1.66 |
ENST00000432298.1
ENST00000422922.1 |
ST7
|
suppression of tumorigenicity 7 |
| chr1_-_203320617 | 1.61 |
ENST00000354955.4
|
FMOD
|
fibromodulin |
| chr1_+_152486950 | 1.59 |
ENST00000368790.3
|
CRCT1
|
cysteine-rich C-terminal 1 |
| chr3_-_148939835 | 1.56 |
ENST00000264613.6
|
CP
|
ceruloplasmin (ferroxidase) |
| chr14_+_56127989 | 1.56 |
ENST00000555573.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr6_-_111804393 | 1.55 |
ENST00000368802.3
ENST00000368805.1 |
REV3L
|
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr1_+_12538594 | 1.53 |
ENST00000543710.1
|
VPS13D
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chrX_-_24665208 | 1.50 |
ENST00000356768.4
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta |
| chrX_+_99839799 | 1.49 |
ENST00000373031.4
|
TNMD
|
tenomodulin |
| chr20_+_62795827 | 1.46 |
ENST00000328439.1
ENST00000536311.1 |
MYT1
|
myelin transcription factor 1 |
| chr9_+_82187487 | 1.45 |
ENST00000435650.1
ENST00000414465.1 ENST00000376537.4 ENST00000376534.4 |
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr16_-_67517716 | 1.42 |
ENST00000290953.2
|
AGRP
|
agouti related protein homolog (mouse) |
| chr1_+_199996702 | 1.36 |
ENST00000367362.3
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr1_+_214161854 | 1.33 |
ENST00000435016.1
|
PROX1
|
prospero homeobox 1 |
| chr2_+_204801471 | 1.30 |
ENST00000316386.6
ENST00000435193.1 |
ICOS
|
inducible T-cell co-stimulator |
| chr4_+_187148556 | 1.27 |
ENST00000264690.6
ENST00000446598.2 ENST00000414291.1 ENST00000513864.1 |
KLKB1
|
kallikrein B, plasma (Fletcher factor) 1 |
| chr14_+_37131058 | 1.24 |
ENST00000361487.6
|
PAX9
|
paired box 9 |
| chr9_+_27109133 | 1.24 |
ENST00000519097.1
ENST00000380036.4 |
TEK
|
TEK tyrosine kinase, endothelial |
| chr10_+_102891048 | 1.23 |
ENST00000467928.2
|
TLX1
|
T-cell leukemia homeobox 1 |
| chr15_+_60296421 | 1.20 |
ENST00000396057.4
|
FOXB1
|
forkhead box B1 |
| chr7_-_81399438 | 1.17 |
ENST00000222390.5
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr6_+_161123270 | 1.15 |
ENST00000366924.2
ENST00000308192.9 ENST00000418964.1 |
PLG
|
plasminogen |
| chr9_+_136287444 | 1.14 |
ENST00000355699.2
ENST00000356589.2 ENST00000371911.3 |
ADAMTS13
|
ADAM metallopeptidase with thrombospondin type 1 motif, 13 |
| chr9_-_98279241 | 1.14 |
ENST00000437951.1
ENST00000375274.2 ENST00000430669.2 ENST00000468211.2 |
PTCH1
|
patched 1 |
| chr4_+_187187098 | 1.12 |
ENST00000403665.2
ENST00000264692.4 |
F11
|
coagulation factor XI |
| chr6_-_89927151 | 1.10 |
ENST00000454853.2
|
GABRR1
|
gamma-aminobutyric acid (GABA) A receptor, rho 1 |
| chr7_+_134832808 | 1.08 |
ENST00000275767.3
|
TMEM140
|
transmembrane protein 140 |
| chr9_-_95186739 | 1.07 |
ENST00000375550.4
|
OMD
|
osteomodulin |
| chr12_-_14849470 | 1.07 |
ENST00000261170.3
|
GUCY2C
|
guanylate cyclase 2C (heat stable enterotoxin receptor) |
| chr3_+_141106643 | 1.06 |
ENST00000514251.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr6_-_161085291 | 1.01 |
ENST00000316300.5
|
LPA
|
lipoprotein, Lp(a) |
| chr8_+_70404996 | 1.00 |
ENST00000402687.4
ENST00000419716.3 |
SULF1
|
sulfatase 1 |
| chr7_-_81399355 | 0.98 |
ENST00000457544.2
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr6_+_135502501 | 0.95 |
ENST00000527615.1
ENST00000420123.2 ENST00000525369.1 ENST00000528774.1 ENST00000534121.1 ENST00000534044.1 ENST00000533624.1 |
MYB
|
v-myb avian myeloblastosis viral oncogene homolog |
| chrX_+_24167746 | 0.93 |
ENST00000428571.1
ENST00000539115.1 |
ZFX
|
zinc finger protein, X-linked |
| chr5_-_127674883 | 0.91 |
ENST00000507835.1
|
FBN2
|
fibrillin 2 |
| chr11_-_26743546 | 0.89 |
ENST00000280467.6
ENST00000396005.3 |
SLC5A12
|
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 12 |
| chr3_+_160394940 | 0.89 |
ENST00000320767.2
|
ARL14
|
ADP-ribosylation factor-like 14 |
| chr7_+_106505696 | 0.88 |
ENST00000440650.2
ENST00000496166.1 ENST00000473541.1 |
PIK3CG
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
| chrX_-_15402498 | 0.85 |
ENST00000297904.3
|
FIGF
|
c-fos induced growth factor (vascular endothelial growth factor D) |
| chr12_+_121416340 | 0.84 |
ENST00000257555.6
ENST00000400024.2 |
HNF1A
|
HNF1 homeobox A |
| chr14_-_64971288 | 0.82 |
ENST00000394715.1
|
ZBTB25
|
zinc finger and BTB domain containing 25 |
| chr7_+_106505912 | 0.82 |
ENST00000359195.3
|
PIK3CG
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
| chr22_-_40929812 | 0.82 |
ENST00000422851.1
|
MKL1
|
megakaryoblastic leukemia (translocation) 1 |
| chr1_+_145883868 | 0.76 |
ENST00000447947.2
|
GPR89C
|
G protein-coupled receptor 89C |
| chr5_-_148758839 | 0.76 |
ENST00000261796.3
|
IL17B
|
interleukin 17B |
| chr8_-_131028869 | 0.76 |
ENST00000518283.1
ENST00000519110.1 |
FAM49B
|
family with sequence similarity 49, member B |
| chr17_-_48546324 | 0.75 |
ENST00000508540.1
|
CHAD
|
chondroadherin |
| chr7_+_95115210 | 0.75 |
ENST00000428113.1
ENST00000325885.5 |
ASB4
|
ankyrin repeat and SOCS box containing 4 |
| chrX_-_124097620 | 0.69 |
ENST00000371130.3
ENST00000422452.2 |
TENM1
|
teneurin transmembrane protein 1 |
| chr7_+_31003621 | 0.67 |
ENST00000326139.2
|
GHRHR
|
growth hormone releasing hormone receptor |
| chr8_-_71316021 | 0.63 |
ENST00000452400.2
|
NCOA2
|
nuclear receptor coactivator 2 |
| chr12_+_103981044 | 0.61 |
ENST00000388887.2
|
STAB2
|
stabilin 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXA1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 12.5 | GO:1900085 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 3.4 | 10.3 | GO:0045658 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 3.1 | 12.4 | GO:0009956 | radial pattern formation(GO:0009956) |
| 2.9 | 11.7 | GO:0018032 | peptide amidation(GO:0001519) protein amidation(GO:0018032) peptide modification(GO:0031179) |
| 2.5 | 22.8 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 2.5 | 7.6 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 2.3 | 11.7 | GO:1902896 | terminal web assembly(GO:1902896) |
| 2.3 | 9.4 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) |
| 2.3 | 7.0 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 2.2 | 8.9 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 2.1 | 6.3 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 2.1 | 14.4 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 2.0 | 6.1 | GO:1904897 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 1.9 | 9.3 | GO:0007619 | courtship behavior(GO:0007619) |
| 1.8 | 9.0 | GO:0021759 | globus pallidus development(GO:0021759) |
| 1.7 | 17.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 1.7 | 6.8 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 1.5 | 6.0 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 1.5 | 4.5 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 1.5 | 4.4 | GO:0050917 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 1.4 | 9.8 | GO:0008218 | bioluminescence(GO:0008218) |
| 1.4 | 5.6 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 1.4 | 4.2 | GO:0035565 | regulation of pronephros size(GO:0035565) renal glucose absorption(GO:0035623) |
| 1.3 | 12.9 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 1.3 | 47.8 | GO:0060706 | cell differentiation involved in embryonic placenta development(GO:0060706) |
| 1.2 | 22.0 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 1.1 | 6.6 | GO:0010193 | response to ozone(GO:0010193) |
| 1.1 | 14.2 | GO:2000680 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 1.0 | 23.0 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 1.0 | 5.1 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.9 | 4.7 | GO:0010520 | meiotic gene conversion(GO:0006311) regulation of reciprocal meiotic recombination(GO:0010520) |
| 0.9 | 10.3 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.9 | 3.7 | GO:0002840 | plasmacytoid dendritic cell activation(GO:0002270) T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) regulation of restriction endodeoxyribonuclease activity(GO:0032072) negative regulation of apoptotic cell clearance(GO:2000426) |
| 0.9 | 4.6 | GO:1904075 | trophectodermal cell proliferation(GO:0001834) regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.9 | 5.4 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.9 | 6.3 | GO:0006642 | triglyceride mobilization(GO:0006642) response to selenium ion(GO:0010269) |
| 0.9 | 3.5 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.9 | 2.6 | GO:0072275 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.8 | 5.9 | GO:2000334 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.8 | 9.9 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.8 | 3.3 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.8 | 4.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.8 | 3.1 | GO:0050883 | negative regulation of sodium:proton antiporter activity(GO:0032416) gastric motility(GO:0035482) gastric emptying(GO:0035483) musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.8 | 2.3 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.7 | 44.2 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.7 | 6.8 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.7 | 5.9 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.7 | 5.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.6 | 3.2 | GO:0072007 | mesangial cell differentiation(GO:0072007) glomerular mesangial cell differentiation(GO:0072008) mesangial cell development(GO:0072143) glomerular mesangial cell development(GO:0072144) |
| 0.6 | 12.5 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.6 | 2.5 | GO:0044010 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.6 | 8.5 | GO:0045779 | negative regulation of bone resorption(GO:0045779) |
| 0.6 | 2.4 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.6 | 2.3 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.5 | 7.6 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.5 | 1.6 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.5 | 9.5 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.5 | 2.4 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.5 | 2.8 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.4 | 3.1 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.4 | 4.7 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.4 | 2.1 | GO:0006696 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.4 | 11.5 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.4 | 1.2 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.4 | 1.9 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) regulation of branching involved in salivary gland morphogenesis(GO:0060693) |
| 0.4 | 1.1 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.4 | 1.5 | GO:0035992 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.4 | 7.2 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.3 | 9.5 | GO:0043555 | regulation of translation in response to stress(GO:0043555) |
| 0.3 | 1.2 | GO:1900154 | regulation of bone trabecula formation(GO:1900154) negative regulation of bone trabecula formation(GO:1900155) |
| 0.3 | 6.0 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.3 | 1.7 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) secretory granule localization(GO:0032252) |
| 0.3 | 0.5 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.3 | 3.2 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.2 | 5.5 | GO:0044849 | estrous cycle(GO:0044849) |
| 0.2 | 5.1 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.2 | 0.7 | GO:0046010 | positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) |
| 0.2 | 5.1 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.2 | 2.3 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.2 | 2.9 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.2 | 3.6 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.2 | 1.2 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.2 | 6.0 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) |
| 0.2 | 1.8 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.2 | 0.8 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.2 | 1.9 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.2 | 0.5 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.2 | 1.4 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.2 | 0.6 | GO:1904016 | response to Thyroglobulin triiodothyronine(GO:1904016) |
| 0.2 | 7.1 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 4.6 | GO:0008211 | glucocorticoid metabolic process(GO:0008211) |
| 0.1 | 1.9 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.1 | 3.1 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.1 | 1.1 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.1 | 5.0 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.1 | 0.8 | GO:2000567 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.1 | 3.8 | GO:1902751 | positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.1 | 0.5 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.1 | 1.0 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.1 | 2.4 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.1 | 0.9 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.1 | 2.1 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 0.9 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.1 | 1.1 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 4.1 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.1 | 7.6 | GO:0006081 | cellular aldehyde metabolic process(GO:0006081) |
| 0.1 | 1.1 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.1 | 1.8 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.1 | 6.7 | GO:0033275 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.1 | 1.6 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 0.7 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.1 | 3.1 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 1.0 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.1 | 0.2 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.1 | 5.0 | GO:0046324 | regulation of glucose import(GO:0046324) |
| 0.1 | 4.6 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.1 | 0.5 | GO:0015705 | iodide transport(GO:0015705) |
| 0.1 | 1.6 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.1 | 0.4 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 0.8 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 1.2 | GO:0042481 | regulation of odontogenesis(GO:0042481) |
| 0.0 | 10.2 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 7.0 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.0 | 2.1 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 6.0 | GO:0021987 | cerebral cortex development(GO:0021987) |
| 0.0 | 1.9 | GO:0010976 | positive regulation of neuron projection development(GO:0010976) |
| 0.0 | 0.5 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 | 2.7 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.4 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.0 | 3.7 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 12.1 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 0.4 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.0 | 6.9 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 3.3 | GO:0050680 | negative regulation of epithelial cell proliferation(GO:0050680) |
| 0.0 | 0.7 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 2.6 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 5.9 | GO:0043687 | post-translational protein modification(GO:0043687) |
| 0.0 | 3.6 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.0 | 1.3 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.0 | 0.3 | GO:0043949 | regulation of cAMP-mediated signaling(GO:0043949) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.8 | 46.6 | GO:1990357 | terminal web(GO:1990357) |
| 5.7 | 22.8 | GO:0071062 | rough endoplasmic reticulum lumen(GO:0048237) alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 1.6 | 4.7 | GO:0032302 | MutSbeta complex(GO:0032302) |
| 1.2 | 10.7 | GO:0005579 | membrane attack complex(GO:0005579) |
| 1.2 | 14.2 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 1.0 | 15.0 | GO:0030478 | actin cap(GO:0030478) |
| 0.9 | 15.5 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.9 | 12.5 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.8 | 14.4 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.8 | 24.9 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.8 | 10.3 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.8 | 3.2 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.8 | 6.3 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.8 | 2.3 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.8 | 4.6 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.7 | 9.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.6 | 6.0 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.6 | 1.7 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.5 | 6.8 | GO:0042555 | MCM complex(GO:0042555) |
| 0.5 | 5.6 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.5 | 2.4 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.4 | 6.7 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.3 | 10.2 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.3 | 4.7 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.3 | 6.8 | GO:0042599 | lamellar body(GO:0042599) |
| 0.3 | 7.0 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.3 | 7.6 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.3 | 4.4 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.2 | 1.2 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.2 | 23.5 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.2 | 13.4 | GO:0005902 | microvillus(GO:0005902) |
| 0.2 | 1.8 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 14.8 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 1.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.1 | 4.1 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 4.1 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 0.7 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 4.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 1.6 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.1 | 2.9 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 5.1 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 5.2 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 0.4 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.1 | 5.0 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 129.8 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 43.2 | GO:0009986 | cell surface(GO:0009986) |
| 0.1 | 3.3 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 7.7 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 7.2 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 6.8 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 1.8 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 2.3 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 1.1 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 2.3 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 3.2 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 1.9 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 3.6 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 1.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 3.6 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 3.0 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 3.9 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 1.5 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 2.4 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.5 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 45.4 | GO:0005654 | nucleoplasm(GO:0005654) |
| 0.0 | 3.3 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 6.8 | GO:0005768 | endosome(GO:0005768) |
| 0.0 | 1.6 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 12.5 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 3.3 | 9.9 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 3.2 | 12.9 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 2.9 | 11.7 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 2.6 | 10.3 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 2.3 | 7.0 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 2.0 | 5.9 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 1.9 | 11.5 | GO:0005499 | vitamin D binding(GO:0005499) |
| 1.7 | 5.2 | GO:0004608 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 1.6 | 6.3 | GO:0035473 | lipase binding(GO:0035473) |
| 1.6 | 4.7 | GO:0000404 | heteroduplex DNA loop binding(GO:0000404) double-strand/single-strand DNA junction binding(GO:0000406) dinucleotide repeat insertion binding(GO:0032181) |
| 1.5 | 7.6 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 1.4 | 22.8 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 1.2 | 7.5 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 1.2 | 9.3 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 1.1 | 6.6 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 1.1 | 4.4 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 1.0 | 5.9 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.9 | 5.4 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.9 | 2.6 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.8 | 14.4 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.8 | 3.1 | GO:0016160 | amylase activity(GO:0016160) |
| 0.8 | 2.3 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.7 | 15.0 | GO:0038187 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.7 | 6.8 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.7 | 3.3 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.6 | 9.6 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.6 | 9.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.6 | 3.7 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.6 | 10.3 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.6 | 6.0 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.6 | 2.3 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.6 | 6.1 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.5 | 15.0 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.5 | 2.1 | GO:0004310 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.5 | 48.7 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.5 | 2.4 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.4 | 3.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.4 | 12.1 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.4 | 1.9 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.4 | 1.8 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.3 | 10.2 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.3 | 21.1 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.3 | 12.4 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.3 | 6.0 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.3 | 48.5 | GO:0005178 | integrin binding(GO:0005178) |
| 0.3 | 5.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.3 | 1.0 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.2 | 1.7 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.2 | 19.0 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.2 | 1.1 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.2 | 1.3 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.2 | 5.6 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.2 | 21.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 0.8 | GO:0023025 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.2 | 15.5 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.2 | 0.9 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.2 | 3.6 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.2 | 1.6 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.2 | 0.7 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.2 | 0.5 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.2 | 6.8 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.1 | 1.2 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.1 | 1.8 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 8.1 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 0.4 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.1 | 1.7 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 3.2 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 2.8 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 6.0 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 0.9 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 7.6 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 4.3 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 6.9 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 0.5 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.1 | 3.8 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 9.2 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 2.1 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 1.1 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.1 | 0.5 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 9.2 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.1 | 1.0 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.1 | 10.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 4.3 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.1 | 1.1 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 5.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 3.8 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.1 | 3.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 1.1 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 1.1 | GO:0004890 | GABA-A receptor activity(GO:0004890) GABA receptor activity(GO:0016917) |
| 0.1 | 0.6 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.1 | 2.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 1.6 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 2.4 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 3.2 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) |
| 0.0 | 0.3 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.0 | 1.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 2.1 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 2.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 6.0 | GO:0035257 | nuclear hormone receptor binding(GO:0035257) |
| 0.0 | 6.5 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 3.7 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 1.0 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 3.6 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 3.1 | GO:0003712 | transcription cofactor activity(GO:0003712) |
| 0.0 | 1.2 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 63.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.8 | 52.3 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.5 | 21.2 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.5 | 15.1 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.4 | 10.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.4 | 23.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 12.2 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.2 | 16.7 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.2 | 16.0 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.2 | 10.6 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 36.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 6.4 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 4.6 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.1 | 4.4 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 3.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 5.8 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 4.7 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 0.9 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.1 | 6.5 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 5.2 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 3.5 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 5.1 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 16.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 2.9 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 1.1 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 1.8 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 1.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 1.7 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.6 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.0 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 1.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 2.1 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.6 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 16.6 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.7 | 12.0 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.6 | 9.4 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.6 | 4.1 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.6 | 9.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.5 | 6.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.5 | 6.8 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.4 | 2.6 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.4 | 9.5 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.4 | 12.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.4 | 6.8 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.4 | 15.5 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.4 | 6.3 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.4 | 14.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.4 | 24.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.4 | 14.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.4 | 9.9 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.3 | 3.1 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.3 | 25.5 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.3 | 7.1 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.3 | 6.0 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.3 | 5.4 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.2 | 7.6 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.2 | 7.6 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.2 | 5.0 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.2 | 10.7 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.2 | 24.7 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.2 | 7.9 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.2 | 5.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.2 | 4.7 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.2 | 3.8 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.2 | 2.1 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 17.0 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 7.0 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.1 | 1.2 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.1 | 1.1 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 2.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 3.1 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 1.7 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.1 | 2.1 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 10.2 | REACTOME DIABETES PATHWAYS | Genes involved in Diabetes pathways |
| 0.1 | 1.6 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 2.6 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 2.3 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.6 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.4 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 1.0 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.0 | 3.3 | REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | Genes involved in RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription |
| 0.0 | 0.7 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 1.3 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.0 | 0.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.6 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.0 | 1.1 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.2 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |