Project
averaged GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
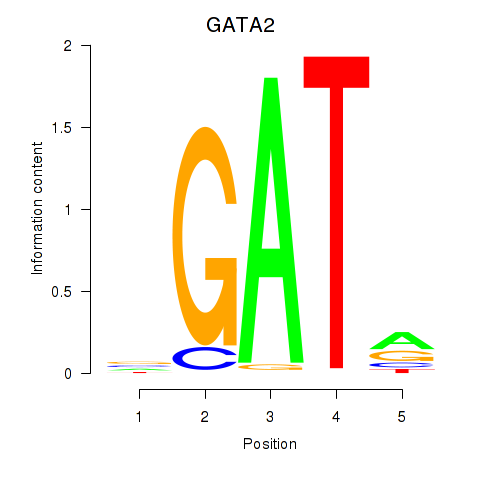
Results for GATA2
Z-value: 1.78
Transcription factors associated with GATA2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GATA2
|
ENSG00000179348.7 | GATA binding protein 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GATA2 | hg19_v2_chr3_-_128206759_128206781 | -0.13 | 6.1e-02 | Click! |
Activity profile of GATA2 motif
Sorted Z-values of GATA2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_125366089 | 35.69 |
ENST00000366139.3
ENST00000278919.3 |
FEZ1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr11_-_111781554 | 34.11 |
ENST00000526167.1
ENST00000528961.1 |
CRYAB
|
crystallin, alpha B |
| chr11_-_111794446 | 32.03 |
ENST00000527950.1
|
CRYAB
|
crystallin, alpha B |
| chr3_+_111718173 | 30.40 |
ENST00000494932.1
|
TAGLN3
|
transgelin 3 |
| chr1_+_160085501 | 28.20 |
ENST00000361216.3
|
ATP1A2
|
ATPase, Na+/K+ transporting, alpha 2 polypeptide |
| chr11_-_111781610 | 27.15 |
ENST00000525823.1
|
CRYAB
|
crystallin, alpha B |
| chr14_-_21945057 | 22.81 |
ENST00000397762.1
|
RAB2B
|
RAB2B, member RAS oncogene family |
| chr11_-_5248294 | 22.56 |
ENST00000335295.4
|
HBB
|
hemoglobin, beta |
| chrX_-_13956737 | 20.64 |
ENST00000454189.2
|
GPM6B
|
glycoprotein M6B |
| chr12_-_91573132 | 20.53 |
ENST00000550563.1
ENST00000546370.1 |
DCN
|
decorin |
| chr11_+_124609823 | 20.23 |
ENST00000412681.2
|
NRGN
|
neurogranin (protein kinase C substrate, RC3) |
| chr2_-_175711133 | 20.10 |
ENST00000409597.1
ENST00000413882.1 |
CHN1
|
chimerin 1 |
| chr10_-_15413035 | 20.08 |
ENST00000378116.4
ENST00000455654.1 |
FAM171A1
|
family with sequence similarity 171, member A1 |
| chrX_-_13835147 | 19.43 |
ENST00000493677.1
ENST00000355135.2 |
GPM6B
|
glycoprotein M6B |
| chr11_-_111782484 | 19.40 |
ENST00000533971.1
|
CRYAB
|
crystallin, alpha B |
| chr16_+_58533951 | 19.10 |
ENST00000566192.1
ENST00000565088.1 ENST00000568640.1 ENST00000563978.1 ENST00000569923.1 ENST00000356752.4 ENST00000563799.1 ENST00000562999.1 ENST00000570248.1 ENST00000562731.1 ENST00000568424.1 |
NDRG4
|
NDRG family member 4 |
| chr16_+_58283814 | 18.88 |
ENST00000443128.2
ENST00000219299.4 |
CCDC113
|
coiled-coil domain containing 113 |
| chr17_-_15165854 | 18.72 |
ENST00000395936.1
ENST00000395938.2 |
PMP22
|
peripheral myelin protein 22 |
| chr2_+_113033164 | 18.59 |
ENST00000409871.1
ENST00000343936.4 |
ZC3H6
|
zinc finger CCCH-type containing 6 |
| chr19_+_41509851 | 18.56 |
ENST00000593831.1
ENST00000330446.5 |
CYP2B6
|
cytochrome P450, family 2, subfamily B, polypeptide 6 |
| chr12_-_91546926 | 18.16 |
ENST00000550758.1
|
DCN
|
decorin |
| chr5_+_140772381 | 17.73 |
ENST00000398604.2
|
PCDHGA8
|
protocadherin gamma subfamily A, 8 |
| chr2_-_99224915 | 17.53 |
ENST00000328709.3
ENST00000409997.1 |
COA5
|
cytochrome c oxidase assembly factor 5 |
| chr11_+_124609742 | 16.95 |
ENST00000284292.6
|
NRGN
|
neurogranin (protein kinase C substrate, RC3) |
| chr8_-_27468842 | 16.88 |
ENST00000523500.1
|
CLU
|
clusterin |
| chr10_+_74451883 | 16.71 |
ENST00000373053.3
ENST00000357157.6 |
MCU
|
mitochondrial calcium uniporter |
| chr3_-_149688655 | 16.59 |
ENST00000461930.1
ENST00000423691.2 ENST00000490975.1 ENST00000461868.1 ENST00000452853.2 |
PFN2
|
profilin 2 |
| chr1_+_151254738 | 16.55 |
ENST00000336715.6
ENST00000324048.5 ENST00000368879.2 |
ZNF687
|
zinc finger protein 687 |
| chr12_-_9268707 | 16.02 |
ENST00000318602.7
|
A2M
|
alpha-2-macroglobulin |
| chr17_-_29624343 | 15.80 |
ENST00000247271.4
|
OMG
|
oligodendrocyte myelin glycoprotein |
| chrX_-_13835461 | 15.56 |
ENST00000316715.4
ENST00000356942.5 |
GPM6B
|
glycoprotein M6B |
| chr7_-_38670957 | 15.45 |
ENST00000325590.5
ENST00000428293.2 |
AMPH
|
amphiphysin |
| chr12_-_91573249 | 15.33 |
ENST00000550099.1
ENST00000546391.1 ENST00000551354.1 |
DCN
|
decorin |
| chr9_-_79307096 | 15.30 |
ENST00000376717.2
ENST00000223609.6 ENST00000443509.2 |
PRUNE2
|
prune homolog 2 (Drosophila) |
| chrX_+_38420623 | 15.26 |
ENST00000378482.2
|
TSPAN7
|
tetraspanin 7 |
| chr5_-_42812143 | 15.18 |
ENST00000514985.1
|
SEPP1
|
selenoprotein P, plasma, 1 |
| chrM_+_4431 | 15.16 |
ENST00000361453.3
|
MT-ND2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr7_-_38671098 | 15.07 |
ENST00000356264.2
|
AMPH
|
amphiphysin |
| chr12_+_121088291 | 14.92 |
ENST00000351200.2
|
CABP1
|
calcium binding protein 1 |
| chr12_+_79439405 | 14.71 |
ENST00000552744.1
|
SYT1
|
synaptotagmin I |
| chr12_-_12491608 | 14.49 |
ENST00000545735.1
|
MANSC1
|
MANSC domain containing 1 |
| chr11_+_117073850 | 14.47 |
ENST00000529622.1
|
TAGLN
|
transgelin |
| chr5_+_36608422 | 14.42 |
ENST00000381918.3
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr12_+_79258547 | 14.23 |
ENST00000457153.2
|
SYT1
|
synaptotagmin I |
| chr5_+_71403061 | 14.03 |
ENST00000512974.1
ENST00000296755.7 |
MAP1B
|
microtubule-associated protein 1B |
| chr8_-_27468945 | 13.90 |
ENST00000405140.3
|
CLU
|
clusterin |
| chr9_-_99417562 | 13.71 |
ENST00000375234.3
ENST00000446045.1 |
AAED1
|
AhpC/TSA antioxidant enzyme domain containing 1 |
| chr16_+_15528332 | 13.67 |
ENST00000566490.1
|
C16orf45
|
chromosome 16 open reading frame 45 |
| chr1_+_171810606 | 13.66 |
ENST00000358155.4
ENST00000367733.2 ENST00000355305.5 ENST00000367731.1 |
DNM3
|
dynamin 3 |
| chr15_+_74422585 | 13.58 |
ENST00000561740.1
ENST00000435464.1 ENST00000453268.2 |
ISLR2
|
immunoglobulin superfamily containing leucine-rich repeat 2 |
| chr15_-_23932437 | 13.51 |
ENST00000331837.4
|
NDN
|
necdin, melanoma antigen (MAGE) family member |
| chr7_+_20686946 | 13.24 |
ENST00000443026.2
ENST00000406935.1 |
ABCB5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr5_+_140220769 | 13.16 |
ENST00000531613.1
ENST00000378123.3 |
PCDHA8
|
protocadherin alpha 8 |
| chr12_+_7023491 | 13.12 |
ENST00000541477.1
ENST00000229277.1 |
ENO2
|
enolase 2 (gamma, neuronal) |
| chr11_-_76155618 | 12.62 |
ENST00000530759.1
|
RP11-111M22.3
|
RP11-111M22.3 |
| chr10_+_64133934 | 12.44 |
ENST00000395254.3
ENST00000395255.3 ENST00000410046.3 |
ZNF365
|
zinc finger protein 365 |
| chr6_-_46293378 | 12.36 |
ENST00000330430.6
|
RCAN2
|
regulator of calcineurin 2 |
| chr1_+_10292308 | 12.34 |
ENST00000377081.1
|
KIF1B
|
kinesin family member 1B |
| chr2_-_100939195 | 11.94 |
ENST00000393437.3
|
LONRF2
|
LON peptidase N-terminal domain and ring finger 2 |
| chr15_+_80733570 | 11.94 |
ENST00000533983.1
ENST00000527771.1 ENST00000525103.1 |
ARNT2
|
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr10_-_99531709 | 11.88 |
ENST00000266066.3
|
SFRP5
|
secreted frizzled-related protein 5 |
| chr17_-_74163159 | 11.76 |
ENST00000591615.1
|
RNF157
|
ring finger protein 157 |
| chr11_-_123525289 | 11.61 |
ENST00000392770.2
ENST00000299333.3 ENST00000530277.1 |
SCN3B
|
sodium channel, voltage-gated, type III, beta subunit |
| chr4_+_113970772 | 11.36 |
ENST00000504454.1
ENST00000394537.3 ENST00000357077.4 ENST00000264366.6 |
ANK2
|
ankyrin 2, neuronal |
| chr4_+_154074217 | 11.08 |
ENST00000437508.2
|
TRIM2
|
tripartite motif containing 2 |
| chr4_-_46911223 | 10.87 |
ENST00000396533.1
|
COX7B2
|
cytochrome c oxidase subunit VIIb2 |
| chr2_-_68384603 | 10.78 |
ENST00000406245.2
ENST00000409164.1 ENST00000295121.6 |
WDR92
|
WD repeat domain 92 |
| chr10_-_90712520 | 10.60 |
ENST00000224784.6
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta |
| chr7_+_24323782 | 10.54 |
ENST00000242152.2
ENST00000407573.1 |
NPY
|
neuropeptide Y |
| chr12_-_91572278 | 10.38 |
ENST00000425043.1
ENST00000420120.2 ENST00000441303.2 ENST00000456569.2 |
DCN
|
decorin |
| chr12_-_71182695 | 10.29 |
ENST00000342084.4
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr16_+_55542910 | 10.25 |
ENST00000262134.5
|
LPCAT2
|
lysophosphatidylcholine acyltransferase 2 |
| chr7_-_5553369 | 10.15 |
ENST00000453700.3
ENST00000382368.3 |
FBXL18
|
F-box and leucine-rich repeat protein 18 |
| chr11_-_76155700 | 10.02 |
ENST00000572035.1
|
RP11-111M22.3
|
RP11-111M22.3 |
| chr18_-_21891460 | 9.98 |
ENST00000357041.4
|
OSBPL1A
|
oxysterol binding protein-like 1A |
| chrX_-_67653614 | 9.80 |
ENST00000355520.5
|
OPHN1
|
oligophrenin 1 |
| chr4_+_128554081 | 9.76 |
ENST00000335251.6
ENST00000296461.5 |
INTU
|
inturned planar cell polarity protein |
| chr6_+_42847348 | 9.70 |
ENST00000493763.1
ENST00000304734.5 |
RPL7L1
|
ribosomal protein L7-like 1 |
| chr5_+_140753444 | 9.68 |
ENST00000517434.1
|
PCDHGA6
|
protocadherin gamma subfamily A, 6 |
| chr2_-_26205340 | 9.56 |
ENST00000264712.3
|
KIF3C
|
kinesin family member 3C |
| chr3_+_101546827 | 9.44 |
ENST00000461724.1
ENST00000483180.1 ENST00000394054.2 |
NFKBIZ
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta |
| chr12_-_124457257 | 9.35 |
ENST00000545891.1
|
CCDC92
|
coiled-coil domain containing 92 |
| chr11_-_117186946 | 9.25 |
ENST00000313005.6
ENST00000528053.1 |
BACE1
|
beta-site APP-cleaving enzyme 1 |
| chr1_+_183774240 | 9.13 |
ENST00000360851.3
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1 |
| chr20_+_44036900 | 9.12 |
ENST00000443296.1
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr1_-_161277210 | 9.11 |
ENST00000491222.2
|
MPZ
|
myelin protein zero |
| chr8_+_85097110 | 9.07 |
ENST00000517638.1
ENST00000522647.1 |
RALYL
|
RALY RNA binding protein-like |
| chr7_+_65338230 | 8.86 |
ENST00000360768.3
|
VKORC1L1
|
vitamin K epoxide reductase complex, subunit 1-like 1 |
| chr15_+_51973550 | 8.64 |
ENST00000220478.3
|
SCG3
|
secretogranin III |
| chr10_-_79397391 | 8.62 |
ENST00000286628.8
ENST00000406533.3 ENST00000354353.5 ENST00000404857.1 |
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr3_-_15140629 | 8.60 |
ENST00000507357.1
ENST00000449050.1 ENST00000253699.3 ENST00000435849.3 ENST00000476527.2 |
ZFYVE20
|
zinc finger, FYVE domain containing 20 |
| chr6_-_33239712 | 8.58 |
ENST00000436044.2
|
VPS52
|
vacuolar protein sorting 52 homolog (S. cerevisiae) |
| chr20_+_5892037 | 8.56 |
ENST00000378961.4
|
CHGB
|
chromogranin B (secretogranin 1) |
| chr6_+_7107830 | 8.56 |
ENST00000379933.3
|
RREB1
|
ras responsive element binding protein 1 |
| chr19_-_4723761 | 8.52 |
ENST00000597849.1
ENST00000598800.1 ENST00000602161.1 ENST00000597726.1 ENST00000601130.1 ENST00000262960.9 |
DPP9
|
dipeptidyl-peptidase 9 |
| chr19_+_9296279 | 8.49 |
ENST00000344248.2
|
OR7D2
|
olfactory receptor, family 7, subfamily D, member 2 |
| chr5_+_150404904 | 8.42 |
ENST00000521632.1
|
GPX3
|
glutathione peroxidase 3 (plasma) |
| chr4_-_17513851 | 8.41 |
ENST00000281243.5
|
QDPR
|
quinoid dihydropteridine reductase |
| chr13_+_53226963 | 8.39 |
ENST00000343788.6
ENST00000535397.1 ENST00000310528.8 |
SUGT1
|
SGT1, suppressor of G2 allele of SKP1 (S. cerevisiae) |
| chr4_+_156680153 | 8.31 |
ENST00000502959.1
ENST00000505764.1 ENST00000507146.1 ENST00000264424.8 ENST00000503520.1 |
GUCY1B3
|
guanylate cyclase 1, soluble, beta 3 |
| chr1_-_85462623 | 8.22 |
ENST00000370608.3
|
MCOLN2
|
mucolipin 2 |
| chr2_-_201936302 | 8.20 |
ENST00000453765.1
ENST00000452799.1 ENST00000446678.1 ENST00000418596.3 |
FAM126B
|
family with sequence similarity 126, member B |
| chr4_-_87281196 | 8.19 |
ENST00000359221.3
|
MAPK10
|
mitogen-activated protein kinase 10 |
| chr7_+_139025875 | 8.15 |
ENST00000297534.6
|
C7orf55
|
chromosome 7 open reading frame 55 |
| chr6_-_80657292 | 7.96 |
ENST00000369816.4
|
ELOVL4
|
ELOVL fatty acid elongase 4 |
| chrX_-_48858667 | 7.93 |
ENST00000376423.4
ENST00000376441.1 |
GRIPAP1
|
GRIP1 associated protein 1 |
| chr16_+_55512742 | 7.92 |
ENST00000568715.1
ENST00000219070.4 |
MMP2
|
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) |
| chr8_+_10530155 | 7.87 |
ENST00000521818.1
|
C8orf74
|
chromosome 8 open reading frame 74 |
| chr11_-_66115032 | 7.87 |
ENST00000311181.4
|
B3GNT1
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 1 |
| chr1_-_163172625 | 7.83 |
ENST00000527988.1
ENST00000531476.1 ENST00000530507.1 |
RGS5
|
regulator of G-protein signaling 5 |
| chr19_+_18794470 | 7.82 |
ENST00000321949.8
ENST00000338797.6 |
CRTC1
|
CREB regulated transcription coactivator 1 |
| chr17_-_10017864 | 7.76 |
ENST00000323816.4
|
GAS7
|
growth arrest-specific 7 |
| chr18_-_4455283 | 7.72 |
ENST00000315677.3
|
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1 |
| chr6_+_7108210 | 7.60 |
ENST00000467782.1
ENST00000334984.6 ENST00000349384.6 |
RREB1
|
ras responsive element binding protein 1 |
| chr4_-_87281224 | 7.59 |
ENST00000395169.3
ENST00000395161.2 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr9_-_21351377 | 7.55 |
ENST00000380210.1
|
IFNA6
|
interferon, alpha 6 |
| chr5_+_140186647 | 7.54 |
ENST00000512229.2
ENST00000356878.4 ENST00000530339.1 |
PCDHA4
|
protocadherin alpha 4 |
| chr1_-_177133818 | 7.54 |
ENST00000424564.2
ENST00000361833.2 |
ASTN1
|
astrotactin 1 |
| chr18_-_5396271 | 7.53 |
ENST00000579951.1
|
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3 |
| chr17_+_12692774 | 7.50 |
ENST00000379672.5
ENST00000340825.3 |
ARHGAP44
|
Rho GTPase activating protein 44 |
| chr17_-_4167142 | 7.47 |
ENST00000570535.1
ENST00000574367.1 ENST00000341657.4 ENST00000433651.1 |
ANKFY1
|
ankyrin repeat and FYVE domain containing 1 |
| chr6_+_126102292 | 7.44 |
ENST00000368357.3
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr4_-_90758118 | 7.44 |
ENST00000420646.2
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr16_+_29674277 | 7.43 |
ENST00000395389.2
|
SPN
|
sialophorin |
| chr2_-_152118352 | 7.39 |
ENST00000331426.5
|
RBM43
|
RNA binding motif protein 43 |
| chr14_-_31926701 | 7.38 |
ENST00000310850.4
|
DTD2
|
D-tyrosyl-tRNA deacylase 2 (putative) |
| chr1_-_160001737 | 7.37 |
ENST00000368090.2
|
PIGM
|
phosphatidylinositol glycan anchor biosynthesis, class M |
| chr5_+_140213815 | 7.35 |
ENST00000525929.1
ENST00000378125.3 |
PCDHA7
|
protocadherin alpha 7 |
| chr4_+_156680518 | 7.31 |
ENST00000513437.1
|
GUCY1B3
|
guanylate cyclase 1, soluble, beta 3 |
| chr8_+_21911054 | 7.27 |
ENST00000519850.1
ENST00000381470.3 |
DMTN
|
dematin actin binding protein |
| chr1_+_27719148 | 7.25 |
ENST00000374024.3
|
GPR3
|
G protein-coupled receptor 3 |
| chrX_-_49056635 | 7.25 |
ENST00000472598.1
ENST00000538567.1 ENST00000479808.1 ENST00000263233.4 |
SYP
|
synaptophysin |
| chr20_+_44036620 | 7.23 |
ENST00000372710.3
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr8_-_57358432 | 7.17 |
ENST00000517415.1
ENST00000314922.3 |
PENK
|
proenkephalin |
| chr1_+_202995611 | 7.14 |
ENST00000367240.2
|
PPFIA4
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 4 |
| chr17_-_34257731 | 7.13 |
ENST00000431884.2
ENST00000425909.3 ENST00000394528.3 ENST00000430160.2 |
RDM1
|
RAD52 motif 1 |
| chr16_-_30023615 | 7.07 |
ENST00000564979.1
ENST00000563378.1 |
DOC2A
|
double C2-like domains, alpha |
| chr8_-_18666360 | 7.06 |
ENST00000286485.8
|
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chr5_+_140767452 | 7.05 |
ENST00000519479.1
|
PCDHGB4
|
protocadherin gamma subfamily B, 4 |
| chr18_-_53303123 | 7.05 |
ENST00000569357.1
ENST00000565124.1 ENST00000398339.1 |
TCF4
|
transcription factor 4 |
| chr13_-_96296944 | 7.01 |
ENST00000361396.2
ENST00000376829.2 |
DZIP1
|
DAZ interacting zinc finger protein 1 |
| chr1_+_10271674 | 6.92 |
ENST00000377086.1
|
KIF1B
|
kinesin family member 1B |
| chr4_+_84377115 | 6.85 |
ENST00000295491.4
ENST00000507019.1 |
MRPS18C
|
mitochondrial ribosomal protein S18C |
| chr1_-_177134024 | 6.84 |
ENST00000367654.3
|
ASTN1
|
astrotactin 1 |
| chr19_-_55866104 | 6.82 |
ENST00000326529.4
|
COX6B2
|
cytochrome c oxidase subunit VIb polypeptide 2 (testis) |
| chr14_-_24584138 | 6.79 |
ENST00000558280.1
ENST00000561028.1 |
NRL
|
neural retina leucine zipper |
| chr5_+_175792459 | 6.79 |
ENST00000310389.5
|
ARL10
|
ADP-ribosylation factor-like 10 |
| chr4_-_57547454 | 6.75 |
ENST00000556376.2
|
HOPX
|
HOP homeobox |
| chr10_-_28571015 | 6.73 |
ENST00000375719.3
ENST00000375732.1 |
MPP7
|
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr5_+_172483347 | 6.72 |
ENST00000522692.1
ENST00000296953.2 ENST00000540014.1 ENST00000520420.1 |
CREBRF
|
CREB3 regulatory factor |
| chr14_-_50065882 | 6.71 |
ENST00000539688.1
|
AL139099.1
|
Full-length cDNA clone CS0DK012YO09 of HeLa cells of Homo sapiens (human); Uncharacterized protein |
| chr4_+_20255123 | 6.68 |
ENST00000504154.1
ENST00000273739.5 |
SLIT2
|
slit homolog 2 (Drosophila) |
| chr3_+_197476621 | 6.65 |
ENST00000241502.4
|
FYTTD1
|
forty-two-three domain containing 1 |
| chr8_+_119294456 | 6.64 |
ENST00000366457.2
|
AC023590.1
|
Uncharacterized protein |
| chr20_-_43729750 | 6.62 |
ENST00000537075.1
ENST00000306117.1 |
KCNS1
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 1 |
| chr19_+_18723660 | 6.60 |
ENST00000262817.3
|
TMEM59L
|
transmembrane protein 59-like |
| chr9_-_14180778 | 6.59 |
ENST00000380924.1
ENST00000543693.1 |
NFIB
|
nuclear factor I/B |
| chr7_-_8302207 | 6.56 |
ENST00000407906.1
|
ICA1
|
islet cell autoantigen 1, 69kDa |
| chr6_-_33168391 | 6.52 |
ENST00000374685.4
ENST00000413614.2 ENST00000374680.3 |
RXRB
|
retinoid X receptor, beta |
| chr7_+_94023873 | 6.48 |
ENST00000297268.6
|
COL1A2
|
collagen, type I, alpha 2 |
| chr5_-_138775177 | 6.47 |
ENST00000302060.5
|
DNAJC18
|
DnaJ (Hsp40) homolog, subfamily C, member 18 |
| chr8_-_18744528 | 6.42 |
ENST00000523619.1
|
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chr4_-_57547870 | 6.42 |
ENST00000381260.3
ENST00000420433.1 ENST00000554144.1 ENST00000557328.1 |
HOPX
|
HOP homeobox |
| chr7_+_43152191 | 6.42 |
ENST00000395891.2
|
HECW1
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1 |
| chr13_+_32838801 | 6.41 |
ENST00000542859.1
|
FRY
|
furry homolog (Drosophila) |
| chr11_+_46958248 | 6.40 |
ENST00000536126.1
ENST00000278460.7 ENST00000378618.2 ENST00000395460.2 ENST00000378615.3 ENST00000543718.1 |
C11orf49
|
chromosome 11 open reading frame 49 |
| chr11_+_76571911 | 6.40 |
ENST00000534206.1
ENST00000532485.1 ENST00000526597.1 ENST00000533873.1 ENST00000538157.1 |
ACER3
|
alkaline ceramidase 3 |
| chr5_-_19988339 | 6.38 |
ENST00000382275.1
|
CDH18
|
cadherin 18, type 2 |
| chr5_+_157170703 | 6.33 |
ENST00000286307.5
|
LSM11
|
LSM11, U7 small nuclear RNA associated |
| chr15_-_51397473 | 6.32 |
ENST00000327536.5
|
TNFAIP8L3
|
tumor necrosis factor, alpha-induced protein 8-like 3 |
| chr5_-_20575959 | 6.29 |
ENST00000507958.1
|
CDH18
|
cadherin 18, type 2 |
| chr18_+_39766626 | 6.27 |
ENST00000593234.1
ENST00000585627.1 ENST00000591199.1 ENST00000586990.1 ENST00000593051.1 ENST00000593316.1 ENST00000591381.1 ENST00000585639.1 ENST00000589068.1 |
LINC00907
|
long intergenic non-protein coding RNA 907 |
| chr6_+_69345166 | 6.26 |
ENST00000370598.1
|
BAI3
|
brain-specific angiogenesis inhibitor 3 |
| chr6_+_17393888 | 6.25 |
ENST00000493172.1
ENST00000465994.1 |
CAP2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr5_+_140588269 | 6.24 |
ENST00000541609.1
ENST00000239450.2 |
PCDHB12
|
protocadherin beta 12 |
| chr12_+_6930813 | 6.24 |
ENST00000428545.2
|
GPR162
|
G protein-coupled receptor 162 |
| chr11_-_82708519 | 6.23 |
ENST00000534301.1
|
RAB30
|
RAB30, member RAS oncogene family |
| chr19_-_11591848 | 6.23 |
ENST00000359227.3
|
ELAVL3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr8_-_87755878 | 6.21 |
ENST00000320005.5
|
CNGB3
|
cyclic nucleotide gated channel beta 3 |
| chr11_+_92085262 | 6.15 |
ENST00000298047.6
ENST00000409404.2 ENST00000541502.1 |
FAT3
|
FAT atypical cadherin 3 |
| chr1_+_180601139 | 6.13 |
ENST00000367590.4
ENST00000367589.3 |
XPR1
|
xenotropic and polytropic retrovirus receptor 1 |
| chr12_-_12509929 | 6.13 |
ENST00000381800.2
|
LOH12CR2
|
loss of heterozygosity, 12, chromosomal region 2 (non-protein coding) |
| chr7_+_87563557 | 6.11 |
ENST00000439864.1
ENST00000412441.1 ENST00000398201.4 ENST00000265727.7 ENST00000315984.7 ENST00000398209.3 |
ADAM22
|
ADAM metallopeptidase domain 22 |
| chr11_+_126225789 | 6.09 |
ENST00000530591.1
ENST00000534083.1 |
ST3GAL4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chr2_+_131769256 | 6.08 |
ENST00000355771.3
|
ARHGEF4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr17_-_33905521 | 6.06 |
ENST00000225873.4
|
PEX12
|
peroxisomal biogenesis factor 12 |
| chr19_-_17799008 | 6.06 |
ENST00000519716.2
|
UNC13A
|
unc-13 homolog A (C. elegans) |
| chr5_+_55033845 | 6.01 |
ENST00000353507.5
ENST00000514278.2 ENST00000505374.1 ENST00000506511.1 |
DDX4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chrX_+_69672136 | 6.00 |
ENST00000374355.3
|
DLG3
|
discs, large homolog 3 (Drosophila) |
| chr16_-_29910853 | 5.98 |
ENST00000308713.5
|
SEZ6L2
|
seizure related 6 homolog (mouse)-like 2 |
| chr1_+_207226574 | 5.93 |
ENST00000367080.3
ENST00000367079.2 |
PFKFB2
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 |
| chr7_+_142829162 | 5.91 |
ENST00000291009.3
|
PIP
|
prolactin-induced protein |
| chr10_-_50970322 | 5.84 |
ENST00000374103.4
|
OGDHL
|
oxoglutarate dehydrogenase-like |
| chr16_+_28834303 | 5.81 |
ENST00000340394.8
ENST00000325215.6 ENST00000395547.2 ENST00000336783.4 ENST00000382686.4 ENST00000564304.1 |
ATXN2L
|
ataxin 2-like |
| chr10_-_123274693 | 5.72 |
ENST00000429361.1
|
FGFR2
|
fibroblast growth factor receptor 2 |
| chr8_-_99306611 | 5.72 |
ENST00000341166.3
|
NIPAL2
|
NIPA-like domain containing 2 |
| chr12_+_107168342 | 5.70 |
ENST00000392837.4
|
RIC8B
|
RIC8 guanine nucleotide exchange factor B |
| chr19_+_49999631 | 5.70 |
ENST00000270625.2
ENST00000596873.1 ENST00000594493.1 ENST00000599561.1 |
RPS11
|
ribosomal protein S11 |
| chr7_+_134430212 | 5.68 |
ENST00000436461.2
|
CALD1
|
caldesmon 1 |
| chr3_-_73673991 | 5.68 |
ENST00000308537.4
ENST00000263666.4 |
PDZRN3
|
PDZ domain containing ring finger 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of GATA2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.6 | 28.9 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 6.9 | 20.6 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 6.7 | 67.0 | GO:0051610 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 6.4 | 19.3 | GO:1904647 | response to rotenone(GO:1904647) |
| 5.9 | 35.7 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 5.8 | 103.7 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 5.6 | 28.2 | GO:0051946 | regulation of amino acid uptake involved in synaptic transmission(GO:0051941) regulation of glutamate uptake involved in transmission of nerve impulse(GO:0051946) regulation of L-glutamate import(GO:1900920) |
| 5.6 | 22.6 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 5.4 | 64.4 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 5.1 | 30.8 | GO:0061517 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 4.6 | 13.7 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 4.0 | 16.2 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 3.8 | 11.4 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 3.3 | 13.2 | GO:0048749 | compound eye development(GO:0048749) |
| 3.1 | 12.4 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 3.0 | 11.9 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 2.9 | 8.6 | GO:0048611 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 2.8 | 8.4 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 2.8 | 8.4 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 2.8 | 16.7 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 2.7 | 13.7 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 2.6 | 7.8 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 2.6 | 10.2 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 2.5 | 5.0 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 2.4 | 19.6 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 2.3 | 6.9 | GO:0021823 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) regulation of negative chemotaxis(GO:0050923) |
| 2.2 | 6.7 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 2.1 | 10.6 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 2.1 | 10.6 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 2.0 | 2.0 | GO:0099612 | protein localization to axon(GO:0099612) |
| 2.0 | 15.8 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 1.9 | 5.7 | GO:0060615 | fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) fibroblast growth factor receptor signaling pathway involved in mammary gland specification(GO:0060595) mammary gland bud formation(GO:0060615) branch elongation involved in salivary gland morphogenesis(GO:0060667) mesenchymal cell differentiation involved in lung development(GO:0060915) |
| 1.9 | 3.8 | GO:0003402 | planar cell polarity pathway involved in axis elongation(GO:0003402) |
| 1.9 | 7.5 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 1.9 | 7.4 | GO:0002884 | regulation of type IV hypersensitivity(GO:0001807) negative regulation of hypersensitivity(GO:0002884) |
| 1.8 | 5.5 | GO:1990637 | response to prolactin(GO:1990637) |
| 1.7 | 6.9 | GO:0001519 | peptide amidation(GO:0001519) protein amidation(GO:0018032) peptide modification(GO:0031179) |
| 1.7 | 10.3 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 1.7 | 5.1 | GO:1903487 | regulation of lactation(GO:1903487) |
| 1.7 | 5.1 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 1.7 | 6.8 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 1.7 | 5.1 | GO:0071109 | superior temporal gyrus development(GO:0071109) |
| 1.7 | 3.4 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 1.7 | 10.0 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 1.7 | 11.6 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 1.6 | 4.9 | GO:1904404 | cellular response to vitamin B1(GO:0071301) response to formaldehyde(GO:1904404) |
| 1.6 | 7.9 | GO:1905244 | regulation of modification of synaptic structure(GO:1905244) |
| 1.6 | 4.8 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 1.5 | 16.7 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 1.5 | 6.1 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 1.5 | 16.6 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 1.5 | 19.6 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 1.5 | 10.5 | GO:0032100 | positive regulation of response to food(GO:0032097) positive regulation of appetite(GO:0032100) |
| 1.5 | 4.4 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 1.5 | 19.1 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 1.5 | 4.4 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 1.5 | 7.3 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 1.4 | 7.0 | GO:0097338 | response to clozapine(GO:0097338) |
| 1.3 | 17.5 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 1.3 | 4.0 | GO:0032900 | regulation of nucleoside transport(GO:0032242) negative regulation of neurotrophin production(GO:0032900) |
| 1.3 | 4.0 | GO:2000824 | negative regulation of androgen receptor activity(GO:2000824) |
| 1.3 | 37.2 | GO:1900273 | positive regulation of long-term synaptic potentiation(GO:1900273) |
| 1.3 | 6.3 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 1.2 | 7.5 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 1.2 | 7.3 | GO:0018343 | protein farnesylation(GO:0018343) |
| 1.2 | 4.8 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 1.2 | 3.5 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 1.2 | 14.0 | GO:0061339 | establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 1.2 | 4.7 | GO:0035900 | response to isolation stress(GO:0035900) |
| 1.2 | 4.6 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 1.1 | 3.4 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 1.1 | 3.3 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 1.1 | 6.6 | GO:0052203 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 1.1 | 9.8 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 1.1 | 7.5 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) protein localization to juxtaparanode region of axon(GO:0071205) |
| 1.1 | 3.2 | GO:0097476 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 1.1 | 7.5 | GO:0099563 | negative regulation of Rac protein signal transduction(GO:0035021) modification of synaptic structure(GO:0099563) |
| 1.1 | 6.4 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 1.0 | 7.3 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 1.0 | 3.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 1.0 | 6.1 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 1.0 | 13.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 1.0 | 16.1 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 1.0 | 3.0 | GO:0042737 | drug catabolic process(GO:0042737) |
| 1.0 | 5.9 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 1.0 | 6.7 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 1.0 | 15.2 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.9 | 6.6 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.9 | 3.7 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.9 | 18.7 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.9 | 2.7 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.9 | 17.2 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.9 | 8.9 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.9 | 2.6 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.9 | 4.3 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.8 | 2.5 | GO:0014859 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.8 | 3.4 | GO:0046502 | uroporphyrinogen III biosynthetic process(GO:0006780) uroporphyrinogen III metabolic process(GO:0046502) |
| 0.8 | 2.5 | GO:0070101 | positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen(GO:0002585) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) detection of peptidoglycan(GO:0032499) positive regulation of chemokine-mediated signaling pathway(GO:0070101) |
| 0.8 | 3.3 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.8 | 9.7 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.8 | 3.2 | GO:0060434 | bronchus morphogenesis(GO:0060434) |
| 0.8 | 23.8 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.8 | 2.4 | GO:0001983 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.8 | 4.7 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.8 | 4.6 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.8 | 1.5 | GO:2001179 | interleukin-10 secretion(GO:0072608) regulation of interleukin-10 secretion(GO:2001179) negative regulation of interleukin-10 secretion(GO:2001180) negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.8 | 6.0 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.7 | 2.2 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.7 | 4.4 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.7 | 5.1 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.7 | 10.2 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.7 | 7.2 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.7 | 2.2 | GO:0050787 | antibiotic metabolic process(GO:0016999) detoxification of mercury ion(GO:0050787) response to antineoplastic agent(GO:0097327) |
| 0.7 | 3.6 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.7 | 13.3 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.7 | 2.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.7 | 9.8 | GO:0021513 | spinal cord dorsal/ventral patterning(GO:0021513) |
| 0.7 | 6.9 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.7 | 2.0 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.7 | 3.4 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.7 | 106.4 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.6 | 6.5 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.6 | 10.3 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.6 | 1.9 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.6 | 6.4 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.6 | 6.4 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) sphingoid biosynthetic process(GO:0046520) |
| 0.6 | 20.1 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.6 | 3.7 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.6 | 3.6 | GO:1990822 | L-cystine transport(GO:0015811) basic amino acid transmembrane transport(GO:1990822) |
| 0.6 | 2.4 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.6 | 1.8 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.6 | 2.4 | GO:0021860 | pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) |
| 0.6 | 5.3 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.6 | 2.9 | GO:0042182 | ketone catabolic process(GO:0042182) |
| 0.6 | 13.5 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.6 | 9.2 | GO:0006705 | mineralocorticoid biosynthetic process(GO:0006705) mineralocorticoid metabolic process(GO:0008212) |
| 0.6 | 1.7 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.6 | 2.3 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.6 | 4.5 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.6 | 2.2 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.5 | 2.7 | GO:0018210 | peptidyl-threonine phosphorylation(GO:0018107) peptidyl-threonine modification(GO:0018210) |
| 0.5 | 4.3 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.5 | 2.7 | GO:0014827 | intestine smooth muscle contraction(GO:0014827) |
| 0.5 | 4.7 | GO:0015747 | urate transport(GO:0015747) |
| 0.5 | 3.1 | GO:0010909 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.5 | 2.1 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.5 | 5.1 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.5 | 6.6 | GO:0038042 | dimeric G-protein coupled receptor signaling pathway(GO:0038042) |
| 0.5 | 5.0 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.5 | 2.0 | GO:0010816 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.5 | 6.8 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.5 | 1.9 | GO:0036294 | cellular response to decreased oxygen levels(GO:0036294) cellular response to hypoxia(GO:0071456) |
| 0.5 | 1.4 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.5 | 2.4 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.5 | 1.9 | GO:0044026 | DNA hypermethylation(GO:0044026) |
| 0.5 | 1.4 | GO:0071810 | regulation of fever generation by regulation of prostaglandin secretion(GO:0071810) positive regulation of fever generation by positive regulation of prostaglandin secretion(GO:0071812) positive regulation of ERK1 and ERK2 cascade via TNFSF11-mediated signaling(GO:0071848) regulation of fever generation by prostaglandin secretion(GO:0100009) |
| 0.5 | 3.2 | GO:0003096 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.5 | 11.3 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.5 | 0.9 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 0.5 | 2.3 | GO:0048682 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) metanephric glomerular epithelium development(GO:0072244) metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 0.4 | 1.3 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.4 | 5.8 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.4 | 4.5 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.4 | 1.3 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.4 | 1.7 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.4 | 1.3 | GO:0098758 | response to interleukin-8(GO:0098758) cellular response to interleukin-8(GO:0098759) |
| 0.4 | 5.6 | GO:0045008 | depyrimidination(GO:0045008) |
| 0.4 | 0.9 | GO:0048867 | stem cell fate determination(GO:0048867) |
| 0.4 | 2.1 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 0.4 | 3.8 | GO:0019732 | antifungal humoral response(GO:0019732) |
| 0.4 | 4.2 | GO:0050672 | negative regulation of mononuclear cell proliferation(GO:0032945) negative regulation of lymphocyte proliferation(GO:0050672) negative regulation of leukocyte proliferation(GO:0070664) |
| 0.4 | 2.1 | GO:0051447 | negative regulation of meiotic cell cycle(GO:0051447) |
| 0.4 | 2.1 | GO:0014050 | negative regulation of glutamate secretion(GO:0014050) |
| 0.4 | 3.3 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.4 | 7.3 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.4 | 2.0 | GO:0033274 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.4 | 1.2 | GO:0051138 | positive regulation of NK T cell differentiation(GO:0051138) |
| 0.4 | 2.8 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.4 | 19.0 | GO:0050771 | negative regulation of axonogenesis(GO:0050771) |
| 0.4 | 1.1 | GO:0021558 | trochlear nerve development(GO:0021558) |
| 0.4 | 1.9 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.4 | 3.4 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.4 | 8.3 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.4 | 4.9 | GO:0021794 | thalamus development(GO:0021794) |
| 0.4 | 4.5 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.4 | 2.6 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.4 | 7.4 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.4 | 13.6 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.4 | 4.0 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.4 | 1.8 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.4 | 3.9 | GO:0030705 | cytoskeleton-dependent intracellular transport(GO:0030705) |
| 0.3 | 3.8 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.3 | 6.3 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.3 | 1.0 | GO:0006427 | histidyl-tRNA aminoacylation(GO:0006427) |
| 0.3 | 0.3 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.3 | 1.0 | GO:0086092 | regulation of the force of heart contraction by cardiac conduction(GO:0086092) |
| 0.3 | 5.4 | GO:0008228 | opsonization(GO:0008228) |
| 0.3 | 3.6 | GO:1904261 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.3 | 8.5 | GO:0097503 | sialylation(GO:0097503) |
| 0.3 | 4.9 | GO:0061436 | regulation of water loss via skin(GO:0033561) establishment of skin barrier(GO:0061436) |
| 0.3 | 5.8 | GO:0001702 | gastrulation with mouth forming second(GO:0001702) |
| 0.3 | 1.9 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.3 | 1.6 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.3 | 0.6 | GO:0050720 | interleukin-1 beta biosynthetic process(GO:0050720) |
| 0.3 | 12.4 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.3 | 2.5 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.3 | 7.0 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.3 | 1.2 | GO:0034343 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.3 | 8.1 | GO:0098743 | cell aggregation(GO:0098743) |
| 0.3 | 0.6 | GO:1901624 | negative regulation of lymphocyte chemotaxis(GO:1901624) |
| 0.3 | 3.9 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.3 | 3.0 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.3 | 4.7 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.3 | 1.2 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.3 | 2.3 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.3 | 4.3 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) |
| 0.3 | 6.6 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.3 | 2.3 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.3 | 2.3 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.3 | 0.6 | GO:0014004 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.3 | 5.6 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.3 | 6.6 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.3 | 0.6 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.3 | 0.8 | GO:2000328 | regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.3 | 1.6 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.3 | 6.0 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.3 | 3.9 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.3 | 1.8 | GO:1901374 | epinephrine transport(GO:0048241) acetate ester transport(GO:1901374) |
| 0.3 | 4.0 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.3 | 5.0 | GO:0040020 | regulation of meiotic nuclear division(GO:0040020) |
| 0.2 | 0.7 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.2 | 1.5 | GO:0032252 | secretory granule localization(GO:0032252) |
| 0.2 | 1.2 | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
| 0.2 | 7.1 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.2 | 5.9 | GO:0070233 | negative regulation of T cell apoptotic process(GO:0070233) |
| 0.2 | 4.5 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.2 | 0.7 | GO:1902159 | transepithelial water transport(GO:0035377) regulation of cyclic nucleotide-gated ion channel activity(GO:1902159) positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) |
| 0.2 | 0.7 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.2 | 11.4 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.2 | 8.0 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.2 | 8.4 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.2 | 3.6 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.2 | 6.0 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.2 | 4.2 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.2 | 8.2 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.2 | 13.3 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.2 | 10.9 | GO:0042308 | negative regulation of protein import into nucleus(GO:0042308) negative regulation of protein import(GO:1904590) |
| 0.2 | 0.8 | GO:0060025 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) regulation of synaptic activity(GO:0060025) |
| 0.2 | 0.6 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.2 | 3.5 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.2 | 5.4 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.2 | 0.8 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) positive regulation of blood vessel remodeling(GO:2000504) |
| 0.2 | 1.0 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) |
| 0.2 | 2.0 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.2 | 6.2 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.2 | 3.8 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.2 | 0.7 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.2 | 0.6 | GO:0002784 | regulation of antimicrobial peptide production(GO:0002784) regulation of antibacterial peptide production(GO:0002786) |
| 0.2 | 2.2 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.2 | 0.9 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.2 | 1.0 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.2 | 1.7 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) negative regulation of telomerase activity(GO:0051974) |
| 0.2 | 1.0 | GO:0051450 | myoblast proliferation(GO:0051450) |
| 0.2 | 1.2 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.2 | 2.6 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.2 | 3.2 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.2 | 4.9 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.2 | 1.4 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.2 | 1.8 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.2 | 1.7 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.1 | 0.7 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.1 | 2.4 | GO:0045725 | positive regulation of glycogen biosynthetic process(GO:0045725) |
| 0.1 | 0.7 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.1 | 2.1 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.1 | 12.1 | GO:0045921 | positive regulation of exocytosis(GO:0045921) |
| 0.1 | 1.0 | GO:0019064 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.1 | 6.8 | GO:0051220 | cytoplasmic sequestering of protein(GO:0051220) |
| 0.1 | 1.6 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) negative regulation of biomineral tissue development(GO:0070168) |
| 0.1 | 1.9 | GO:1900181 | negative regulation of protein localization to nucleus(GO:1900181) |
| 0.1 | 4.5 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.1 | 2.6 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) positive regulation of neutrophil migration(GO:1902624) |
| 0.1 | 8.4 | GO:0072384 | organelle transport along microtubule(GO:0072384) |
| 0.1 | 1.4 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.1 | 1.0 | GO:0042364 | water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.1 | 0.9 | GO:0035878 | nail development(GO:0035878) |
| 0.1 | 0.4 | GO:2001190 | positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.1 | 1.8 | GO:0034599 | cellular response to oxidative stress(GO:0034599) |
| 0.1 | 0.2 | GO:0014831 | gastro-intestinal system smooth muscle contraction(GO:0014831) |
| 0.1 | 1.8 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.1 | 1.2 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.1 | 2.4 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 4.6 | GO:0042733 | embryonic digit morphogenesis(GO:0042733) |
| 0.1 | 0.7 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.1 | 1.2 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.1 | 1.8 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 5.1 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 4.6 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.1 | 2.0 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.1 | 2.2 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 1.4 | GO:0035584 | calcium-mediated signaling using intracellular calcium source(GO:0035584) |
| 0.1 | 3.8 | GO:0031018 | endocrine pancreas development(GO:0031018) |
| 0.1 | 3.7 | GO:0016055 | Wnt signaling pathway(GO:0016055) cell-cell signaling by wnt(GO:0198738) |
| 0.1 | 0.5 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.1 | 12.9 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.1 | 1.0 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 2.2 | GO:0035635 | entry of bacterium into host cell(GO:0035635) |
| 0.1 | 2.0 | GO:1901661 | quinone metabolic process(GO:1901661) |
| 0.1 | 0.1 | GO:0034163 | regulation of toll-like receptor 9 signaling pathway(GO:0034163) positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) |
| 0.1 | 3.8 | GO:0048599 | oocyte development(GO:0048599) |
| 0.1 | 1.0 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 6.1 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.1 | 0.8 | GO:0042268 | regulation of cytolysis(GO:0042268) |
| 0.1 | 1.3 | GO:0032354 | response to follicle-stimulating hormone(GO:0032354) |
| 0.1 | 1.3 | GO:0030220 | platelet formation(GO:0030220) |
| 0.1 | 0.7 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.1 | 0.5 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 | 2.8 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.1 | 0.2 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.1 | 3.3 | GO:0002548 | monocyte chemotaxis(GO:0002548) |
| 0.1 | 1.5 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.1 | 2.5 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.1 | 4.7 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 3.5 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.1 | 0.8 | GO:0051647 | nucleus localization(GO:0051647) |
| 0.1 | 0.6 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 1.3 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.1 | 0.9 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 | 1.8 | GO:0051923 | sulfation(GO:0051923) |
| 0.1 | 2.1 | GO:0021954 | central nervous system neuron development(GO:0021954) |
| 0.1 | 1.2 | GO:0030947 | regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030947) |
| 0.1 | 1.1 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.1 | 2.6 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.1 | 1.3 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.1 | 1.5 | GO:0001764 | neuron migration(GO:0001764) |
| 0.1 | 5.0 | GO:0071805 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.1 | 1.5 | GO:0007140 | male meiosis(GO:0007140) |
| 0.1 | 1.3 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
| 0.1 | 2.9 | GO:0071806 | protein transmembrane transport(GO:0071806) |
| 0.1 | 0.4 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.1 | 0.9 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 6.2 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.1 | 0.4 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 1.6 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.1 | GO:0046449 | creatinine metabolic process(GO:0046449) |
| 0.0 | 4.0 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 0.3 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 1.1 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.2 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.8 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 12.7 | GO:0098916 | chemical synaptic transmission(GO:0007268) anterograde trans-synaptic signaling(GO:0098916) trans-synaptic signaling(GO:0099537) |
| 0.0 | 0.3 | GO:0044241 | lipid digestion(GO:0044241) |
| 0.0 | 0.7 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 1.9 | GO:0098773 | skin epidermis development(GO:0098773) |
| 0.0 | 1.0 | GO:0014911 | positive regulation of smooth muscle cell migration(GO:0014911) |
| 0.0 | 0.5 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 1.0 | GO:0051058 | negative regulation of Ras protein signal transduction(GO:0046580) negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.0 | 0.3 | GO:0051930 | regulation of sensory perception of pain(GO:0051930) regulation of sensory perception(GO:0051931) |
| 0.0 | 0.4 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.7 | GO:0002763 | positive regulation of myeloid leukocyte differentiation(GO:0002763) |
| 0.0 | 2.8 | GO:0021915 | neural tube development(GO:0021915) |
| 0.0 | 0.6 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.1 | GO:0035150 | regulation of tube size(GO:0035150) regulation of blood vessel size(GO:0050880) |
| 0.0 | 1.6 | GO:1902108 | regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902108) |
| 0.0 | 0.4 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 0.2 | GO:0060536 | cartilage morphogenesis(GO:0060536) |
| 0.0 | 2.7 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.5 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 2.2 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.0 | 2.7 | GO:0072659 | protein localization to plasma membrane(GO:0072659) |
| 0.0 | 1.2 | GO:0001525 | angiogenesis(GO:0001525) |
| 0.0 | 0.6 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.0 | 0.9 | GO:0007267 | cell-cell signaling(GO:0007267) |
| 0.0 | 0.1 | GO:0072174 | thyroid-stimulating hormone signaling pathway(GO:0038194) pronephric field specification(GO:0039003) pattern specification involved in pronephros development(GO:0039017) thyroid-stimulating hormone secretion(GO:0070460) kidney rudiment formation(GO:0072003) kidney field specification(GO:0072004) regulation of mesenchymal cell apoptotic process involved in nephron morphogenesis(GO:0072039) negative regulation of mesenchymal cell apoptotic process involved in nephron morphogenesis(GO:0072040) comma-shaped body morphogenesis(GO:0072049) metanephric tubule formation(GO:0072174) metanephric comma-shaped body morphogenesis(GO:0072278) metanephric nephron tubule formation(GO:0072289) regulation of mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:0072304) negative regulation of mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:0072305) mesenchymal stem cell maintenance involved in metanephric nephron morphogenesis(GO:0072309) mesenchymal cell apoptotic process involved in metanephros development(GO:1900200) apoptotic process involved in metanephric collecting duct development(GO:1900204) apoptotic process involved in metanephric nephron tubule development(GO:1900205) regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900211) negative regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900212) regulation of apoptotic process involved in metanephric collecting duct development(GO:1900214) negative regulation of apoptotic process involved in metanephric collecting duct development(GO:1900215) regulation of apoptotic process involved in metanephric nephron tubule development(GO:1900217) negative regulation of apoptotic process involved in metanephric nephron tubule development(GO:1900218) mesenchymal cell apoptotic process involved in nephron morphogenesis(GO:1901145) mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:1901147) negative regulation of apoptotic process involved in morphogenesis(GO:1902338) negative regulation of somatic stem cell population maintenance(GO:1904673) negative regulation of apoptotic process involved in development(GO:1904746) regulation of metanephric DCT cell differentiation(GO:2000592) positive regulation of metanephric DCT cell differentiation(GO:2000594) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.0 | 0.2 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.0 | 0.5 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 0.3 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 0.4 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.3 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.7 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.1 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 1.3 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.2 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.5 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 1.3 | GO:0006836 | neurotransmitter transport(GO:0006836) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.0 | 112.7 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 6.0 | 23.9 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 5.8 | 28.9 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 4.6 | 64.4 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 4.2 | 50.7 | GO:0044327 | dendritic spine head(GO:0044327) |
| 4.2 | 16.7 | GO:1990246 | uniplex complex(GO:1990246) |
| 2.6 | 10.6 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 2.3 | 6.8 | GO:0044609 | DBIRD complex(GO:0044609) |
| 2.2 | 30.8 | GO:0042583 | chromaffin granule(GO:0042583) |
| 2.2 | 13.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 2.2 | 8.7 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 2.1 | 8.6 | GO:1990745 | EARP complex(GO:1990745) |
| 2.1 | 14.6 | GO:0070852 | cell body fiber(GO:0070852) |
| 2.0 | 6.1 | GO:0044305 | calyx of Held(GO:0044305) |
| 2.0 | 2.0 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 2.0 | 5.9 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 1.7 | 10.3 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) DNA ligase IV complex(GO:0032807) |
| 1.7 | 11.9 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 1.6 | 11.1 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 1.6 | 7.9 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 1.6 | 28.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 1.5 | 7.5 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 1.4 | 7.2 | GO:0045240 | dihydrolipoyl dehydrogenase complex(GO:0045240) |
| 1.2 | 7.3 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 1.2 | 6.0 | GO:0071546 | pi-body(GO:0071546) |
| 1.2 | 8.4 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.9 | 1.8 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.9 | 4.5 | GO:0044308 | axonal spine(GO:0044308) |
| 0.9 | 7.0 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.9 | 25.3 | GO:0043218 | compact myelin(GO:0043218) |
| 0.9 | 11.9 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.8 | 7.5 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.8 | 20.6 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.8 | 3.2 | GO:0038038 | G-protein coupled receptor homodimeric complex(GO:0038038) |
| 0.8 | 2.4 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.8 | 6.3 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.8 | 5.5 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.8 | 5.5 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.8 | 66.9 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.8 | 3.8 | GO:0089701 | U2AF(GO:0089701) |
| 0.8 | 11.3 | GO:0030478 | actin cap(GO:0030478) |
| 0.8 | 2.3 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.7 | 3.0 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.7 | 18.9 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.7 | 4.8 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.7 | 2.8 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.7 | 4.0 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.7 | 6.6 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.6 | 23.8 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.6 | 4.2 | GO:0030315 | T-tubule(GO:0030315) |
| 0.6 | 4.0 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.6 | 2.3 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.6 | 6.7 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.6 | 3.3 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.5 | 3.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.5 | 2.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.5 | 4.7 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.5 | 3.1 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.5 | 16.5 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.5 | 2.0 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.5 | 1.5 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.5 | 2.4 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.5 | 3.4 | GO:1903440 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.5 | 3.8 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.5 | 5.5 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.5 | 7.3 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.4 | 1.8 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.4 | 2.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.4 | 2.0 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.4 | 1.6 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.4 | 7.4 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.4 | 3.5 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.4 | 27.7 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.4 | 7.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.4 | 2.9 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.4 | 5.5 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.4 | 2.5 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.4 | 2.2 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.4 | 7.5 | GO:0097546 | ciliary base(GO:0097546) |
| 0.4 | 20.7 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.3 | 4.2 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.3 | 3.4 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.3 | 1.0 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.3 | 2.3 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.3 | 3.3 | GO:0016011 | dystroglycan complex(GO:0016011) |
| 0.3 | 77.4 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.3 | 41.1 | GO:0043204 | perikaryon(GO:0043204) |
| 0.3 | 2.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.3 | 5.1 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.3 | 0.9 | GO:0070081 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.3 | 24.5 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.3 | 5.1 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.3 | 1.7 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.3 | 14.4 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.3 | 1.9 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.3 | 1.8 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.2 | 1.0 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.2 | 5.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.2 | 8.1 | GO:0043034 | costamere(GO:0043034) |
| 0.2 | 3.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.2 | 6.1 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.2 | 2.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.2 | 4.6 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.2 | 1.3 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.2 | 1.9 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.2 | 16.4 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.2 | 3.5 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.2 | 1.2 | GO:0002177 | manchette(GO:0002177) |
| 0.2 | 3.2 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.2 | 11.1 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.2 | 9.7 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.1 | 28.0 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 1.5 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 0.1 | 36.0 | GO:0030424 | axon(GO:0030424) |
| 0.1 | 0.7 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.1 | 1.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 7.9 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.1 | 3.4 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 1.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 2.5 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 36.9 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 8.0 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.1 | 0.7 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 12.5 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.1 | 4.8 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 0.4 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 1.5 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 1.2 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 6.8 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.1 | 7.7 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.1 | 1.6 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.1 | 1.3 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.1 | 1.0 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 11.8 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.1 | 2.5 | GO:0034703 | cation channel complex(GO:0034703) |
| 0.1 | 4.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 1.1 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.5 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 72.5 | GO:0031226 | intrinsic component of plasma membrane(GO:0031226) |
| 0.0 | 1.3 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.6 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 30.2 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.3 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 1.0 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 2.0 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 2.1 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 1.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 3.6 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 1.2 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 36.1 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 3.1 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 0.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.2 | GO:0005795 | Golgi stack(GO:0005795) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.8 | 28.9 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 5.8 | 17.3 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 4.7 | 14.2 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 4.0 | 23.9 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 3.7 | 114.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 3.1 | 9.2 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 3.0 | 9.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 2.8 | 19.6 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 2.8 | 19.4 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 2.6 | 15.8 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 2.4 | 7.3 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 2.4 | 7.2 | GO:0003826 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 2.4 | 16.7 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 2.3 | 11.6 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 2.3 | 11.4 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 2.2 | 11.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 2.2 | 8.9 | GO:0016900 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 2.2 | 13.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 2.1 | 6.3 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 2.1 | 12.4 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 2.1 | 8.2 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 2.0 | 28.1 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 1.8 | 7.3 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 1.8 | 8.9 | GO:0004945 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 1.8 | 12.4 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 1.8 | 5.3 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 1.7 | 38.4 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 1.7 | 6.9 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 1.7 | 6.9 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 1.7 | 13.7 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 1.7 | 8.4 | GO:0070404 | NADH binding(GO:0070404) |
| 1.7 | 10.0 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 1.7 | 33.1 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 1.6 | 4.9 | GO:0061663 | NEDD8 ligase activity(GO:0061663) |
| 1.6 | 6.5 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 1.6 | 49.8 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 1.6 | 6.4 | GO:0017040 | ceramidase activity(GO:0017040) |
| 1.6 | 4.7 | GO:0047783 | steroid 11-beta-monooxygenase activity(GO:0004507) corticosterone 18-monooxygenase activity(GO:0047783) |
| 1.6 | 4.7 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 1.4 | 5.6 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 1.3 | 7.9 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 1.3 | 6.4 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 1.3 | 6.3 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 1.3 | 3.8 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 1.2 | 7.5 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 1.2 | 22.3 | GO:0008391 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 1.2 | 15.1 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 1.1 | 2.3 | GO:0030109 | HLA-B specific inhibitory MHC class I receptor activity(GO:0030109) |
| 1.0 | 4.0 | GO:0030760 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 1.0 | 66.5 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 1.0 | 5.7 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 1.0 | 13.3 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.9 | 6.5 | GO:0030345 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.9 | 4.4 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.9 | 6.2 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.9 | 15.6 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.8 | 20.6 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.8 | 2.5 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.8 | 4.0 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.8 | 2.4 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.8 | 12.6 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.8 | 4.6 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.8 | 8.4 | GO:0008430 | selenium binding(GO:0008430) |
| 0.8 | 3.8 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.7 | 3.7 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 0.7 | 8.2 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.7 | 2.9 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.7 | 10.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.7 | 3.6 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.7 | 5.0 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.7 | 2.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.7 | 3.4 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.7 | 4.7 | GO:0005497 | androgen binding(GO:0005497) |
| 0.7 | 17.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.7 | 4.0 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.6 | 1.9 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.6 | 3.6 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.6 | 2.4 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.6 | 5.7 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.6 | 2.3 | GO:0042020 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.6 | 6.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.5 | 1.6 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.5 | 1.6 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.5 | 5.5 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.5 | 4.7 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.5 | 26.8 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.5 | 4.0 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.5 | 4.5 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.5 | 6.6 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.5 | 2.8 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.5 | 8.3 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.5 | 2.7 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.5 | 1.8 | GO:0005277 | acetylcholine transmembrane transporter activity(GO:0005277) secondary active organic cation transmembrane transporter activity(GO:0008513) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.4 | 6.3 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.4 | 10.0 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.4 | 1.3 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.4 | 18.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.4 | 5.6 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.4 | 2.1 | GO:0070905 | serine binding(GO:0070905) |
| 0.4 | 11.1 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.4 | 3.7 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.4 | 2.0 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.4 | 18.0 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.4 | 16.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.4 | 11.6 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.4 | 5.4 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.4 | 1.5 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.4 | 8.9 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.4 | 2.9 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.4 | 5.5 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.4 | 2.5 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.4 | 3.2 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.4 | 11.9 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.3 | 1.0 | GO:0004821 | histidine-tRNA ligase activity(GO:0004821) |
| 0.3 | 7.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.3 | 2.0 | GO:0008443 | 6-phosphofructokinase activity(GO:0003872) phosphofructokinase activity(GO:0008443) |
| 0.3 | 6.1 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.3 | 3.7 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.3 | 5.6 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.3 | 3.5 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.3 | 8.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.3 | 6.6 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.3 | 5.1 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.3 | 0.9 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.3 | 1.2 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.3 | 8.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.3 | 15.8 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.3 | 5.6 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.3 | 7.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.3 | 9.4 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.3 | 2.6 | GO:0016918 | retinal binding(GO:0016918) |
| 0.3 | 6.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.3 | 2.3 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.3 | 3.3 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.2 | 5.0 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.2 | 8.4 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.2 | 1.7 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.2 | 7.1 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.2 | 7.5 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.2 | 10.2 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.2 | 4.6 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.2 | 4.1 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.2 | 10.7 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.2 | 4.3 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.2 | 5.8 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.2 | 1.8 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.2 | 25.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.2 | 0.9 | GO:0099589 | serotonin receptor activity(GO:0099589) |
| 0.2 | 3.9 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.2 | 7.7 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.2 | 2.5 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.2 | 1.1 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.2 | 0.6 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.2 | 4.8 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.2 | 2.9 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.2 | 3.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.2 | 1.8 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.2 | 3.2 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.2 | 24.9 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.2 | 3.5 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.2 | 5.1 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.2 | 1.1 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.2 | 0.7 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.2 | 1.8 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.2 | 1.6 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.2 | 1.4 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.2 | 0.9 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.2 | 2.6 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.2 | 2.4 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.2 | 6.0 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.2 | 7.5 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.2 | 15.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.2 | 2.0 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.2 | 3.1 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.2 | 2.6 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.2 | 3.2 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.2 | 9.1 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.1 | 0.7 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.1 | 1.0 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.1 | 2.3 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.1 | 2.4 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 29.1 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 0.5 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.1 | 2.4 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 4.8 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 4.2 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 3.1 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 0.7 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.1 | 3.5 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.1 | 2.6 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.1 | 1.4 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 4.0 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 2.0 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 3.6 | GO:0001848 | complement binding(GO:0001848) |
| 0.1 | 2.0 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 2.7 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 2.4 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 3.7 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 3.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 1.6 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.1 | 7.2 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 25.7 | GO:0005543 | phospholipid binding(GO:0005543) |
| 0.1 | 0.6 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.1 | 18.3 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.1 | 3.0 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 0.2 | GO:0015180 | L-alanine transmembrane transporter activity(GO:0015180) alanine transmembrane transporter activity(GO:0022858) |
| 0.1 | 1.1 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.1 | 1.7 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 7.9 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 50.7 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.1 | 3.8 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 3.4 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.1 | 3.6 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 0.8 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.1 | 0.2 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 0.5 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.1 | 1.1 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 0.3 | GO:0004771 | sterol esterase activity(GO:0004771) methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.1 | 2.3 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 0.5 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 0.7 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.1 | 3.3 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 1.2 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.1 | 0.7 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 0.4 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 1.7 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 4.8 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 10.7 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.1 | 0.4 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.1 | 1.5 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 3.5 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 2.4 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.3 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 3.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.4 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.7 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 2.1 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.3 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 0.3 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 8.6 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 1.3 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.4 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.6 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 1.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 3.1 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 0.2 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.0 | 0.6 | GO:0015144 | carbohydrate transmembrane transporter activity(GO:0015144) carbohydrate transporter activity(GO:1901476) |
| 0.0 | 0.2 | GO:0008143 | poly(A) binding(GO:0008143) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 65.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.8 | 46.6 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.7 | 6.1 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.4 | 17.3 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.4 | 13.0 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.4 | 5.7 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.3 | 6.3 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.3 | 32.7 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.3 | 5.5 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.3 | 16.9 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.3 | 22.2 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.3 | 5.8 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.3 | 21.4 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.3 | 12.4 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.2 | 14.5 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.2 | 9.8 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.2 | 7.9 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.2 | 11.7 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.2 | 3.7 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.2 | 7.0 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.2 | 10.3 | PID ATM PATHWAY | ATM pathway |
| 0.2 | 5.0 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.2 | 2.8 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.2 | 9.4 | PID FGF PATHWAY | FGF signaling pathway |
| 0.2 | 2.7 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.2 | 15.4 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.2 | 4.8 | ST ADRENERGIC | Adrenergic Pathway |
| 0.2 | 7.8 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.2 | 11.9 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.2 | 1.3 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.2 | 14.3 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.2 | 1.5 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.2 | 6.7 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.2 | 2.2 | PID EPO PATHWAY | EPO signaling pathway |
| 0.2 | 0.8 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.2 | 6.3 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.2 | 6.2 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 5.2 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.1 | 8.7 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 11.4 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 2.4 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 0.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 2.8 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 1.3 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 3.7 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 9.0 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.1 | 1.5 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.1 | 0.4 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 1.2 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 19.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 1.5 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 1.4 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 1.0 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 19.2 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 1.7 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 9.6 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.0 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 0.5 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.8 | NABA CORE MATRISOME | Ensemble of genes encoding core extracellular matrix including ECM glycoproteins, collagens and proteoglycans |
| 0.0 | 1.7 | NABA MATRISOME | Ensemble of genes encoding extracellular matrix and extracellular matrix-associated proteins |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 38.8 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 2.1 | 64.4 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 1.3 | 22.8 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.9 | 17.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.8 | 15.8 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.6 | 32.5 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.6 | 32.5 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.6 | 6.6 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.6 | 19.1 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.6 | 7.4 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.6 | 10.8 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.5 | 13.6 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.5 | 14.2 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |
| 0.5 | 15.0 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.4 | 10.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.4 | 13.3 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.4 | 2.0 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.4 | 14.5 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.4 | 6.4 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.4 | 8.1 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.3 | 6.3 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.3 | 8.0 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.3 | 11.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.3 | 5.1 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.3 | 4.4 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.3 | 6.5 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.3 | 7.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.3 | 5.6 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.3 | 17.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.3 | 4.8 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.3 | 6.2 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.3 | 5.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.3 | 11.3 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.3 | 11.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.3 | 15.7 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.3 | 4.6 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.3 | 12.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.3 | 16.9 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.3 | 4.9 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.2 | 23.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.2 | 1.5 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.2 | 7.9 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.2 | 3.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.2 | 4.7 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.2 | 11.1 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.2 | 6.2 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.2 | 42.4 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.2 | 25.8 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.2 | 23.7 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.2 | 2.5 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.2 | 6.6 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.2 | 4.7 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.2 | 6.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.2 | 1.8 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 1.6 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.1 | 4.2 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 5.2 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.1 | 11.8 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 1.4 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.1 | 4.0 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 1.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 0.9 | REACTOME ION CHANNEL TRANSPORT | Genes involved in Ion channel transport |
| 0.1 | 3.7 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.1 | 1.3 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 0.5 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 1.7 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.1 | 9.8 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 1.2 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.1 | 1.7 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 1.1 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 1.9 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.1 | 1.2 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.1 | 1.8 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 1.5 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 1.9 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.1 | 1.9 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 0.9 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.7 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 1.6 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.9 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.7 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 1.0 | REACTOME PHOSPHOLIPID METABOLISM | Genes involved in Phospholipid metabolism |
| 0.0 | 1.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.5 | REACTOME LIPID DIGESTION MOBILIZATION AND TRANSPORT | Genes involved in Lipid digestion, mobilization, and transport |
| 0.0 | 0.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 2.0 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.5 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.2 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.9 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.2 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.4 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |