Project
averaged GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
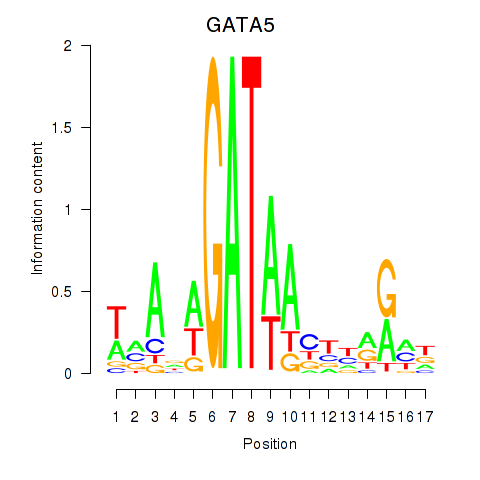
Results for GATA5
Z-value: 0.78
Transcription factors associated with GATA5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GATA5
|
ENSG00000130700.6 | GATA binding protein 5 |
Activity profile of GATA5 motif
Sorted Z-values of GATA5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_66874502 | 11.78 |
ENST00000558797.1
|
RP11-321F6.1
|
HCG2003567; Uncharacterized protein |
| chr1_-_160681593 | 10.42 |
ENST00000368045.3
ENST00000368046.3 |
CD48
|
CD48 molecule |
| chr6_-_32557610 | 9.53 |
ENST00000360004.5
|
HLA-DRB1
|
major histocompatibility complex, class II, DR beta 1 |
| chr6_+_33043703 | 7.64 |
ENST00000418931.2
ENST00000535465.1 |
HLA-DPB1
|
major histocompatibility complex, class II, DP beta 1 |
| chr13_-_46756351 | 7.55 |
ENST00000323076.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr4_+_155484155 | 7.05 |
ENST00000509493.1
|
FGB
|
fibrinogen beta chain |
| chr19_+_35820064 | 6.90 |
ENST00000341773.6
ENST00000600131.1 ENST00000270311.6 ENST00000595780.1 ENST00000597916.1 ENST00000593867.1 ENST00000600424.1 ENST00000599811.1 ENST00000536635.2 ENST00000085219.5 ENST00000544992.2 ENST00000419549.2 |
CD22
|
CD22 molecule |
| chr4_+_175204818 | 6.66 |
ENST00000503780.1
|
CEP44
|
centrosomal protein 44kDa |
| chr6_+_32709119 | 6.52 |
ENST00000374940.3
|
HLA-DQA2
|
major histocompatibility complex, class II, DQ alpha 2 |
| chr15_+_99791567 | 6.27 |
ENST00000558879.1
ENST00000301981.3 ENST00000422500.2 ENST00000447360.2 ENST00000442993.2 |
LRRC28
|
leucine rich repeat containing 28 |
| chr1_-_213031418 | 6.23 |
ENST00000356684.3
ENST00000426161.1 ENST00000424044.1 |
FLVCR1-AS1
|
FLVCR1 antisense RNA 1 (head to head) |
| chr1_+_174844645 | 5.92 |
ENST00000486220.1
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr8_+_9046503 | 5.41 |
ENST00000512942.2
|
RP11-10A14.5
|
RP11-10A14.5 |
| chr10_+_7745232 | 5.11 |
ENST00000358415.4
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr10_+_7745303 | 4.90 |
ENST00000429820.1
ENST00000379587.4 |
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr4_+_155484103 | 4.79 |
ENST00000302068.4
|
FGB
|
fibrinogen beta chain |
| chr6_-_32908765 | 4.53 |
ENST00000416244.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr11_+_128563652 | 3.96 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr6_-_24936170 | 3.90 |
ENST00000538035.1
|
FAM65B
|
family with sequence similarity 65, member B |
| chr8_-_6837602 | 3.79 |
ENST00000382692.2
|
DEFA1
|
defensin, alpha 1 |
| chr12_-_45270077 | 3.77 |
ENST00000551601.1
ENST00000549027.1 ENST00000452445.2 |
NELL2
|
NEL-like 2 (chicken) |
| chr8_-_6875778 | 3.75 |
ENST00000535841.1
ENST00000327857.2 |
DEFA1B
DEFA3
|
defensin, alpha 1B defensin, alpha 3, neutrophil-specific |
| chr12_-_45270151 | 3.74 |
ENST00000429094.2
|
NELL2
|
NEL-like 2 (chicken) |
| chr1_-_158656488 | 3.74 |
ENST00000368147.4
|
SPTA1
|
spectrin, alpha, erythrocytic 1 (elliptocytosis 2) |
| chr11_+_121461097 | 3.73 |
ENST00000527934.1
|
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing |
| chr12_-_45269430 | 3.68 |
ENST00000395487.2
|
NELL2
|
NEL-like 2 (chicken) |
| chr9_-_35650900 | 3.55 |
ENST00000259608.3
|
SIT1
|
signaling threshold regulating transmembrane adaptor 1 |
| chr11_-_33891362 | 3.36 |
ENST00000395833.3
|
LMO2
|
LIM domain only 2 (rhombotin-like 1) |
| chr6_+_31916733 | 3.16 |
ENST00000483004.1
|
CFB
|
complement factor B |
| chr16_+_31539183 | 3.11 |
ENST00000302312.4
|
AHSP
|
alpha hemoglobin stabilizing protein |
| chr2_+_102615416 | 3.03 |
ENST00000393414.2
|
IL1R2
|
interleukin 1 receptor, type II |
| chr4_-_72649763 | 2.93 |
ENST00000513476.1
|
GC
|
group-specific component (vitamin D binding protein) |
| chr6_-_133079022 | 2.87 |
ENST00000525289.1
ENST00000326499.6 |
VNN2
|
vanin 2 |
| chr12_+_54892550 | 2.83 |
ENST00000545638.2
|
NCKAP1L
|
NCK-associated protein 1-like |
| chr7_+_134331550 | 2.79 |
ENST00000344924.3
ENST00000418040.1 ENST00000393132.2 |
BPGM
|
2,3-bisphosphoglycerate mutase |
| chr1_+_115572415 | 2.75 |
ENST00000256592.1
|
TSHB
|
thyroid stimulating hormone, beta |
| chr9_+_91933407 | 2.73 |
ENST00000375807.3
ENST00000339901.4 |
SECISBP2
|
SECIS binding protein 2 |
| chr1_-_17445930 | 2.67 |
ENST00000375486.4
ENST00000375481.1 ENST00000444885.2 |
PADI2
|
peptidyl arginine deiminase, type II |
| chr4_-_110723335 | 2.63 |
ENST00000394634.2
|
CFI
|
complement factor I |
| chr8_+_28196157 | 2.50 |
ENST00000522209.1
|
PNOC
|
prepronociceptin |
| chr4_-_110723194 | 2.44 |
ENST00000394635.3
|
CFI
|
complement factor I |
| chr8_+_117778736 | 2.44 |
ENST00000309822.2
ENST00000357148.3 ENST00000517814.1 ENST00000517820.1 |
UTP23
|
UTP23, small subunit (SSU) processome component, homolog (yeast) |
| chr1_+_156123318 | 2.43 |
ENST00000368285.3
|
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr19_+_9296279 | 2.41 |
ENST00000344248.2
|
OR7D2
|
olfactory receptor, family 7, subfamily D, member 2 |
| chr12_-_9268707 | 2.40 |
ENST00000318602.7
|
A2M
|
alpha-2-macroglobulin |
| chr12_+_9102632 | 2.35 |
ENST00000539240.1
|
KLRG1
|
killer cell lectin-like receptor subfamily G, member 1 |
| chr1_-_203274418 | 2.33 |
ENST00000457348.1
|
RP11-134P9.1
|
long intergenic non-protein coding RNA 1136 |
| chr13_-_107214291 | 2.32 |
ENST00000375926.1
|
ARGLU1
|
arginine and glutamate rich 1 |
| chr19_-_14889349 | 2.31 |
ENST00000315576.3
ENST00000392967.2 ENST00000346057.1 ENST00000353876.1 ENST00000353005.1 |
EMR2
|
egf-like module containing, mucin-like, hormone receptor-like 2 |
| chr6_-_133055896 | 2.30 |
ENST00000367927.5
ENST00000425515.2 ENST00000207771.3 ENST00000392393.3 ENST00000450865.2 ENST00000392394.2 |
VNN3
|
vanin 3 |
| chr5_-_150537279 | 2.28 |
ENST00000517486.1
ENST00000377751.5 ENST00000356496.5 ENST00000521512.1 ENST00000517757.1 ENST00000354546.5 |
ANXA6
|
annexin A6 |
| chr9_-_6645628 | 2.27 |
ENST00000321612.6
|
GLDC
|
glycine dehydrogenase (decarboxylating) |
| chr17_-_61996192 | 2.26 |
ENST00000392824.4
|
CSHL1
|
chorionic somatomammotropin hormone-like 1 |
| chr19_-_52133588 | 2.23 |
ENST00000570106.2
|
SIGLEC5
|
sialic acid binding Ig-like lectin 5 |
| chr14_+_76618242 | 2.20 |
ENST00000557542.1
ENST00000557263.1 ENST00000557207.1 ENST00000312858.5 ENST00000261530.7 |
GPATCH2L
|
G patch domain containing 2-like |
| chr16_-_33647696 | 2.19 |
ENST00000558425.1
ENST00000569103.2 |
RP11-812E19.9
|
Uncharacterized protein |
| chr16_+_31366455 | 2.11 |
ENST00000268296.4
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit) |
| chr17_-_61996160 | 2.07 |
ENST00000458650.2
ENST00000351388.4 ENST00000323322.5 |
GH1
|
growth hormone 1 |
| chr19_-_10450328 | 2.01 |
ENST00000160262.5
|
ICAM3
|
intercellular adhesion molecule 3 |
| chr7_+_26332645 | 1.94 |
ENST00000396376.1
|
SNX10
|
sorting nexin 10 |
| chr6_-_135271260 | 1.92 |
ENST00000265605.2
|
ALDH8A1
|
aldehyde dehydrogenase 8 family, member A1 |
| chr6_-_135271219 | 1.89 |
ENST00000367847.2
ENST00000367845.2 |
ALDH8A1
|
aldehyde dehydrogenase 8 family, member A1 |
| chr16_+_31366536 | 1.88 |
ENST00000562522.1
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit) |
| chr18_-_5419797 | 1.86 |
ENST00000542146.1
ENST00000427684.2 |
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3 |
| chr4_-_110723134 | 1.85 |
ENST00000510800.1
ENST00000512148.1 |
CFI
|
complement factor I |
| chr3_+_178276488 | 1.78 |
ENST00000432997.1
ENST00000455865.1 |
KCNMB2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
| chr6_-_52710893 | 1.73 |
ENST00000284562.2
|
GSTA5
|
glutathione S-transferase alpha 5 |
| chr13_-_99910673 | 1.71 |
ENST00000397473.2
ENST00000397470.2 |
GPR18
|
G protein-coupled receptor 18 |
| chr4_-_47983519 | 1.68 |
ENST00000358519.4
ENST00000544810.1 ENST00000402813.3 |
CNGA1
|
cyclic nucleotide gated channel alpha 1 |
| chr15_-_20193370 | 1.68 |
ENST00000558565.2
|
IGHV3OR15-7
|
immunoglobulin heavy variable 3/OR15-7 (pseudogene) |
| chr10_-_98031265 | 1.64 |
ENST00000224337.5
ENST00000371176.2 |
BLNK
|
B-cell linker |
| chr19_+_42580274 | 1.57 |
ENST00000359044.4
|
ZNF574
|
zinc finger protein 574 |
| chr14_+_61995722 | 1.56 |
ENST00000556347.1
|
RP11-47I22.4
|
RP11-47I22.4 |
| chr12_+_62860581 | 1.51 |
ENST00000393632.2
ENST00000393630.3 ENST00000280379.6 ENST00000546600.1 ENST00000552738.1 ENST00000393629.2 ENST00000552115.1 |
MON2
|
MON2 homolog (S. cerevisiae) |
| chr16_+_77224732 | 1.50 |
ENST00000569610.1
ENST00000248248.3 ENST00000567291.1 ENST00000320859.6 ENST00000563612.1 ENST00000563279.1 |
MON1B
|
MON1 secretory trafficking family member B |
| chr10_+_43916052 | 1.44 |
ENST00000442526.2
|
RP11-517P14.2
|
RP11-517P14.2 |
| chr5_+_140261703 | 1.43 |
ENST00000409494.1
ENST00000289272.2 |
PCDHA13
|
protocadherin alpha 13 |
| chrX_-_65253506 | 1.42 |
ENST00000427538.1
|
VSIG4
|
V-set and immunoglobulin domain containing 4 |
| chr17_-_61996136 | 1.38 |
ENST00000342364.4
|
GH1
|
growth hormone 1 |
| chr16_+_67927147 | 1.35 |
ENST00000291041.5
|
PSKH1
|
protein serine kinase H1 |
| chr6_+_131571535 | 1.34 |
ENST00000474850.2
|
AKAP7
|
A kinase (PRKA) anchor protein 7 |
| chr17_+_53344945 | 1.32 |
ENST00000575345.1
|
HLF
|
hepatic leukemia factor |
| chr18_+_39766626 | 1.32 |
ENST00000593234.1
ENST00000585627.1 ENST00000591199.1 ENST00000586990.1 ENST00000593051.1 ENST00000593316.1 ENST00000591381.1 ENST00000585639.1 ENST00000589068.1 |
LINC00907
|
long intergenic non-protein coding RNA 907 |
| chr2_+_29336855 | 1.30 |
ENST00000404424.1
|
CLIP4
|
CAP-GLY domain containing linker protein family, member 4 |
| chrX_-_55020511 | 1.28 |
ENST00000375006.3
ENST00000374992.2 |
PFKFB1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr6_+_31783291 | 1.27 |
ENST00000375651.5
ENST00000608703.1 ENST00000458062.2 |
HSPA1A
|
heat shock 70kDa protein 1A |
| chr6_-_33290580 | 1.26 |
ENST00000446511.1
ENST00000446403.1 ENST00000414083.2 ENST00000266000.6 ENST00000374542.5 |
DAXX
|
death-domain associated protein |
| chr1_+_196857144 | 1.25 |
ENST00000367416.2
ENST00000251424.4 ENST00000367418.2 |
CFHR4
|
complement factor H-related 4 |
| chr10_+_15085936 | 1.23 |
ENST00000378217.3
|
OLAH
|
oleoyl-ACP hydrolase |
| chr5_-_39270725 | 1.21 |
ENST00000512138.1
ENST00000512982.1 ENST00000540520.1 |
FYB
|
FYN binding protein |
| chr6_-_154677900 | 1.20 |
ENST00000265198.4
ENST00000520261.1 |
IPCEF1
|
interaction protein for cytohesin exchange factors 1 |
| chr2_+_69201705 | 1.19 |
ENST00000377938.2
|
GKN1
|
gastrokine 1 |
| chr2_+_143635067 | 1.19 |
ENST00000264170.4
|
KYNU
|
kynureninase |
| chr1_-_182921119 | 1.19 |
ENST00000423786.1
|
SHCBP1L
|
SHC SH2-domain binding protein 1-like |
| chr19_-_14952689 | 1.17 |
ENST00000248058.1
|
OR7A10
|
olfactory receptor, family 7, subfamily A, member 10 |
| chr14_+_21785693 | 1.17 |
ENST00000382933.4
ENST00000557351.1 |
RPGRIP1
|
retinitis pigmentosa GTPase regulator interacting protein 1 |
| chr14_-_82000140 | 1.14 |
ENST00000555824.1
ENST00000557372.1 ENST00000336735.4 |
SEL1L
|
sel-1 suppressor of lin-12-like (C. elegans) |
| chr2_-_109605663 | 1.12 |
ENST00000409271.1
ENST00000258443.2 ENST00000376651.1 |
EDAR
|
ectodysplasin A receptor |
| chr15_-_63674034 | 1.11 |
ENST00000344366.3
ENST00000422263.2 |
CA12
|
carbonic anhydrase XII |
| chr15_+_41245160 | 1.09 |
ENST00000444189.2
ENST00000446533.3 |
CHAC1
|
ChaC, cation transport regulator homolog 1 (E. coli) |
| chr15_+_100106126 | 1.07 |
ENST00000558812.1
ENST00000338042.6 |
MEF2A
|
myocyte enhancer factor 2A |
| chr10_+_53806501 | 1.06 |
ENST00000373975.2
|
PRKG1
|
protein kinase, cGMP-dependent, type I |
| chr11_+_30252549 | 1.06 |
ENST00000254122.3
ENST00000417547.1 |
FSHB
|
follicle stimulating hormone, beta polypeptide |
| chr18_+_21032781 | 1.03 |
ENST00000339486.3
|
RIOK3
|
RIO kinase 3 |
| chr15_+_100106155 | 0.95 |
ENST00000557785.1
ENST00000558049.1 ENST00000449277.2 |
MEF2A
|
myocyte enhancer factor 2A |
| chr11_+_118754475 | 0.92 |
ENST00000292174.4
|
CXCR5
|
chemokine (C-X-C motif) receptor 5 |
| chr6_-_90529418 | 0.91 |
ENST00000439638.1
ENST00000369393.3 ENST00000428876.1 |
MDN1
|
MDN1, midasin homolog (yeast) |
| chr22_+_23412479 | 0.90 |
ENST00000248996.4
|
GNAZ
|
guanine nucleotide binding protein (G protein), alpha z polypeptide |
| chr5_-_75919253 | 0.90 |
ENST00000296641.4
|
F2RL2
|
coagulation factor II (thrombin) receptor-like 2 |
| chr1_+_150229554 | 0.87 |
ENST00000369111.4
|
CA14
|
carbonic anhydrase XIV |
| chr10_+_15085895 | 0.85 |
ENST00000378228.3
|
OLAH
|
oleoyl-ACP hydrolase |
| chr15_-_34628951 | 0.83 |
ENST00000397707.2
ENST00000560611.1 |
SLC12A6
|
solute carrier family 12 (potassium/chloride transporter), member 6 |
| chr6_-_29395509 | 0.81 |
ENST00000377147.2
|
OR11A1
|
olfactory receptor, family 11, subfamily A, member 1 |
| chrX_+_11777671 | 0.81 |
ENST00000380693.3
ENST00000380692.2 |
MSL3
|
male-specific lethal 3 homolog (Drosophila) |
| chr4_+_71570430 | 0.79 |
ENST00000417478.2
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr14_+_39735411 | 0.78 |
ENST00000603904.1
|
RP11-407N17.3
|
cTAGE family member 5 isoform 4 |
| chr3_+_46283916 | 0.78 |
ENST00000395940.2
|
CCR3
|
chemokine (C-C motif) receptor 3 |
| chr3_-_137851220 | 0.77 |
ENST00000236709.3
|
A4GNT
|
alpha-1,4-N-acetylglucosaminyltransferase |
| chr12_-_54582655 | 0.69 |
ENST00000504338.1
ENST00000514685.1 ENST00000504797.1 ENST00000513838.1 ENST00000505128.1 ENST00000337581.3 ENST00000503306.1 ENST00000243112.5 ENST00000514196.1 ENST00000506169.1 ENST00000507904.1 ENST00000508394.2 |
SMUG1
|
single-strand-selective monofunctional uracil-DNA glycosylase 1 |
| chr19_-_14992264 | 0.68 |
ENST00000327462.2
|
OR7A17
|
olfactory receptor, family 7, subfamily A, member 17 |
| chr5_-_75919217 | 0.67 |
ENST00000504899.1
|
F2RL2
|
coagulation factor II (thrombin) receptor-like 2 |
| chr1_+_104159999 | 0.67 |
ENST00000414303.2
ENST00000423678.1 |
AMY2A
|
amylase, alpha 2A (pancreatic) |
| chr7_+_136553370 | 0.66 |
ENST00000445907.2
|
CHRM2
|
cholinergic receptor, muscarinic 2 |
| chr4_-_48782259 | 0.63 |
ENST00000507711.1
ENST00000358350.4 ENST00000537810.1 ENST00000264319.7 |
FRYL
|
FRY-like |
| chr4_-_83769996 | 0.62 |
ENST00000511338.1
|
SEC31A
|
SEC31 homolog A (S. cerevisiae) |
| chr8_-_18666360 | 0.60 |
ENST00000286485.8
|
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chr3_+_108541545 | 0.59 |
ENST00000295756.6
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1 |
| chr18_-_3874752 | 0.54 |
ENST00000534970.1
|
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1 |
| chr3_+_108541608 | 0.53 |
ENST00000426646.1
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1 |
| chr10_+_103911926 | 0.53 |
ENST00000605788.1
ENST00000405356.1 |
NOLC1
|
nucleolar and coiled-body phosphoprotein 1 |
| chr14_-_61748550 | 0.52 |
ENST00000555868.1
|
TMEM30B
|
transmembrane protein 30B |
| chr9_-_95244781 | 0.52 |
ENST00000375544.3
ENST00000375543.1 ENST00000395538.3 ENST00000450139.2 |
ASPN
|
asporin |
| chr1_+_196743943 | 0.50 |
ENST00000471440.2
ENST00000391985.3 |
CFHR3
|
complement factor H-related 3 |
| chr15_+_100106244 | 0.49 |
ENST00000557942.1
|
MEF2A
|
myocyte enhancer factor 2A |
| chr1_+_57320437 | 0.49 |
ENST00000361249.3
|
C8A
|
complement component 8, alpha polypeptide |
| chr2_+_191792376 | 0.48 |
ENST00000409428.1
ENST00000409215.1 |
GLS
|
glutaminase |
| chr7_-_81399411 | 0.48 |
ENST00000423064.2
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr3_-_183145873 | 0.47 |
ENST00000447025.2
ENST00000414362.2 ENST00000328913.3 |
MCF2L2
|
MCF.2 cell line derived transforming sequence-like 2 |
| chr1_+_79115503 | 0.46 |
ENST00000370747.4
ENST00000438486.1 ENST00000545124.1 |
IFI44
|
interferon-induced protein 44 |
| chr17_+_41323204 | 0.42 |
ENST00000542611.1
ENST00000590996.1 ENST00000389312.4 ENST00000589872.1 |
NBR1
|
neighbor of BRCA1 gene 1 |
| chr16_-_87466740 | 0.40 |
ENST00000561928.1
|
ZCCHC14
|
zinc finger, CCHC domain containing 14 |
| chr3_-_183145765 | 0.40 |
ENST00000473233.1
|
MCF2L2
|
MCF.2 cell line derived transforming sequence-like 2 |
| chr6_+_153019023 | 0.39 |
ENST00000367245.5
ENST00000529453.1 |
MYCT1
|
myc target 1 |
| chr17_-_72772462 | 0.37 |
ENST00000582870.1
ENST00000581136.1 ENST00000357814.3 ENST00000579218.1 ENST00000583476.1 ENST00000580301.1 ENST00000583757.1 ENST00000582524.1 |
NAT9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chrX_+_1733876 | 0.37 |
ENST00000381241.3
|
ASMT
|
acetylserotonin O-methyltransferase |
| chr3_+_46283863 | 0.35 |
ENST00000545097.1
ENST00000541018.1 |
CCR3
|
chemokine (C-C motif) receptor 3 |
| chr3_-_178984759 | 0.34 |
ENST00000349697.2
ENST00000497599.1 |
KCNMB3
|
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr2_+_201997676 | 0.31 |
ENST00000462763.1
ENST00000479953.2 |
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr12_-_68619586 | 0.31 |
ENST00000229134.4
|
IL26
|
interleukin 26 |
| chr2_-_136594740 | 0.30 |
ENST00000264162.2
|
LCT
|
lactase |
| chrX_+_1734051 | 0.29 |
ENST00000381229.4
ENST00000381233.3 |
ASMT
|
acetylserotonin O-methyltransferase |
| chr3_-_182817297 | 0.29 |
ENST00000539926.1
ENST00000476176.1 |
MCCC1
|
methylcrotonoyl-CoA carboxylase 1 (alpha) |
| chr16_-_28503357 | 0.27 |
ENST00000333496.9
ENST00000561505.1 ENST00000567963.1 ENST00000354630.5 ENST00000355477.5 ENST00000357076.5 ENST00000565688.1 ENST00000359984.7 |
CLN3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr5_-_58652788 | 0.17 |
ENST00000405755.2
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr11_+_124824000 | 0.17 |
ENST00000529051.1
ENST00000344762.5 |
CCDC15
|
coiled-coil domain containing 15 |
| chr3_+_118905564 | 0.14 |
ENST00000460625.1
|
UPK1B
|
uroplakin 1B |
| chr12_+_75874580 | 0.13 |
ENST00000456650.3
|
GLIPR1
|
GLI pathogenesis-related 1 |
| chr6_-_55739542 | 0.11 |
ENST00000446683.2
|
BMP5
|
bone morphogenetic protein 5 |
| chr4_+_110834033 | 0.10 |
ENST00000509793.1
ENST00000265171.5 |
EGF
|
epidermal growth factor |
| chr15_-_63674218 | 0.08 |
ENST00000178638.3
|
CA12
|
carbonic anhydrase XII |
| chr3_+_155860751 | 0.08 |
ENST00000471742.1
|
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr1_+_171454639 | 0.05 |
ENST00000392078.3
ENST00000426496.2 |
PRRC2C
|
proline-rich coiled-coil 2C |
| chr2_+_166150541 | 0.02 |
ENST00000283256.6
|
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit |
| chr6_+_126102292 | 0.01 |
ENST00000368357.3
|
NCOA7
|
nuclear receptor coactivator 7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of GATA5
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 14.1 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 1.3 | 3.8 | GO:0035915 | pore formation in membrane of other organism(GO:0035915) |
| 1.2 | 3.7 | GO:1902769 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 1.2 | 11.8 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.9 | 2.7 | GO:1901624 | negative regulation of lymphocyte chemotaxis(GO:1901624) |
| 0.8 | 3.0 | GO:0032690 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.8 | 10.6 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.7 | 2.8 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) actin polymerization-dependent cell motility(GO:0070358) |
| 0.7 | 2.8 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.5 | 5.2 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.5 | 7.6 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.5 | 2.3 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.4 | 3.4 | GO:0010536 | positive regulation of activation of Janus kinase activity(GO:0010536) |
| 0.4 | 2.5 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.4 | 2.4 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.4 | 1.2 | GO:0097052 | tryptophan catabolic process to acetyl-CoA(GO:0019442) L-kynurenine metabolic process(GO:0097052) |
| 0.4 | 1.5 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.3 | 3.8 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.3 | 1.3 | GO:0070434 | positive regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070426) positive regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070434) |
| 0.3 | 1.3 | GO:1903935 | response to sodium arsenite(GO:1903935) cellular response to sodium arsenite(GO:1903936) |
| 0.3 | 2.7 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.3 | 1.9 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.3 | 1.0 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.2 | 3.7 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.2 | 0.7 | GO:0030186 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.2 | 1.3 | GO:0033133 | fructose 2,6-bisphosphate metabolic process(GO:0006003) positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.2 | 5.1 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.2 | 1.2 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.2 | 1.7 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |
| 0.2 | 2.4 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.2 | 4.0 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.2 | 0.5 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.2 | 0.8 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.2 | 10.2 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.2 | 3.1 | GO:0020027 | hemoglobin metabolic process(GO:0020027) |
| 0.2 | 3.4 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.2 | 1.1 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.1 | 1.1 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.1 | 1.0 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.1 | 0.7 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.1 | 14.2 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.1 | 2.9 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.1 | 2.7 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.1 | 7.3 | GO:0002260 | lymphocyte homeostasis(GO:0002260) |
| 0.1 | 0.5 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.1 | 2.3 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.1 | 1.1 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.1 | 0.9 | GO:0048535 | lymph node development(GO:0048535) |
| 0.1 | 10.2 | GO:0002819 | regulation of adaptive immune response(GO:0002819) |
| 0.1 | 1.2 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 1.9 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.1 | 2.1 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.1 | 1.5 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 5.9 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.1 | 8.2 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.1 | 1.1 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.1 | 1.6 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.1 | 1.1 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.1 | 0.3 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.1 | 0.5 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.1 | 1.5 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.1 | 3.2 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 1.3 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 0.7 | GO:0045008 | depyrimidination(GO:0045008) |
| 0.1 | 4.0 | GO:0034113 | heterotypic cell-cell adhesion(GO:0034113) |
| 0.1 | 0.1 | GO:0021502 | neural fold elevation formation(GO:0021502) allantois development(GO:1905069) |
| 0.1 | 0.5 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 3.5 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 1.1 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 1.7 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 0.6 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.8 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 1.2 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.6 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.3 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 1.1 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.8 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 2.5 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.9 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.2 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 1.3 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 1.4 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 1.6 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 0.9 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.5 | GO:0099536 | chemical synaptic transmission(GO:0007268) anterograde trans-synaptic signaling(GO:0098916) synaptic signaling(GO:0099536) trans-synaptic signaling(GO:0099537) |
| 0.0 | 1.3 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.0 | 0.8 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.9 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 | 0.5 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 28.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 1.1 | 3.4 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.9 | 11.8 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.8 | 2.3 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.6 | 0.6 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.5 | 3.7 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.5 | 1.9 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.4 | 7.6 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.2 | 3.7 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.2 | 1.1 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.2 | 3.1 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.2 | 2.8 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.2 | 0.7 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.1 | 1.2 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.1 | 1.0 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 1.9 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.1 | 0.8 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 1.7 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 10.2 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.1 | 15.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 18.9 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 4.0 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 2.4 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.1 | 0.3 | GO:0002169 | 3-methylcrotonyl-CoA carboxylase complex, mitochondrial(GO:0002169) methylcrotonoyl-CoA carboxylase complex(GO:1905202) |
| 0.0 | 0.7 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 6.1 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 1.1 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.9 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 1.2 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 2.2 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 2.3 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 5.8 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 1.9 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 1.6 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 1.3 | GO:0070603 | SWI/SNF superfamily-type complex(GO:0070603) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.4 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 1.0 | 5.2 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.8 | 2.4 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.7 | 2.8 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.7 | 2.1 | GO:0016297 | oleoyl-[acyl-carrier-protein] hydrolase activity(GO:0004320) myristoyl-[acyl-carrier-protein] hydrolase activity(GO:0016295) palmitoyl-[acyl-carrier-protein] hydrolase activity(GO:0016296) acyl-[acyl-carrier-protein] hydrolase activity(GO:0016297) |
| 0.6 | 3.0 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.6 | 17.2 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.5 | 2.7 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.5 | 3.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.5 | 6.5 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.5 | 2.7 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.4 | 2.3 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.4 | 2.5 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.3 | 3.8 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.3 | 2.4 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.3 | 1.2 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.3 | 1.6 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.3 | 1.3 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.2 | 3.7 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.2 | 1.1 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.2 | 1.3 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.2 | 0.7 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.2 | 4.5 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.2 | 0.7 | GO:0016160 | amylase activity(GO:0016160) |
| 0.2 | 2.3 | GO:0016594 | glycine binding(GO:0016594) |
| 0.1 | 2.8 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 2.4 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.7 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 0.9 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.1 | 1.7 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.1 | 2.1 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 6.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 1.2 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 11.8 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.1 | 3.7 | GO:0001848 | complement binding(GO:0001848) |
| 0.1 | 1.1 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 0.3 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.1 | 0.5 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 1.9 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 1.0 | GO:0089720 | caspase binding(GO:0089720) |
| 0.1 | 9.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.7 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.1 | 0.9 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 0.8 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 5.9 | GO:0001190 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.1 | 3.2 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 1.3 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.1 | 0.3 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.1 | 1.5 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 1.3 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 1.1 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.5 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 5.2 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 2.3 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 4.0 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 5.9 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 1.7 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 7.5 | GO:0051020 | GTPase binding(GO:0051020) |
| 0.0 | 2.9 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.0 | 0.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 5.9 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.5 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 1.2 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.8 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.8 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 17.8 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.2 | 9.2 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 1.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 3.2 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 8.5 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.1 | 2.3 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 1.6 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 2.7 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 0.9 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 3.0 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 1.7 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 10.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 2.1 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 2.4 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 1.1 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 6.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 4.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.2 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.5 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 1.3 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.5 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 3.9 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.1 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 23.7 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.7 | 11.8 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.4 | 7.5 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.4 | 10.6 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.3 | 3.8 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.2 | 3.4 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 2.1 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 2.4 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 3.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 14.4 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.1 | 2.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 1.6 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.1 | 1.0 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 1.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 2.9 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 1.1 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.1 | 2.5 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 2.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 3.2 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 0.9 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.1 | 3.0 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 1.1 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 1.6 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 1.4 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 1.2 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.7 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.5 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 6.9 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 1.3 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 2.0 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 2.3 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.5 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |