Project
averaged GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
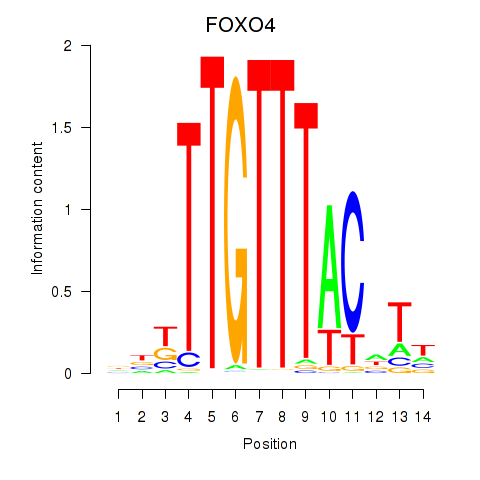
Results for FOXO4
Z-value: 1.34
Transcription factors associated with FOXO4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXO4
|
ENSG00000184481.12 | forkhead box O4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXO4 | hg19_v2_chrX_+_70316005_70316047 | 0.69 | 9.4e-32 | Click! |
Activity profile of FOXO4 motif
Sorted Z-values of FOXO4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_42812143 | 27.62 |
ENST00000514985.1
|
SEPP1
|
selenoprotein P, plasma, 1 |
| chr5_-_42811986 | 27.49 |
ENST00000511224.1
ENST00000507920.1 ENST00000510965.1 |
SEPP1
|
selenoprotein P, plasma, 1 |
| chr1_-_182360918 | 23.86 |
ENST00000339526.4
|
GLUL
|
glutamate-ammonia ligase |
| chr1_-_57045228 | 22.44 |
ENST00000371250.3
|
PPAP2B
|
phosphatidic acid phosphatase type 2B |
| chr1_+_6845384 | 21.52 |
ENST00000303635.7
|
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr16_+_6069586 | 21.38 |
ENST00000547372.1
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr14_-_60097297 | 20.58 |
ENST00000395090.1
|
RTN1
|
reticulon 1 |
| chr1_-_182360498 | 19.92 |
ENST00000417584.2
|
GLUL
|
glutamate-ammonia ligase |
| chr3_-_112127981 | 19.83 |
ENST00000486726.2
|
RP11-231E6.1
|
RP11-231E6.1 |
| chr19_-_36523709 | 19.76 |
ENST00000592017.1
ENST00000360535.4 |
CLIP3
|
CAP-GLY domain containing linker protein 3 |
| chr2_-_71454185 | 19.63 |
ENST00000244221.8
|
PAIP2B
|
poly(A) binding protein interacting protein 2B |
| chr11_-_62476965 | 18.43 |
ENST00000405837.1
ENST00000531524.1 |
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr8_-_134309823 | 18.10 |
ENST00000414097.2
|
NDRG1
|
N-myc downstream regulated 1 |
| chr11_-_62477041 | 17.70 |
ENST00000433053.1
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr16_+_6069072 | 15.93 |
ENST00000547605.1
ENST00000550418.1 ENST00000553186.1 |
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr8_-_134309335 | 15.53 |
ENST00000522890.1
ENST00000323851.7 ENST00000518176.1 ENST00000354944.5 ENST00000537882.1 ENST00000522476.1 ENST00000518066.1 ENST00000521544.1 ENST00000518480.1 ENST00000523892.1 |
NDRG1
|
N-myc downstream regulated 1 |
| chr20_+_44035200 | 15.40 |
ENST00000372717.1
ENST00000360981.4 |
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr10_-_90751038 | 15.25 |
ENST00000458159.1
ENST00000415557.1 ENST00000458208.1 |
ACTA2
|
actin, alpha 2, smooth muscle, aorta |
| chr17_-_17875688 | 15.13 |
ENST00000379504.3
ENST00000318094.10 ENST00000540946.1 ENST00000542206.1 ENST00000395739.4 ENST00000581396.1 ENST00000535933.1 ENST00000579586.1 |
TOM1L2
|
target of myb1-like 2 (chicken) |
| chr14_-_103987679 | 14.53 |
ENST00000553610.1
|
CKB
|
creatine kinase, brain |
| chr6_+_101846664 | 13.66 |
ENST00000421544.1
ENST00000413795.1 ENST00000369138.1 ENST00000358361.3 |
GRIK2
|
glutamate receptor, ionotropic, kainate 2 |
| chr19_-_37019136 | 13.56 |
ENST00000592282.1
|
ZNF260
|
zinc finger protein 260 |
| chr6_-_152489484 | 13.00 |
ENST00000354674.4
ENST00000539504.1 |
SYNE1
|
spectrin repeat containing, nuclear envelope 1 |
| chr5_-_64920115 | 12.88 |
ENST00000381018.3
ENST00000274327.7 |
TRIM23
|
tripartite motif containing 23 |
| chr3_-_28390581 | 12.68 |
ENST00000479665.1
|
AZI2
|
5-azacytidine induced 2 |
| chr2_+_233527443 | 12.64 |
ENST00000410095.1
|
EFHD1
|
EF-hand domain family, member D1 |
| chr20_+_44035847 | 12.62 |
ENST00000372712.2
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr2_-_37193606 | 12.44 |
ENST00000379213.2
ENST00000263918.4 |
STRN
|
striatin, calmodulin binding protein |
| chr17_+_17876127 | 12.39 |
ENST00000582416.1
ENST00000313838.8 ENST00000411504.2 ENST00000581264.1 ENST00000399187.1 ENST00000479684.2 ENST00000584166.1 ENST00000585108.1 ENST00000399182.1 ENST00000579977.1 |
LRRC48
|
leucine rich repeat containing 48 |
| chr19_+_50380682 | 11.99 |
ENST00000221543.5
|
TBC1D17
|
TBC1 domain family, member 17 |
| chr11_-_114466477 | 11.94 |
ENST00000375478.3
|
NXPE4
|
neurexophilin and PC-esterase domain family, member 4 |
| chr19_+_50380917 | 11.61 |
ENST00000535102.2
|
TBC1D17
|
TBC1 domain family, member 17 |
| chr16_-_4852915 | 11.30 |
ENST00000322048.7
|
ROGDI
|
rogdi homolog (Drosophila) |
| chr10_-_73848086 | 10.89 |
ENST00000536168.1
|
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr14_+_70918874 | 10.19 |
ENST00000603540.1
|
ADAM21
|
ADAM metallopeptidase domain 21 |
| chr1_+_43855560 | 10.07 |
ENST00000562955.1
|
SZT2
|
seizure threshold 2 homolog (mouse) |
| chr12_-_12491608 | 9.89 |
ENST00000545735.1
|
MANSC1
|
MANSC domain containing 1 |
| chr9_+_27109133 | 9.33 |
ENST00000519097.1
ENST00000380036.4 |
TEK
|
TEK tyrosine kinase, endothelial |
| chr11_-_114466471 | 9.29 |
ENST00000424261.2
|
NXPE4
|
neurexophilin and PC-esterase domain family, member 4 |
| chr3_-_48956818 | 9.21 |
ENST00000408959.2
|
ARIH2OS
|
ariadne homolog 2 opposite strand |
| chr5_-_115872142 | 9.19 |
ENST00000510263.1
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr9_+_108006880 | 9.05 |
ENST00000374723.1
ENST00000374720.3 ENST00000374724.1 |
SLC44A1
|
solute carrier family 44 (choline transporter), member 1 |
| chr10_+_111985713 | 9.03 |
ENST00000239007.7
|
MXI1
|
MAX interactor 1, dimerization protein |
| chr18_-_21852143 | 8.96 |
ENST00000399443.3
|
OSBPL1A
|
oxysterol binding protein-like 1A |
| chr18_-_53303123 | 8.72 |
ENST00000569357.1
ENST00000565124.1 ENST00000398339.1 |
TCF4
|
transcription factor 4 |
| chr11_+_46639071 | 8.60 |
ENST00000580238.1
ENST00000581416.1 ENST00000529655.1 ENST00000533325.1 ENST00000581438.1 ENST00000583249.1 ENST00000530500.1 ENST00000526508.1 ENST00000578626.1 ENST00000577256.1 ENST00000524625.1 ENST00000582547.1 ENST00000359513.4 ENST00000528494.1 |
ATG13
|
autophagy related 13 |
| chr1_+_150229554 | 8.55 |
ENST00000369111.4
|
CA14
|
carbonic anhydrase XIV |
| chr12_-_10251576 | 8.35 |
ENST00000315330.4
|
CLEC1A
|
C-type lectin domain family 1, member A |
| chr3_+_138068051 | 8.26 |
ENST00000474559.1
|
MRAS
|
muscle RAS oncogene homolog |
| chr5_+_64920543 | 8.23 |
ENST00000399438.3
ENST00000510585.2 |
TRAPPC13
CTC-534A2.2
|
trafficking protein particle complex 13 CDNA FLJ26957 fis, clone SLV00486; Uncharacterized protein |
| chr7_-_37026108 | 8.16 |
ENST00000396045.3
|
ELMO1
|
engulfment and cell motility 1 |
| chr19_-_40732594 | 8.15 |
ENST00000430325.2
ENST00000433940.1 |
CNTD2
|
cyclin N-terminal domain containing 2 |
| chr14_-_75518129 | 8.09 |
ENST00000556257.1
ENST00000557648.1 ENST00000553263.1 ENST00000355774.2 ENST00000380968.2 ENST00000238662.7 |
MLH3
|
mutL homolog 3 |
| chr14_+_94577074 | 8.08 |
ENST00000444961.1
ENST00000448882.1 ENST00000557098.1 ENST00000554800.1 ENST00000556544.1 ENST00000298902.5 ENST00000555819.1 ENST00000557634.1 ENST00000555744.1 |
IFI27
|
interferon, alpha-inducible protein 27 |
| chr15_+_43985725 | 8.04 |
ENST00000413453.2
|
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr19_-_58662139 | 8.00 |
ENST00000598312.1
|
ZNF329
|
zinc finger protein 329 |
| chr11_+_12115543 | 7.95 |
ENST00000537344.1
ENST00000532179.1 ENST00000526065.1 |
MICAL2
|
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
| chr12_-_10251603 | 7.76 |
ENST00000457018.2
|
CLEC1A
|
C-type lectin domain family 1, member A |
| chr4_+_88754069 | 7.73 |
ENST00000395102.4
ENST00000497649.2 |
MEPE
|
matrix extracellular phosphoglycoprotein |
| chr7_-_143059780 | 7.72 |
ENST00000409578.1
ENST00000409346.1 |
FAM131B
|
family with sequence similarity 131, member B |
| chr11_+_46638805 | 7.68 |
ENST00000434074.1
ENST00000312040.4 ENST00000451945.1 |
ATG13
|
autophagy related 13 |
| chr6_+_123110465 | 7.67 |
ENST00000539041.1
|
SMPDL3A
|
sphingomyelin phosphodiesterase, acid-like 3A |
| chr11_+_57365150 | 7.61 |
ENST00000457869.1
ENST00000340687.6 ENST00000378323.4 ENST00000378324.2 ENST00000403558.1 |
SERPING1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 |
| chr15_+_43985084 | 7.61 |
ENST00000434505.1
ENST00000411750.1 |
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr21_-_32931290 | 7.52 |
ENST00000286827.3
|
TIAM1
|
T-cell lymphoma invasion and metastasis 1 |
| chr6_-_46889694 | 7.43 |
ENST00000283296.7
ENST00000362015.4 ENST00000456426.2 |
GPR116
|
G protein-coupled receptor 116 |
| chr1_+_162039558 | 7.39 |
ENST00000530878.1
ENST00000361897.5 |
NOS1AP
|
nitric oxide synthase 1 (neuronal) adaptor protein |
| chr2_+_157292859 | 7.31 |
ENST00000438166.2
|
GPD2
|
glycerol-3-phosphate dehydrogenase 2 (mitochondrial) |
| chr4_-_186696425 | 7.24 |
ENST00000430503.1
ENST00000319454.6 ENST00000450341.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr7_-_143059845 | 7.20 |
ENST00000443739.2
|
FAM131B
|
family with sequence similarity 131, member B |
| chr15_+_43885252 | 7.17 |
ENST00000453782.1
ENST00000300283.6 ENST00000437924.1 ENST00000450086.2 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr2_+_175260451 | 6.78 |
ENST00000458563.1
ENST00000409673.3 ENST00000272732.6 ENST00000435964.1 |
SCRN3
|
secernin 3 |
| chr4_+_88754113 | 6.72 |
ENST00000560249.1
ENST00000540395.1 ENST00000511670.1 ENST00000361056.3 |
MEPE
|
matrix extracellular phosphoglycoprotein |
| chr19_+_1205740 | 6.48 |
ENST00000326873.7
|
STK11
|
serine/threonine kinase 11 |
| chr10_-_52645379 | 6.39 |
ENST00000395489.2
|
A1CF
|
APOBEC1 complementation factor |
| chr5_+_156712372 | 6.38 |
ENST00000541131.1
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2 |
| chr3_+_148447887 | 6.38 |
ENST00000475347.1
ENST00000474935.1 ENST00000461609.1 |
AGTR1
|
angiotensin II receptor, type 1 |
| chr11_-_94226964 | 6.31 |
ENST00000538923.1
ENST00000540013.1 ENST00000407439.3 ENST00000393241.4 |
MRE11A
|
MRE11 meiotic recombination 11 homolog A (S. cerevisiae) |
| chr4_-_186697044 | 6.30 |
ENST00000437304.2
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr2_+_32502952 | 6.25 |
ENST00000238831.4
|
YIPF4
|
Yip1 domain family, member 4 |
| chr13_+_32838801 | 6.22 |
ENST00000542859.1
|
FRY
|
furry homolog (Drosophila) |
| chr11_-_55703876 | 6.16 |
ENST00000301532.3
|
OR5I1
|
olfactory receptor, family 5, subfamily I, member 1 |
| chr5_-_131132658 | 6.13 |
ENST00000514667.1
ENST00000511848.1 ENST00000510461.1 |
CTC-432M15.3
FNIP1
|
Folliculin-interacting protein 1 folliculin interacting protein 1 |
| chr6_+_72596604 | 6.00 |
ENST00000348717.5
ENST00000517960.1 ENST00000518273.1 ENST00000522291.1 ENST00000521978.1 ENST00000520567.1 ENST00000264839.7 |
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr5_-_107703556 | 5.98 |
ENST00000496714.1
|
FBXL17
|
F-box and leucine-rich repeat protein 17 |
| chr10_-_61900762 | 5.96 |
ENST00000355288.2
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr17_-_8263538 | 5.90 |
ENST00000535173.1
|
AC135178.1
|
HCG1985372; Uncharacterized protein; cDNA FLJ37541 fis, clone BRCAN2026340 |
| chr19_+_30863271 | 5.83 |
ENST00000355537.3
|
ZNF536
|
zinc finger protein 536 |
| chr18_+_77160282 | 5.78 |
ENST00000318065.5
ENST00000545796.1 ENST00000592223.1 ENST00000329101.4 ENST00000586434.1 |
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 |
| chr9_+_27109392 | 5.74 |
ENST00000406359.4
|
TEK
|
TEK tyrosine kinase, endothelial |
| chr4_-_90757364 | 5.67 |
ENST00000508895.1
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr3_+_169490606 | 5.66 |
ENST00000349841.5
|
MYNN
|
myoneurin |
| chr10_-_62332357 | 5.63 |
ENST00000503366.1
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr8_-_42698433 | 5.62 |
ENST00000345117.2
ENST00000254250.3 |
THAP1
|
THAP domain containing, apoptosis associated protein 1 |
| chr5_-_133706695 | 5.60 |
ENST00000521755.1
ENST00000523054.1 ENST00000435240.2 ENST00000609654.1 ENST00000536186.1 ENST00000609383.1 |
CDKL3
|
cyclin-dependent kinase-like 3 |
| chr15_+_34310428 | 5.56 |
ENST00000557872.1
|
CHRM5
|
cholinergic receptor, muscarinic 5 |
| chr13_-_41240717 | 5.51 |
ENST00000379561.5
|
FOXO1
|
forkhead box O1 |
| chr4_+_124320665 | 5.49 |
ENST00000394339.2
|
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
| chr17_+_72427477 | 5.44 |
ENST00000342648.5
ENST00000481232.1 |
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr6_+_134758827 | 5.39 |
ENST00000431422.1
|
LINC01010
|
long intergenic non-protein coding RNA 1010 |
| chr11_+_112832090 | 5.38 |
ENST00000533760.1
|
NCAM1
|
neural cell adhesion molecule 1 |
| chr1_-_150669604 | 5.31 |
ENST00000427665.1
ENST00000540514.1 |
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr14_+_102276192 | 5.31 |
ENST00000557714.1
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr9_-_15472730 | 5.29 |
ENST00000481862.1
|
PSIP1
|
PC4 and SFRS1 interacting protein 1 |
| chr6_+_32121908 | 5.28 |
ENST00000375143.2
ENST00000424499.1 |
PPT2
|
palmitoyl-protein thioesterase 2 |
| chr22_+_45809560 | 5.24 |
ENST00000342894.3
ENST00000538017.1 |
RIBC2
|
RIB43A domain with coiled-coils 2 |
| chr10_-_52645416 | 5.17 |
ENST00000374001.2
ENST00000373997.3 ENST00000373995.3 ENST00000282641.2 ENST00000395495.1 ENST00000414883.1 |
A1CF
|
APOBEC1 complementation factor |
| chr2_-_101767715 | 5.14 |
ENST00000376840.4
ENST00000409318.1 |
TBC1D8
|
TBC1 domain family, member 8 (with GRAM domain) |
| chr6_+_32121789 | 5.00 |
ENST00000437001.2
ENST00000375137.2 |
PPT2
|
palmitoyl-protein thioesterase 2 |
| chr2_-_166060552 | 4.89 |
ENST00000283254.7
ENST00000453007.1 |
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit |
| chr19_+_15160130 | 4.82 |
ENST00000427043.3
|
CASP14
|
caspase 14, apoptosis-related cysteine peptidase |
| chr2_-_183731882 | 4.79 |
ENST00000295113.4
|
FRZB
|
frizzled-related protein |
| chr12_+_96588279 | 4.77 |
ENST00000552142.1
|
ELK3
|
ELK3, ETS-domain protein (SRF accessory protein 2) |
| chr13_+_98086445 | 4.72 |
ENST00000245304.4
|
RAP2A
|
RAP2A, member of RAS oncogene family |
| chr1_-_13673511 | 4.62 |
ENST00000344998.3
ENST00000334600.6 |
PRAMEF14
|
PRAME family member 14 |
| chr7_+_106505696 | 4.60 |
ENST00000440650.2
ENST00000496166.1 ENST00000473541.1 |
PIK3CG
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
| chr8_-_87755878 | 4.60 |
ENST00000320005.5
|
CNGB3
|
cyclic nucleotide gated channel beta 3 |
| chr3_+_138067666 | 4.50 |
ENST00000475711.1
ENST00000464896.1 |
MRAS
|
muscle RAS oncogene homolog |
| chr17_+_3379284 | 4.46 |
ENST00000263080.2
|
ASPA
|
aspartoacylase |
| chr17_-_40897043 | 4.32 |
ENST00000428826.2
ENST00000592492.1 ENST00000585893.1 ENST00000593214.1 ENST00000590078.1 ENST00000586382.1 ENST00000415827.2 ENST00000592743.1 ENST00000586089.1 ENST00000435174.1 |
EZH1
|
enhancer of zeste homolog 1 (Drosophila) |
| chr6_-_117747015 | 4.29 |
ENST00000368508.3
ENST00000368507.3 |
ROS1
|
c-ros oncogene 1 , receptor tyrosine kinase |
| chr4_-_100242549 | 4.29 |
ENST00000305046.8
ENST00000394887.3 |
ADH1B
|
alcohol dehydrogenase 1B (class I), beta polypeptide |
| chr5_-_10308125 | 4.19 |
ENST00000296658.3
|
CMBL
|
carboxymethylenebutenolidase homolog (Pseudomonas) |
| chr2_-_100721923 | 4.18 |
ENST00000356421.2
|
AFF3
|
AF4/FMR2 family, member 3 |
| chr7_+_106505912 | 4.17 |
ENST00000359195.3
|
PIK3CG
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
| chr12_-_102591604 | 4.12 |
ENST00000329406.4
|
PMCH
|
pro-melanin-concentrating hormone |
| chr1_-_203151933 | 4.11 |
ENST00000404436.2
|
CHI3L1
|
chitinase 3-like 1 (cartilage glycoprotein-39) |
| chr4_-_90756769 | 4.11 |
ENST00000345009.4
ENST00000505199.1 ENST00000502987.1 |
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr8_+_19171128 | 4.07 |
ENST00000265807.3
|
SH2D4A
|
SH2 domain containing 4A |
| chr7_-_95225768 | 4.07 |
ENST00000005178.5
|
PDK4
|
pyruvate dehydrogenase kinase, isozyme 4 |
| chr16_+_1730338 | 3.97 |
ENST00000566691.1
ENST00000382710.4 |
HN1L
|
hematological and neurological expressed 1-like |
| chr16_-_4323015 | 3.94 |
ENST00000204517.6
|
TFAP4
|
transcription factor AP-4 (activating enhancer binding protein 4) |
| chr3_-_101232019 | 3.93 |
ENST00000394095.2
ENST00000394091.1 ENST00000394094.2 ENST00000358203.3 ENST00000348610.3 ENST00000314261.7 |
SENP7
|
SUMO1/sentrin specific peptidase 7 |
| chr4_-_153332886 | 3.91 |
ENST00000603841.1
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr7_+_47834908 | 3.87 |
ENST00000418326.2
|
C7orf69
|
chromosome 7 open reading frame 69 |
| chr1_+_154193325 | 3.86 |
ENST00000428931.1
ENST00000441890.1 ENST00000271877.7 ENST00000412596.1 ENST00000368504.1 ENST00000437652.1 |
UBAP2L
|
ubiquitin associated protein 2-like |
| chr5_-_141338627 | 3.83 |
ENST00000231484.3
|
PCDH12
|
protocadherin 12 |
| chr2_-_166060571 | 3.82 |
ENST00000360093.3
|
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit |
| chr20_-_44993012 | 3.77 |
ENST00000372229.1
ENST00000372230.5 ENST00000543605.1 ENST00000243896.2 ENST00000317734.8 |
SLC35C2
|
solute carrier family 35 (GDP-fucose transporter), member C2 |
| chr2_+_65283506 | 3.74 |
ENST00000377990.2
|
CEP68
|
centrosomal protein 68kDa |
| chr17_-_67138015 | 3.74 |
ENST00000284425.2
ENST00000590645.1 |
ABCA6
|
ATP-binding cassette, sub-family A (ABC1), member 6 |
| chr3_-_185826855 | 3.71 |
ENST00000306376.5
|
ETV5
|
ets variant 5 |
| chr13_+_28712614 | 3.69 |
ENST00000380958.3
|
PAN3
|
PAN3 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr2_-_113012592 | 3.60 |
ENST00000272570.5
ENST00000409573.2 |
ZC3H8
|
zinc finger CCCH-type containing 8 |
| chr16_+_77233294 | 3.48 |
ENST00000378644.4
|
SYCE1L
|
synaptonemal complex central element protein 1-like |
| chr18_-_53069419 | 3.45 |
ENST00000570177.2
|
TCF4
|
transcription factor 4 |
| chr18_+_60382672 | 3.39 |
ENST00000400316.4
ENST00000262719.5 |
PHLPP1
|
PH domain and leucine rich repeat protein phosphatase 1 |
| chr1_+_151739131 | 3.38 |
ENST00000400999.1
|
OAZ3
|
ornithine decarboxylase antizyme 3 |
| chr12_-_71031185 | 3.37 |
ENST00000548122.1
ENST00000551525.1 ENST00000550358.1 |
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chr9_-_95186739 | 3.37 |
ENST00000375550.4
|
OMD
|
osteomodulin |
| chr15_-_31283618 | 3.36 |
ENST00000563714.1
|
MTMR10
|
myotubularin related protein 10 |
| chr1_-_153514241 | 3.35 |
ENST00000368718.1
ENST00000359215.1 |
S100A5
|
S100 calcium binding protein A5 |
| chr17_+_72426891 | 3.32 |
ENST00000392627.1
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr9_+_5510558 | 3.30 |
ENST00000397747.3
|
PDCD1LG2
|
programmed cell death 1 ligand 2 |
| chr14_-_99737822 | 3.23 |
ENST00000345514.2
ENST00000443726.2 |
BCL11B
|
B-cell CLL/lymphoma 11B (zinc finger protein) |
| chr15_-_58571445 | 3.22 |
ENST00000558231.1
|
ALDH1A2
|
aldehyde dehydrogenase 1 family, member A2 |
| chr7_+_134551583 | 3.20 |
ENST00000435928.1
|
CALD1
|
caldesmon 1 |
| chr19_-_40791302 | 3.13 |
ENST00000392038.2
ENST00000578123.1 |
AKT2
|
v-akt murine thymoma viral oncogene homolog 2 |
| chr13_-_99630233 | 3.12 |
ENST00000376460.1
ENST00000442173.1 |
DOCK9
|
dedicator of cytokinesis 9 |
| chr7_+_99425633 | 3.09 |
ENST00000354829.2
ENST00000421837.2 ENST00000417625.1 ENST00000342499.4 ENST00000444905.1 ENST00000415413.1 ENST00000312017.5 ENST00000222382.5 |
CYP3A43
|
cytochrome P450, family 3, subfamily A, polypeptide 43 |
| chr1_+_160765860 | 3.09 |
ENST00000368037.5
|
LY9
|
lymphocyte antigen 9 |
| chr2_+_65283529 | 3.05 |
ENST00000546106.1
ENST00000537589.1 ENST00000260569.4 |
CEP68
|
centrosomal protein 68kDa |
| chr5_+_176853702 | 3.04 |
ENST00000507633.1
ENST00000393576.3 ENST00000355958.5 ENST00000528793.1 ENST00000512684.1 |
GRK6
|
G protein-coupled receptor kinase 6 |
| chr11_-_46638720 | 2.99 |
ENST00000326737.3
|
HARBI1
|
harbinger transposase derived 1 |
| chr15_-_55657428 | 2.97 |
ENST00000568543.1
|
CCPG1
|
cell cycle progression 1 |
| chr5_-_59481406 | 2.97 |
ENST00000546160.1
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr2_+_135809835 | 2.93 |
ENST00000264158.8
ENST00000539493.1 ENST00000442034.1 ENST00000425393.1 |
RAB3GAP1
|
RAB3 GTPase activating protein subunit 1 (catalytic) |
| chr1_+_22778337 | 2.93 |
ENST00000404138.1
ENST00000400239.2 ENST00000375647.4 ENST00000374651.4 |
ZBTB40
|
zinc finger and BTB domain containing 40 |
| chr2_+_33661382 | 2.91 |
ENST00000402538.3
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr7_-_14028488 | 2.89 |
ENST00000405358.4
|
ETV1
|
ets variant 1 |
| chr1_+_160765884 | 2.88 |
ENST00000392203.4
|
LY9
|
lymphocyte antigen 9 |
| chr12_-_71031220 | 2.86 |
ENST00000334414.6
|
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chr17_+_67410832 | 2.83 |
ENST00000590474.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr1_+_160765947 | 2.82 |
ENST00000263285.6
ENST00000368039.2 |
LY9
|
lymphocyte antigen 9 |
| chr8_+_120079478 | 2.81 |
ENST00000332843.2
|
COLEC10
|
collectin sub-family member 10 (C-type lectin) |
| chr1_-_53608249 | 2.80 |
ENST00000371494.4
|
SLC1A7
|
solute carrier family 1 (glutamate transporter), member 7 |
| chr17_+_2240916 | 2.79 |
ENST00000574563.1
|
SGSM2
|
small G protein signaling modulator 2 |
| chr16_+_28858004 | 2.74 |
ENST00000322610.8
|
SH2B1
|
SH2B adaptor protein 1 |
| chr11_-_123612319 | 2.73 |
ENST00000526252.1
ENST00000530393.1 ENST00000533463.1 ENST00000336139.4 ENST00000529691.1 ENST00000528306.1 |
ZNF202
|
zinc finger protein 202 |
| chr10_+_99400443 | 2.72 |
ENST00000370631.3
|
PI4K2A
|
phosphatidylinositol 4-kinase type 2 alpha |
| chr1_-_53608289 | 2.70 |
ENST00000371491.4
|
SLC1A7
|
solute carrier family 1 (glutamate transporter), member 7 |
| chr13_-_36920420 | 2.70 |
ENST00000438666.2
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome) |
| chr1_-_227505826 | 2.70 |
ENST00000334218.5
ENST00000366766.2 ENST00000366764.2 |
CDC42BPA
|
CDC42 binding protein kinase alpha (DMPK-like) |
| chrX_-_131623982 | 2.68 |
ENST00000370844.1
|
MBNL3
|
muscleblind-like splicing regulator 3 |
| chr1_+_10490127 | 2.63 |
ENST00000602787.1
ENST00000602296.1 ENST00000400900.2 |
APITD1
APITD1-CORT
|
apoptosis-inducing, TAF9-like domain 1 APITD1-CORT readthrough |
| chr3_-_18466026 | 2.61 |
ENST00000417717.2
|
SATB1
|
SATB homeobox 1 |
| chr10_+_123923105 | 2.59 |
ENST00000368999.1
|
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr14_-_92572894 | 2.57 |
ENST00000532032.1
ENST00000506466.1 ENST00000555381.1 ENST00000557311.1 ENST00000554592.1 ENST00000554672.1 ENST00000553491.1 ENST00000556220.1 ENST00000502250.1 ENST00000503767.1 ENST00000393287.5 ENST00000340660.6 ENST00000545170.1 ENST00000429774.2 |
ATXN3
|
ataxin 3 |
| chr2_+_157292933 | 2.52 |
ENST00000540309.1
|
GPD2
|
glycerol-3-phosphate dehydrogenase 2 (mitochondrial) |
| chr4_-_70626430 | 2.50 |
ENST00000310613.3
|
SULT1B1
|
sulfotransferase family, cytosolic, 1B, member 1 |
| chr8_-_42623924 | 2.50 |
ENST00000276410.2
|
CHRNA6
|
cholinergic receptor, nicotinic, alpha 6 (neuronal) |
| chr3_+_159570722 | 2.50 |
ENST00000482804.1
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr7_-_38389573 | 2.50 |
ENST00000390344.2
|
TRGV5
|
T cell receptor gamma variable 5 |
| chr1_+_13521973 | 2.44 |
ENST00000327795.5
|
PRAMEF21
|
PRAME family member 21 |
| chr1_+_160765919 | 2.43 |
ENST00000341032.4
ENST00000368041.2 ENST00000368040.1 |
LY9
|
lymphocyte antigen 9 |
| chr1_+_13742808 | 2.42 |
ENST00000602960.1
|
PRAMEF20
|
PRAME family member 20 |
| chr12_-_10251539 | 2.41 |
ENST00000420265.2
|
CLEC1A
|
C-type lectin domain family 1, member A |
| chr16_-_86542455 | 2.40 |
ENST00000595886.1
ENST00000597578.1 ENST00000593604.1 |
FENDRR
|
FOXF1 adjacent non-coding developmental regulatory RNA |
| chr1_+_117297007 | 2.39 |
ENST00000369478.3
ENST00000369477.1 |
CD2
|
CD2 molecule |
| chr3_-_150920979 | 2.38 |
ENST00000309180.5
ENST00000480322.1 |
GPR171
|
G protein-coupled receptor 171 |
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXO4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.9 | 43.8 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 4.0 | 19.8 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 3.6 | 14.5 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 3.3 | 36.8 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 2.9 | 11.6 | GO:0072660 | positive regulation of cell communication by electrical coupling(GO:0010650) maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 2.7 | 8.0 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 2.5 | 15.3 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 2.1 | 16.6 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 2.0 | 9.8 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 2.0 | 9.8 | GO:1903285 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) regulation of peroxidase activity(GO:2000468) positive regulation of peroxidase activity(GO:2000470) |
| 1.9 | 11.6 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 1.8 | 7.4 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 1.8 | 5.5 | GO:0060940 | epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) |
| 1.8 | 8.8 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 1.7 | 40.7 | GO:0034389 | lipid particle organization(GO:0034389) |
| 1.6 | 6.4 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 1.6 | 11.1 | GO:0042426 | choline catabolic process(GO:0042426) |
| 1.6 | 15.7 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 1.5 | 6.1 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 1.5 | 4.5 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 1.3 | 13.0 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 1.3 | 10.3 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 1.3 | 9.0 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 1.3 | 7.5 | GO:1904338 | regulation of dopaminergic neuron differentiation(GO:1904338) |
| 1.2 | 13.7 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 1.1 | 5.6 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 1.1 | 5.5 | GO:1902617 | regulation of gluconeogenesis by regulation of transcription from RNA polymerase II promoter(GO:0035947) response to fluoride(GO:1902617) |
| 0.9 | 2.8 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.9 | 2.7 | GO:0002276 | basophil activation involved in immune response(GO:0002276) |
| 0.9 | 3.6 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.9 | 4.3 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.8 | 8.1 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.8 | 10.4 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.8 | 4.8 | GO:0060029 | convergent extension involved in organogenesis(GO:0060029) |
| 0.8 | 6.4 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.8 | 2.4 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.8 | 3.9 | GO:2000639 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.8 | 6.2 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.8 | 7.7 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.8 | 9.2 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.7 | 3.7 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.7 | 4.7 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.7 | 5.3 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.7 | 2.6 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 0.6 | 3.9 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.6 | 2.6 | GO:1904327 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.6 | 3.0 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.6 | 37.3 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.6 | 11.2 | GO:0072540 | T-helper 17 cell lineage commitment(GO:0072540) |
| 0.6 | 2.3 | GO:1990535 | transformation of host cell by virus(GO:0019087) neuron projection maintenance(GO:1990535) |
| 0.5 | 12.0 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.5 | 3.7 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.5 | 16.2 | GO:0098780 | response to mitochondrial depolarisation(GO:0098780) |
| 0.5 | 10.9 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.5 | 4.6 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.5 | 15.7 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.5 | 1.5 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.5 | 2.4 | GO:0030885 | regulation of myeloid dendritic cell activation(GO:0030885) |
| 0.5 | 1.4 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.4 | 4.3 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.4 | 5.8 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.4 | 1.2 | GO:0044179 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.4 | 6.8 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.4 | 3.8 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) protein O-linked fucosylation(GO:0036066) |
| 0.4 | 3.4 | GO:0002249 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) |
| 0.4 | 4.1 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.4 | 4.7 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.4 | 21.5 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.4 | 1.1 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.4 | 2.1 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.4 | 3.2 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.4 | 26.4 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.4 | 6.0 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.3 | 0.7 | GO:0099543 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) |
| 0.3 | 1.7 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.3 | 2.0 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.3 | 1.0 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.3 | 1.0 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.3 | 1.9 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.3 | 2.2 | GO:0009597 | detection of virus(GO:0009597) |
| 0.3 | 3.5 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.3 | 9.0 | GO:0042994 | cytoplasmic sequestering of transcription factor(GO:0042994) |
| 0.3 | 4.3 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.2 | 16.4 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) |
| 0.2 | 0.5 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.2 | 8.2 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.2 | 9.4 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.2 | 1.9 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.2 | 3.9 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.2 | 12.9 | GO:0046850 | regulation of bone remodeling(GO:0046850) |
| 0.2 | 2.1 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.2 | 1.1 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.2 | 1.9 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.2 | 5.6 | GO:0097484 | dendrite extension(GO:0097484) |
| 0.2 | 9.0 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.2 | 0.6 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
| 0.2 | 3.4 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.2 | 8.7 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.2 | 2.9 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.2 | 8.1 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 0.2 | 1.8 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.2 | 2.6 | GO:0060004 | reflex(GO:0060004) |
| 0.2 | 1.7 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.2 | 4.3 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.2 | 8.6 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.2 | 1.7 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.2 | 12.4 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.2 | 4.0 | GO:0032607 | interferon-alpha production(GO:0032607) |
| 0.2 | 3.5 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.2 | 8.5 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 18.0 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.1 | 0.7 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 2.8 | GO:0006068 | ethanol metabolic process(GO:0006067) ethanol catabolic process(GO:0006068) |
| 0.1 | 11.3 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.1 | 2.4 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.1 | 0.2 | GO:0061739 | protein lipidation involved in autophagosome assembly(GO:0061739) |
| 0.1 | 1.9 | GO:0055022 | negative regulation of cardiac muscle tissue growth(GO:0055022) negative regulation of cardiac muscle cell proliferation(GO:0060044) negative regulation of heart growth(GO:0061117) |
| 0.1 | 0.4 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.1 | 0.8 | GO:0030038 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.1 | 2.4 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.1 | 2.1 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.1 | 3.4 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.1 | 3.2 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.1 | 26.6 | GO:0006469 | negative regulation of protein kinase activity(GO:0006469) |
| 0.1 | 3.8 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.1 | 2.5 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
| 0.1 | 0.2 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.1 | 1.4 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.1 | 3.0 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.1 | 2.3 | GO:0035456 | response to interferon-beta(GO:0035456) |
| 0.1 | 1.9 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 4.1 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 4.1 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.1 | 5.9 | GO:0021987 | cerebral cortex development(GO:0021987) |
| 0.1 | 1.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 4.3 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.1 | 5.4 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.1 | 1.9 | GO:0007632 | visual behavior(GO:0007632) |
| 0.0 | 1.3 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 12.2 | GO:0065004 | protein-DNA complex assembly(GO:0065004) |
| 0.0 | 1.0 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.0 | 4.3 | GO:0001935 | endothelial cell proliferation(GO:0001935) |
| 0.0 | 2.7 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.8 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.0 | 0.5 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 1.3 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 1.3 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.0 | 3.9 | GO:0007188 | adenylate cyclase-modulating G-protein coupled receptor signaling pathway(GO:0007188) |
| 0.0 | 0.8 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.0 | 1.8 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.7 | GO:0036152 | phosphatidylethanolamine acyl-chain remodeling(GO:0036152) |
| 0.0 | 3.0 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 1.5 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 0.4 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.0 | 0.7 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 4.6 | GO:0007601 | visual perception(GO:0007601) sensory perception of light stimulus(GO:0050953) |
| 0.0 | 1.6 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 1.4 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 1.3 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.5 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.3 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 11.6 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 3.8 | 15.3 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 2.9 | 8.8 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 2.7 | 16.3 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 2.3 | 43.8 | GO:0097386 | glial cell projection(GO:0097386) |
| 2.2 | 6.5 | GO:0036398 | TCR signalosome(GO:0036398) |
| 2.0 | 8.1 | GO:0005712 | chiasma(GO:0005712) |
| 1.7 | 10.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 1.6 | 9.8 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 1.5 | 7.4 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 1.2 | 3.7 | GO:0031251 | PAN complex(GO:0031251) |
| 1.1 | 11.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 1.0 | 13.0 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.8 | 58.1 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.7 | 11.6 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.7 | 2.7 | GO:0035838 | growing cell tip(GO:0035838) |
| 0.6 | 8.7 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.6 | 3.9 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.5 | 1.5 | GO:0034657 | GID complex(GO:0034657) |
| 0.5 | 2.3 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.4 | 13.7 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.4 | 9.8 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.3 | 7.5 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.3 | 28.0 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.3 | 12.9 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.3 | 49.0 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.3 | 8.9 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.3 | 2.2 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.3 | 27.4 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.2 | 1.9 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.2 | 0.7 | GO:0031213 | RSF complex(GO:0031213) |
| 0.2 | 2.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.2 | 1.1 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.2 | 1.3 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.2 | 3.9 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.2 | 3.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.2 | 9.0 | GO:0005770 | late endosome(GO:0005770) |
| 0.2 | 2.6 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.2 | 4.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.2 | 1.4 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.2 | 0.8 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.2 | 6.0 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 3.5 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 6.7 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 4.8 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.1 | 2.0 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.1 | 3.5 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.1 | 1.9 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 4.8 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 4.6 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 19.1 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 2.8 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 1.7 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.1 | 15.6 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 1.3 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 12.7 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 2.1 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.1 | 2.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 30.8 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 2.8 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 1.0 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 6.2 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.1 | 17.6 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.1 | 12.2 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.1 | 0.7 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 10.7 | GO:0031968 | organelle outer membrane(GO:0031968) |
| 0.1 | 5.6 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 3.4 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 4.4 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 21.1 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 3.1 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 5.6 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 1.1 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 1.2 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 1.1 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 9.2 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 3.3 | GO:0098794 | postsynapse(GO:0098794) |
| 0.0 | 1.9 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 1.3 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 2.9 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.2 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.2 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.7 | GO:0042383 | sarcolemma(GO:0042383) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.9 | 43.8 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 4.9 | 19.8 | GO:0097001 | ceramide binding(GO:0097001) |
| 3.8 | 30.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 2.9 | 14.4 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 2.5 | 9.8 | GO:0052590 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 2.3 | 9.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 2.1 | 6.4 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 2.1 | 6.2 | GO:0005549 | odorant binding(GO:0005549) |
| 2.0 | 9.8 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 2.0 | 13.7 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 1.7 | 5.0 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 1.5 | 6.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 1.5 | 5.8 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 1.3 | 10.3 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 1.3 | 8.8 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 1.1 | 5.6 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 1.1 | 33.6 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 1.0 | 4.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 1.0 | 19.6 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.9 | 12.2 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.8 | 8.1 | GO:0019237 | satellite DNA binding(GO:0003696) centromeric DNA binding(GO:0019237) |
| 0.8 | 7.7 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.8 | 21.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.7 | 4.3 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.7 | 2.7 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.7 | 5.3 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.7 | 4.6 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.6 | 5.8 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.6 | 4.5 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.6 | 12.4 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.5 | 8.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.5 | 13.5 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.5 | 5.5 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.5 | 3.9 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.5 | 1.9 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.4 | 10.9 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.4 | 3.6 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.4 | 12.8 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.4 | 4.1 | GO:0004568 | chitinase activity(GO:0004568) chitin binding(GO:0008061) |
| 0.4 | 8.7 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.4 | 2.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.4 | 7.4 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.3 | 7.5 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.3 | 2.3 | GO:0043559 | insulin binding(GO:0043559) |
| 0.3 | 8.0 | GO:0071949 | FAD binding(GO:0071949) |
| 0.3 | 4.6 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.3 | 3.2 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.3 | 6.8 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.3 | 5.4 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.3 | 2.6 | GO:1990380 | Lys63-specific deubiquitinase activity(GO:0061578) Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.3 | 1.3 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.3 | 1.8 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.3 | 11.6 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.2 | 6.5 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.2 | 1.0 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.2 | 3.6 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.2 | 6.2 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.2 | 15.1 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.2 | 1.7 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.2 | 4.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.2 | 11.2 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.2 | 0.6 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.2 | 1.9 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.2 | 3.4 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.2 | 2.2 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.2 | 1.2 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.2 | 1.2 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.2 | 3.1 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.2 | 0.5 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.2 | 2.9 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.2 | 13.8 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.1 | 2.8 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 3.5 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 21.2 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 5.8 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.1 | 2.4 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 2.0 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.1 | 1.9 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 32.4 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.1 | 3.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 3.5 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.1 | 3.6 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 2.8 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 1.6 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 1.3 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 6.3 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 3.9 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 2.2 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 8.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 7.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.8 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 1.4 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.1 | 0.3 | GO:0005497 | androgen binding(GO:0005497) |
| 0.1 | 5.4 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 17.0 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 3.8 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 0.7 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.6 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 2.4 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 3.4 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 6.9 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 5.4 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 10.8 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 5.9 | GO:0035257 | nuclear hormone receptor binding(GO:0035257) |
| 0.0 | 12.1 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.8 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 4.1 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 2.9 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.8 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.3 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 2.1 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 7.9 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 1.7 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.2 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.6 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 2.4 | GO:0004518 | nuclease activity(GO:0004518) |
| 0.0 | 0.7 | GO:0030165 | PDZ domain binding(GO:0030165) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.7 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.3 | 8.1 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.3 | 1.9 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.3 | 33.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.3 | 7.5 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.3 | 14.6 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.2 | 5.9 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.2 | 11.6 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.2 | 3.1 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.2 | 6.9 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.2 | 16.3 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.2 | 9.8 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.2 | 28.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.2 | 2.8 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 2.9 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 2.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 6.0 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 5.2 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.1 | 5.4 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 5.8 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 3.9 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 2.9 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 3.1 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 3.9 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 0.6 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 3.1 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 3.2 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.1 | 1.7 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 2.3 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 3.7 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 3.8 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 1.5 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 2.7 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 1.7 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 4.9 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.1 | 14.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 0.3 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.6 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 1.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 8.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.4 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 2.1 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.0 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 43.8 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.7 | 6.5 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.7 | 22.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.6 | 5.3 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.6 | 8.5 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.5 | 13.7 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.5 | 4.3 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.5 | 11.5 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.4 | 9.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.4 | 7.6 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.3 | 11.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.3 | 9.1 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.3 | 18.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.3 | 8.8 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.3 | 15.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.3 | 6.5 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.2 | 4.1 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.2 | 5.8 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.2 | 7.9 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.2 | 3.5 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.2 | 4.7 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.2 | 3.7 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.2 | 3.4 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.2 | 8.9 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.2 | 2.7 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.2 | 25.0 | REACTOME MEIOSIS | Genes involved in Meiosis |
| 0.2 | 8.6 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 7.5 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 3.9 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 2.3 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 5.6 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 3.1 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 10.4 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 5.4 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 2.8 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 2.7 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.1 | 1.3 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 9.0 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 1.7 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 3.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 2.5 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 1.7 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 1.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 10.4 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 3.1 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 1.0 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 10.3 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 3.1 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 1.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.7 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 1.3 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.5 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 1.3 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |