Project
averaged GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
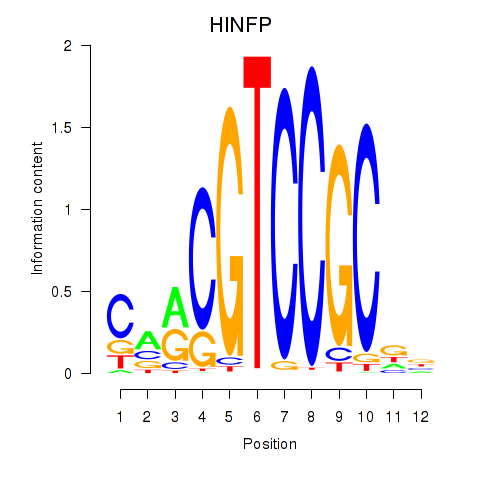
Results for HINFP
Z-value: 0.74
Transcription factors associated with HINFP
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HINFP
|
ENSG00000172273.8 | histone H4 transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HINFP | hg19_v2_chr11_+_118992269_118992334 | 0.05 | 4.5e-01 | Click! |
Activity profile of HINFP motif
Sorted Z-values of HINFP motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_94023873 | 13.67 |
ENST00000297268.6
|
COL1A2
|
collagen, type I, alpha 2 |
| chr8_-_27462822 | 8.97 |
ENST00000522098.1
|
CLU
|
clusterin |
| chr5_-_136834982 | 8.58 |
ENST00000510689.1
ENST00000394945.1 |
SPOCK1
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1 |
| chr1_+_6845384 | 6.60 |
ENST00000303635.7
|
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr11_-_66336060 | 6.53 |
ENST00000310325.5
|
CTSF
|
cathepsin F |
| chr9_+_130965651 | 6.49 |
ENST00000475805.1
ENST00000341179.7 ENST00000372923.3 |
DNM1
|
dynamin 1 |
| chr9_+_130965677 | 6.33 |
ENST00000393594.3
ENST00000486160.1 |
DNM1
|
dynamin 1 |
| chr20_-_35492048 | 6.22 |
ENST00000237536.4
|
SOGA1
|
suppressor of glucose, autophagy associated 1 |
| chr4_-_492891 | 5.91 |
ENST00000338977.5
ENST00000511833.2 |
ZNF721
|
zinc finger protein 721 |
| chr20_-_32031680 | 5.83 |
ENST00000217381.2
|
SNTA1
|
syntrophin, alpha 1 |
| chr15_+_91643442 | 5.81 |
ENST00000394232.1
|
SV2B
|
synaptic vesicle glycoprotein 2B |
| chr3_-_13009168 | 5.80 |
ENST00000273221.4
|
IQSEC1
|
IQ motif and Sec7 domain 1 |
| chr13_-_77460525 | 5.69 |
ENST00000377474.2
ENST00000317765.2 |
KCTD12
|
potassium channel tetramerization domain containing 12 |
| chr4_+_166128735 | 5.57 |
ENST00000226725.6
|
KLHL2
|
kelch-like family member 2 |
| chr1_+_204797749 | 5.47 |
ENST00000367172.4
ENST00000367171.4 ENST00000367170.4 ENST00000338515.6 ENST00000339876.6 ENST00000338586.6 ENST00000539706.1 ENST00000360049.4 ENST00000367169.4 ENST00000446412.1 ENST00000403080.1 |
NFASC
|
neurofascin |
| chr12_+_53773944 | 5.39 |
ENST00000551969.1
ENST00000327443.4 |
SP1
|
Sp1 transcription factor |
| chr10_-_131909071 | 4.86 |
ENST00000456581.1
|
LINC00959
|
long intergenic non-protein coding RNA 959 |
| chr11_-_72353451 | 4.54 |
ENST00000376450.3
|
PDE2A
|
phosphodiesterase 2A, cGMP-stimulated |
| chr11_+_62104897 | 4.47 |
ENST00000415229.2
ENST00000535727.1 ENST00000301776.5 |
ASRGL1
|
asparaginase like 1 |
| chr15_+_43803143 | 4.40 |
ENST00000382031.1
|
MAP1A
|
microtubule-associated protein 1A |
| chr16_+_56225248 | 4.36 |
ENST00000262493.6
|
GNAO1
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O |
| chr19_-_49944806 | 4.32 |
ENST00000221485.3
|
SLC17A7
|
solute carrier family 17 (vesicular glutamate transporter), member 7 |
| chr3_-_16555150 | 4.26 |
ENST00000334133.4
|
RFTN1
|
raftlin, lipid raft linker 1 |
| chr16_+_6069586 | 4.11 |
ENST00000547372.1
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chrX_+_21392529 | 4.07 |
ENST00000425654.2
ENST00000543067.1 |
CNKSR2
|
connector enhancer of kinase suppressor of Ras 2 |
| chr17_+_43971643 | 4.07 |
ENST00000344290.5
ENST00000262410.5 ENST00000351559.5 ENST00000340799.5 ENST00000535772.1 ENST00000347967.5 |
MAPT
|
microtubule-associated protein tau |
| chr18_+_268148 | 4.01 |
ENST00000581677.1
|
RP11-705O1.8
|
RP11-705O1.8 |
| chr11_-_132813566 | 3.96 |
ENST00000331898.7
|
OPCML
|
opioid binding protein/cell adhesion molecule-like |
| chr17_+_43972010 | 3.87 |
ENST00000334239.8
ENST00000446361.3 |
MAPT
|
microtubule-associated protein tau |
| chr3_-_129612394 | 3.86 |
ENST00000505616.1
ENST00000426664.2 |
TMCC1
|
transmembrane and coiled-coil domain family 1 |
| chr13_-_36705425 | 3.86 |
ENST00000255448.4
ENST00000360631.3 ENST00000379892.4 |
DCLK1
|
doublecortin-like kinase 1 |
| chr9_+_100174344 | 3.78 |
ENST00000422139.2
|
TDRD7
|
tudor domain containing 7 |
| chr9_-_139922631 | 3.77 |
ENST00000341511.6
|
ABCA2
|
ATP-binding cassette, sub-family A (ABC1), member 2 |
| chrX_+_21392553 | 3.71 |
ENST00000279451.4
|
CNKSR2
|
connector enhancer of kinase suppressor of Ras 2 |
| chr11_-_6440624 | 3.58 |
ENST00000311051.3
|
APBB1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr11_-_6440283 | 3.56 |
ENST00000299402.6
ENST00000609360.1 ENST00000389906.2 ENST00000532020.2 |
APBB1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr17_-_42277203 | 3.48 |
ENST00000587097.1
|
ATXN7L3
|
ataxin 7-like 3 |
| chr8_-_9760839 | 3.44 |
ENST00000519461.1
ENST00000517675.1 |
LINC00599
|
long intergenic non-protein coding RNA 599 |
| chr20_+_30946106 | 3.42 |
ENST00000375687.4
ENST00000542461.1 |
ASXL1
|
additional sex combs like 1 (Drosophila) |
| chr2_-_97304009 | 3.38 |
ENST00000431828.1
ENST00000435669.1 ENST00000440133.1 ENST00000448075.1 |
KANSL3
|
KAT8 regulatory NSL complex subunit 3 |
| chr10_-_118502070 | 3.35 |
ENST00000369209.3
|
HSPA12A
|
heat shock 70kDa protein 12A |
| chr9_-_139922726 | 3.29 |
ENST00000265662.5
ENST00000371605.3 |
ABCA2
|
ATP-binding cassette, sub-family A (ABC1), member 2 |
| chr17_-_42402138 | 3.26 |
ENST00000592857.1
ENST00000586016.1 ENST00000590194.1 ENST00000377095.5 ENST00000588049.1 ENST00000586633.1 ENST00000537904.2 ENST00000585636.1 ENST00000585523.1 ENST00000225308.8 |
SLC25A39
|
solute carrier family 25, member 39 |
| chr2_-_97304105 | 3.26 |
ENST00000599854.1
ENST00000441706.2 |
KANSL3
|
KAT8 regulatory NSL complex subunit 3 |
| chr19_+_56186606 | 3.19 |
ENST00000085079.7
|
EPN1
|
epsin 1 |
| chr22_+_18593446 | 3.13 |
ENST00000316027.6
|
TUBA8
|
tubulin, alpha 8 |
| chr6_-_139695757 | 3.06 |
ENST00000367651.2
|
CITED2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
| chr22_-_22221658 | 2.98 |
ENST00000544786.1
|
MAPK1
|
mitogen-activated protein kinase 1 |
| chr6_+_42847348 | 2.97 |
ENST00000493763.1
ENST00000304734.5 |
RPL7L1
|
ribosomal protein L7-like 1 |
| chr9_-_139094988 | 2.96 |
ENST00000371746.3
|
LHX3
|
LIM homeobox 3 |
| chr19_-_14016877 | 2.87 |
ENST00000454313.1
ENST00000591586.1 ENST00000346736.2 |
C19orf57
|
chromosome 19 open reading frame 57 |
| chr7_-_17980091 | 2.86 |
ENST00000409389.1
ENST00000409604.1 ENST00000428135.3 |
SNX13
|
sorting nexin 13 |
| chr3_+_49449636 | 2.86 |
ENST00000273590.3
|
TCTA
|
T-cell leukemia translocation altered |
| chr9_+_139921916 | 2.84 |
ENST00000314330.2
|
C9orf139
|
chromosome 9 open reading frame 139 |
| chr1_-_92952433 | 2.76 |
ENST00000294702.5
|
GFI1
|
growth factor independent 1 transcription repressor |
| chr7_+_128095900 | 2.69 |
ENST00000435296.2
|
HILPDA
|
hypoxia inducible lipid droplet-associated |
| chr13_+_111806055 | 2.66 |
ENST00000218789.5
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr9_+_137533615 | 2.62 |
ENST00000371817.3
|
COL5A1
|
collagen, type V, alpha 1 |
| chr9_-_122131696 | 2.58 |
ENST00000373964.2
ENST00000265922.3 |
BRINP1
|
bone morphogenetic protein/retinoic acid inducible neural-specific 1 |
| chr20_+_57267669 | 2.53 |
ENST00000356091.6
|
NPEPL1
|
aminopeptidase-like 1 |
| chr11_-_2162162 | 2.52 |
ENST00000381389.1
|
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chr13_+_21714653 | 2.49 |
ENST00000382533.4
|
SAP18
|
Sin3A-associated protein, 18kDa |
| chr16_-_2185899 | 2.49 |
ENST00000262304.4
ENST00000423118.1 |
PKD1
|
polycystic kidney disease 1 (autosomal dominant) |
| chrX_-_38186811 | 2.48 |
ENST00000318842.7
|
RPGR
|
retinitis pigmentosa GTPase regulator |
| chr2_+_157292859 | 2.47 |
ENST00000438166.2
|
GPD2
|
glycerol-3-phosphate dehydrogenase 2 (mitochondrial) |
| chr22_-_28197486 | 2.46 |
ENST00000302326.4
|
MN1
|
meningioma (disrupted in balanced translocation) 1 |
| chr11_-_2162468 | 2.45 |
ENST00000434045.2
|
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chr8_+_26240414 | 2.45 |
ENST00000380629.2
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like |
| chr4_+_123747979 | 2.44 |
ENST00000608478.1
|
FGF2
|
fibroblast growth factor 2 (basic) |
| chr6_+_35310391 | 2.41 |
ENST00000337400.2
ENST00000311565.4 ENST00000540939.1 |
PPARD
|
peroxisome proliferator-activated receptor delta |
| chr17_-_1420182 | 2.40 |
ENST00000421807.2
|
INPP5K
|
inositol polyphosphate-5-phosphatase K |
| chr11_+_3876859 | 2.39 |
ENST00000300737.4
|
STIM1
|
stromal interaction molecule 1 |
| chr19_-_16653226 | 2.34 |
ENST00000198939.6
|
CHERP
|
calcium homeostasis endoplasmic reticulum protein |
| chr13_+_21714711 | 2.28 |
ENST00000607003.1
ENST00000492245.1 |
SAP18
|
Sin3A-associated protein, 18kDa |
| chr9_-_89562104 | 2.27 |
ENST00000298743.7
|
GAS1
|
growth arrest-specific 1 |
| chr13_-_41240717 | 2.25 |
ENST00000379561.5
|
FOXO1
|
forkhead box O1 |
| chr6_+_42847649 | 2.21 |
ENST00000424341.2
ENST00000602561.1 |
RPL7L1
|
ribosomal protein L7-like 1 |
| chr11_-_73309228 | 2.19 |
ENST00000356467.4
ENST00000064778.4 |
FAM168A
|
family with sequence similarity 168, member A |
| chr19_+_45754505 | 2.18 |
ENST00000262891.4
ENST00000300843.4 |
MARK4
|
MAP/microtubule affinity-regulating kinase 4 |
| chr9_+_100174232 | 2.16 |
ENST00000355295.4
|
TDRD7
|
tudor domain containing 7 |
| chr6_-_33267101 | 2.14 |
ENST00000497454.1
|
RGL2
|
ral guanine nucleotide dissociation stimulator-like 2 |
| chr7_-_134143841 | 2.13 |
ENST00000285930.4
|
AKR1B1
|
aldo-keto reductase family 1, member B1 (aldose reductase) |
| chr9_+_37650945 | 2.11 |
ENST00000377765.3
|
FRMPD1
|
FERM and PDZ domain containing 1 |
| chr19_-_39523165 | 2.09 |
ENST00000509137.2
ENST00000292853.4 |
FBXO27
|
F-box protein 27 |
| chr22_-_22221900 | 2.08 |
ENST00000215832.6
ENST00000398822.3 |
MAPK1
|
mitogen-activated protein kinase 1 |
| chr3_-_49377499 | 2.08 |
ENST00000265560.4
|
USP4
|
ubiquitin specific peptidase 4 (proto-oncogene) |
| chr19_-_16653325 | 2.05 |
ENST00000546361.2
|
CHERP
|
calcium homeostasis endoplasmic reticulum protein |
| chr17_-_1420006 | 2.03 |
ENST00000320345.6
ENST00000406424.4 |
INPP5K
|
inositol polyphosphate-5-phosphatase K |
| chr3_-_49131473 | 2.02 |
ENST00000430979.1
ENST00000357496.2 ENST00000437939.1 |
QRICH1
|
glutamine-rich 1 |
| chr17_-_1419878 | 1.99 |
ENST00000449479.1
ENST00000477910.1 ENST00000542125.1 ENST00000575172.1 |
INPP5K
|
inositol polyphosphate-5-phosphatase K |
| chr19_-_45004556 | 1.98 |
ENST00000587047.1
ENST00000391956.4 ENST00000221327.4 ENST00000586637.1 ENST00000591064.1 ENST00000592529.1 |
ZNF180
|
zinc finger protein 180 |
| chr6_+_35310312 | 1.97 |
ENST00000448077.2
ENST00000360694.3 ENST00000418635.2 ENST00000444397.1 |
PPARD
|
peroxisome proliferator-activated receptor delta |
| chr4_+_123747834 | 1.95 |
ENST00000264498.3
|
FGF2
|
fibroblast growth factor 2 (basic) |
| chr17_-_4545170 | 1.92 |
ENST00000576394.1
ENST00000574640.1 |
ALOX15
|
arachidonate 15-lipoxygenase |
| chr11_-_1593150 | 1.92 |
ENST00000397374.3
|
DUSP8
|
dual specificity phosphatase 8 |
| chr7_+_148959262 | 1.91 |
ENST00000434415.1
|
ZNF783
|
zinc finger family member 783 |
| chr9_-_101017862 | 1.91 |
ENST00000375064.1
ENST00000342112.5 |
TBC1D2
|
TBC1 domain family, member 2 |
| chr5_-_132166579 | 1.91 |
ENST00000378679.3
|
SHROOM1
|
shroom family member 1 |
| chr11_+_1411129 | 1.88 |
ENST00000308219.9
ENST00000528841.1 ENST00000531197.1 ENST00000308230.5 |
BRSK2
|
BR serine/threonine kinase 2 |
| chr3_-_48936272 | 1.87 |
ENST00000544097.1
ENST00000430379.1 ENST00000319017.4 |
SLC25A20
|
solute carrier family 25 (carnitine/acylcarnitine translocase), member 20 |
| chr12_+_7282795 | 1.86 |
ENST00000266546.6
|
CLSTN3
|
calsyntenin 3 |
| chr17_+_29158962 | 1.85 |
ENST00000321990.4
|
ATAD5
|
ATPase family, AAA domain containing 5 |
| chr5_-_127873496 | 1.85 |
ENST00000508989.1
|
FBN2
|
fibrillin 2 |
| chr8_-_144241664 | 1.85 |
ENST00000342752.4
|
LY6H
|
lymphocyte antigen 6 complex, locus H |
| chr11_+_119076745 | 1.80 |
ENST00000264033.4
|
CBL
|
Cbl proto-oncogene, E3 ubiquitin protein ligase |
| chr17_-_1419914 | 1.79 |
ENST00000397335.3
ENST00000574561.1 |
INPP5K
|
inositol polyphosphate-5-phosphatase K |
| chr6_+_116692102 | 1.78 |
ENST00000359564.2
|
DSE
|
dermatan sulfate epimerase |
| chrX_-_38186775 | 1.75 |
ENST00000339363.3
ENST00000309513.3 ENST00000338898.3 ENST00000342811.3 ENST00000378505.2 |
RPGR
|
retinitis pigmentosa GTPase regulator |
| chr5_-_132073210 | 1.73 |
ENST00000378735.1
ENST00000378746.4 |
KIF3A
|
kinesin family member 3A |
| chr8_-_144241432 | 1.71 |
ENST00000430474.2
|
LY6H
|
lymphocyte antigen 6 complex, locus H |
| chr17_-_46115122 | 1.69 |
ENST00000006101.4
|
COPZ2
|
coatomer protein complex, subunit zeta 2 |
| chr6_+_44355257 | 1.67 |
ENST00000371477.3
|
CDC5L
|
cell division cycle 5-like |
| chr12_+_69633317 | 1.65 |
ENST00000435070.2
|
CPSF6
|
cleavage and polyadenylation specific factor 6, 68kDa |
| chr17_-_4544960 | 1.65 |
ENST00000293761.3
|
ALOX15
|
arachidonate 15-lipoxygenase |
| chr13_+_111806083 | 1.64 |
ENST00000375736.4
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr3_-_49377446 | 1.63 |
ENST00000351842.4
ENST00000416417.1 ENST00000415188.1 |
USP4
|
ubiquitin specific peptidase 4 (proto-oncogene) |
| chr2_-_73053126 | 1.62 |
ENST00000272427.6
ENST00000410104.1 |
EXOC6B
|
exocyst complex component 6B |
| chr1_-_40157345 | 1.62 |
ENST00000372844.3
|
HPCAL4
|
hippocalcin like 4 |
| chr18_-_268019 | 1.57 |
ENST00000261600.6
|
THOC1
|
THO complex 1 |
| chr3_+_110790590 | 1.57 |
ENST00000485303.1
|
PVRL3
|
poliovirus receptor-related 3 |
| chr2_-_175547571 | 1.57 |
ENST00000409415.3
ENST00000359761.3 ENST00000272746.5 |
WIPF1
|
WAS/WASL interacting protein family, member 1 |
| chr22_-_43010928 | 1.57 |
ENST00000348657.2
ENST00000252115.5 |
POLDIP3
|
polymerase (DNA-directed), delta interacting protein 3 |
| chr19_+_36545833 | 1.56 |
ENST00000401500.2
ENST00000270301.7 ENST00000427823.2 |
WDR62
|
WD repeat domain 62 |
| chrX_-_53711064 | 1.56 |
ENST00000342160.3
ENST00000446750.1 |
HUWE1
|
HECT, UBA and WWE domain containing 1, E3 ubiquitin protein ligase |
| chr22_+_32870661 | 1.55 |
ENST00000266087.7
|
FBXO7
|
F-box protein 7 |
| chr1_-_212208842 | 1.54 |
ENST00000366992.3
ENST00000366993.3 ENST00000440600.2 ENST00000366994.3 |
INTS7
|
integrator complex subunit 7 |
| chr4_-_149363662 | 1.54 |
ENST00000355292.3
ENST00000358102.3 |
NR3C2
|
nuclear receptor subfamily 3, group C, member 2 |
| chr11_+_28131821 | 1.54 |
ENST00000379199.2
ENST00000303459.6 |
METTL15
|
methyltransferase like 15 |
| chr12_-_90102594 | 1.53 |
ENST00000428670.3
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr17_+_74380683 | 1.52 |
ENST00000592299.1
ENST00000590959.1 ENST00000323374.4 |
SPHK1
|
sphingosine kinase 1 |
| chrX_-_119695279 | 1.49 |
ENST00000336592.6
|
CUL4B
|
cullin 4B |
| chr6_-_89673280 | 1.47 |
ENST00000369475.3
ENST00000369485.4 ENST00000538899.1 ENST00000265607.6 |
RNGTT
|
RNA guanylyltransferase and 5'-phosphatase |
| chr10_-_104179682 | 1.47 |
ENST00000406432.1
|
PSD
|
pleckstrin and Sec7 domain containing |
| chr8_+_32405785 | 1.46 |
ENST00000287842.3
|
NRG1
|
neuregulin 1 |
| chr11_+_1411503 | 1.43 |
ENST00000526678.1
|
BRSK2
|
BR serine/threonine kinase 2 |
| chr22_+_18593507 | 1.41 |
ENST00000330423.3
|
TUBA8
|
tubulin, alpha 8 |
| chr3_-_156272924 | 1.40 |
ENST00000467789.1
ENST00000265044.2 |
SSR3
|
signal sequence receptor, gamma (translocon-associated protein gamma) |
| chr19_-_14117074 | 1.40 |
ENST00000588885.1
ENST00000254325.4 |
RFX1
|
regulatory factor X, 1 (influences HLA class II expression) |
| chr12_-_117175819 | 1.38 |
ENST00000261318.3
ENST00000536380.1 |
C12orf49
|
chromosome 12 open reading frame 49 |
| chr5_-_132073111 | 1.37 |
ENST00000403231.1
|
KIF3A
|
kinesin family member 3A |
| chr22_+_17082732 | 1.35 |
ENST00000558085.2
ENST00000592918.1 ENST00000400593.2 ENST00000592107.1 ENST00000426585.1 ENST00000591299.1 |
TPTEP1
|
transmembrane phosphatase with tensin homology pseudogene 1 |
| chr8_+_32406179 | 1.35 |
ENST00000405005.3
|
NRG1
|
neuregulin 1 |
| chr5_-_142782862 | 1.32 |
ENST00000415690.2
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr18_-_44336998 | 1.31 |
ENST00000315087.7
|
ST8SIA5
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 5 |
| chr19_+_45596218 | 1.31 |
ENST00000421905.1
ENST00000221462.4 |
PPP1R37
|
protein phosphatase 1, regulatory subunit 37 |
| chr13_+_25875662 | 1.27 |
ENST00000381736.3
ENST00000463407.1 ENST00000381718.3 |
NUPL1
|
nucleoporin like 1 |
| chr19_+_45596398 | 1.26 |
ENST00000544069.2
|
PPP1R37
|
protein phosphatase 1, regulatory subunit 37 |
| chr1_-_153919128 | 1.26 |
ENST00000361217.4
|
DENND4B
|
DENN/MADD domain containing 4B |
| chr16_+_84002234 | 1.25 |
ENST00000305202.4
|
NECAB2
|
N-terminal EF-hand calcium binding protein 2 |
| chr9_-_101017900 | 1.24 |
ENST00000375066.5
|
TBC1D2
|
TBC1 domain family, member 2 |
| chr1_+_22351977 | 1.23 |
ENST00000420503.1
ENST00000416769.1 ENST00000404210.2 |
LINC00339
|
long intergenic non-protein coding RNA 339 |
| chr15_-_35047166 | 1.23 |
ENST00000290374.4
|
GJD2
|
gap junction protein, delta 2, 36kDa |
| chr18_+_43914159 | 1.21 |
ENST00000588679.1
ENST00000269439.7 ENST00000543885.1 |
RNF165
|
ring finger protein 165 |
| chr2_+_26915584 | 1.19 |
ENST00000302909.3
|
KCNK3
|
potassium channel, subfamily K, member 3 |
| chr10_-_62761188 | 1.18 |
ENST00000357917.4
|
RHOBTB1
|
Rho-related BTB domain containing 1 |
| chr2_-_25564750 | 1.16 |
ENST00000321117.5
|
DNMT3A
|
DNA (cytosine-5-)-methyltransferase 3 alpha |
| chr20_-_62258394 | 1.16 |
ENST00000370077.1
|
GMEB2
|
glucocorticoid modulatory element binding protein 2 |
| chr6_-_144329531 | 1.15 |
ENST00000429150.1
ENST00000392309.1 ENST00000416623.1 ENST00000392307.1 |
PLAGL1
|
pleiomorphic adenoma gene-like 1 |
| chr2_+_120770581 | 1.13 |
ENST00000263713.5
|
EPB41L5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr2_+_120770645 | 1.11 |
ENST00000443902.2
|
EPB41L5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr15_+_101459420 | 1.10 |
ENST00000388948.3
ENST00000284395.5 ENST00000534045.1 ENST00000532029.2 |
LRRK1
|
leucine-rich repeat kinase 1 |
| chr17_-_7518145 | 1.10 |
ENST00000250113.7
ENST00000571597.1 |
FXR2
|
fragile X mental retardation, autosomal homolog 2 |
| chr6_+_146350622 | 1.07 |
ENST00000507907.1
|
GRM1
|
glutamate receptor, metabotropic 1 |
| chr2_+_10442993 | 1.05 |
ENST00000423674.1
ENST00000307845.3 |
HPCAL1
|
hippocalcin-like 1 |
| chr5_-_142783175 | 1.01 |
ENST00000231509.3
ENST00000394464.2 |
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr8_-_30670053 | 1.01 |
ENST00000518564.1
|
PPP2CB
|
protein phosphatase 2, catalytic subunit, beta isozyme |
| chr1_+_28995258 | 1.00 |
ENST00000361872.4
ENST00000294409.2 |
GMEB1
|
glucocorticoid modulatory element binding protein 1 |
| chr1_-_16563641 | 0.98 |
ENST00000375599.3
|
RSG1
|
REM2 and RAB-like small GTPase 1 |
| chr9_+_34458771 | 0.95 |
ENST00000437363.1
ENST00000242317.4 |
DNAI1
|
dynein, axonemal, intermediate chain 1 |
| chr5_+_113697983 | 0.95 |
ENST00000264773.3
|
KCNN2
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2 |
| chr1_-_200379129 | 0.94 |
ENST00000367353.1
|
ZNF281
|
zinc finger protein 281 |
| chr17_+_17206635 | 0.91 |
ENST00000389022.4
|
NT5M
|
5',3'-nucleotidase, mitochondrial |
| chr12_+_57482665 | 0.90 |
ENST00000300131.3
|
NAB2
|
NGFI-A binding protein 2 (EGR1 binding protein 2) |
| chr8_+_42995548 | 0.89 |
ENST00000458501.2
ENST00000379644.4 |
HGSNAT
|
heparan-alpha-glucosaminide N-acetyltransferase |
| chr17_-_66453562 | 0.88 |
ENST00000262139.5
ENST00000546360.1 |
WIPI1
|
WD repeat domain, phosphoinositide interacting 1 |
| chr2_-_43453734 | 0.88 |
ENST00000282388.3
|
ZFP36L2
|
ZFP36 ring finger protein-like 2 |
| chr17_+_74381343 | 0.85 |
ENST00000392496.3
|
SPHK1
|
sphingosine kinase 1 |
| chr8_+_32405728 | 0.85 |
ENST00000523079.1
ENST00000338921.4 ENST00000356819.4 ENST00000287845.5 ENST00000341377.5 |
NRG1
|
neuregulin 1 |
| chr1_-_51796226 | 0.83 |
ENST00000451380.1
ENST00000371747.3 ENST00000439482.1 |
TTC39A
|
tetratricopeptide repeat domain 39A |
| chr3_-_38691119 | 0.81 |
ENST00000333535.4
ENST00000413689.1 ENST00000443581.1 ENST00000425664.1 ENST00000451551.2 |
SCN5A
|
sodium channel, voltage-gated, type V, alpha subunit |
| chr3_+_152552685 | 0.77 |
ENST00000305097.3
|
P2RY1
|
purinergic receptor P2Y, G-protein coupled, 1 |
| chr6_-_35464817 | 0.77 |
ENST00000338863.7
|
TEAD3
|
TEA domain family member 3 |
| chr1_-_92351769 | 0.77 |
ENST00000212355.4
|
TGFBR3
|
transforming growth factor, beta receptor III |
| chr19_+_14017116 | 0.77 |
ENST00000589606.1
|
CC2D1A
|
coiled-coil and C2 domain containing 1A |
| chr12_-_125052010 | 0.76 |
ENST00000458234.1
|
NCOR2
|
nuclear receptor corepressor 2 |
| chr12_+_113659234 | 0.76 |
ENST00000551096.1
ENST00000551099.1 ENST00000335509.6 ENST00000552897.1 ENST00000550785.1 ENST00000549279.1 |
TPCN1
|
two pore segment channel 1 |
| chr13_+_25875785 | 0.75 |
ENST00000381747.3
|
NUPL1
|
nucleoporin like 1 |
| chr17_-_42276574 | 0.75 |
ENST00000589805.1
|
ATXN7L3
|
ataxin 7-like 3 |
| chr17_+_46125707 | 0.72 |
ENST00000584137.1
ENST00000362042.3 ENST00000585291.1 ENST00000357480.5 |
NFE2L1
|
nuclear factor, erythroid 2-like 1 |
| chr7_-_140624499 | 0.71 |
ENST00000288602.6
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B |
| chr5_-_114598548 | 0.69 |
ENST00000379615.3
ENST00000419445.1 |
PGGT1B
|
protein geranylgeranyltransferase type I, beta subunit |
| chr6_-_35464727 | 0.67 |
ENST00000402886.3
|
TEAD3
|
TEA domain family member 3 |
| chr5_-_141257954 | 0.67 |
ENST00000456271.1
ENST00000394536.3 ENST00000503492.1 ENST00000287008.3 |
PCDH1
|
protocadherin 1 |
| chr22_-_19132154 | 0.66 |
ENST00000252137.6
|
DGCR14
|
DiGeorge syndrome critical region gene 14 |
| chr19_-_15543368 | 0.65 |
ENST00000599686.3
|
WIZ
|
widely interspaced zinc finger motifs |
| chr19_-_460996 | 0.65 |
ENST00000264554.6
|
SHC2
|
SHC (Src homology 2 domain containing) transforming protein 2 |
| chr4_+_154387480 | 0.64 |
ENST00000409663.3
ENST00000440693.1 ENST00000409959.3 |
KIAA0922
|
KIAA0922 |
| chr20_+_3451650 | 0.63 |
ENST00000262919.5
|
ATRN
|
attractin |
| chr20_+_3451732 | 0.60 |
ENST00000446916.2
|
ATRN
|
attractin |
| chr14_-_92572894 | 0.56 |
ENST00000532032.1
ENST00000506466.1 ENST00000555381.1 ENST00000557311.1 ENST00000554592.1 ENST00000554672.1 ENST00000553491.1 ENST00000556220.1 ENST00000502250.1 ENST00000503767.1 ENST00000393287.5 ENST00000340660.6 ENST00000545170.1 ENST00000429774.2 |
ATXN3
|
ataxin 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HINFP
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.3 | 12.8 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 2.4 | 7.1 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 2.1 | 8.2 | GO:2001151 | regulation of renal water transport(GO:2001151) positive regulation of renal water transport(GO:2001153) |
| 1.7 | 5.1 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 1.6 | 7.9 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 1.5 | 4.5 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 1.5 | 9.0 | GO:0061518 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 1.4 | 4.3 | GO:0042137 | sequestering of neurotransmitter(GO:0042137) |
| 1.4 | 7.1 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 1.2 | 3.6 | GO:2001303 | cellular response to interleukin-13(GO:0035963) lipoxin biosynthetic process(GO:2001301) lipoxin A4 metabolic process(GO:2001302) lipoxin A4 biosynthetic process(GO:2001303) |
| 1.1 | 3.4 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 1.1 | 2.2 | GO:0045739 | positive regulation of DNA repair(GO:0045739) |
| 0.9 | 2.8 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.9 | 5.5 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.9 | 13.7 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.8 | 2.5 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.8 | 2.4 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.8 | 2.4 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.8 | 3.1 | GO:0061428 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.7 | 4.5 | GO:1900239 | phenotypic switching(GO:0036166) regulation of phenotypic switching(GO:1900239) |
| 0.7 | 2.2 | GO:0003383 | apical constriction(GO:0003383) mesoderm migration involved in gastrulation(GO:0007509) |
| 0.7 | 2.9 | GO:0072674 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.7 | 2.1 | GO:0006059 | hexitol metabolic process(GO:0006059) response to methylglyoxal(GO:0051595) |
| 0.7 | 2.6 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) |
| 0.6 | 4.4 | GO:1904849 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.6 | 4.4 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.6 | 2.5 | GO:0072237 | metanephric proximal tubule development(GO:0072237) |
| 0.6 | 4.3 | GO:0032596 | protein transport within lipid bilayer(GO:0032594) protein transport into membrane raft(GO:0032596) |
| 0.6 | 2.3 | GO:1902617 | response to fluoride(GO:1902617) |
| 0.6 | 4.5 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.6 | 1.7 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.6 | 5.0 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.5 | 3.5 | GO:0038129 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.5 | 3.0 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.5 | 2.5 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.5 | 4.4 | GO:0032966 | negative regulation of collagen biosynthetic process(GO:0032966) |
| 0.4 | 6.6 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.4 | 0.4 | GO:0086100 | endothelin receptor signaling pathway(GO:0086100) |
| 0.4 | 5.2 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.4 | 4.3 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.4 | 1.3 | GO:0060168 | positive regulation of adenosine receptor signaling pathway(GO:0060168) |
| 0.4 | 6.2 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.4 | 1.6 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.4 | 2.4 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.4 | 1.6 | GO:0045829 | negative regulation of isotype switching(GO:0045829) |
| 0.4 | 5.8 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.4 | 7.3 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.3 | 1.9 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.3 | 3.1 | GO:0072386 | plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.3 | 1.5 | GO:1901660 | calcium ion export(GO:1901660) calcium ion export from cell(GO:1990034) |
| 0.3 | 0.9 | GO:1902949 | positive regulation of tau-protein kinase activity(GO:1902949) |
| 0.3 | 0.9 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.3 | 2.3 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.3 | 1.0 | GO:1990868 | response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) |
| 0.3 | 1.8 | GO:0090649 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.3 | 0.8 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.2 | 1.4 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.2 | 3.3 | GO:1904152 | regulation of retrograde protein transport, ER to cytosol(GO:1904152) |
| 0.2 | 5.9 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.2 | 1.5 | GO:0098501 | polynucleotide dephosphorylation(GO:0098501) |
| 0.2 | 1.3 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.2 | 1.8 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.2 | 1.2 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.2 | 1.5 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.2 | 2.1 | GO:1903599 | negative regulation of oxidative stress-induced neuron death(GO:1903204) positive regulation of mitophagy(GO:1903599) |
| 0.2 | 0.7 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.2 | 0.7 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.2 | 0.8 | GO:0086047 | membrane depolarization during Purkinje myocyte cell action potential(GO:0086047) |
| 0.2 | 0.8 | GO:0060406 | positive regulation of penile erection(GO:0060406) |
| 0.2 | 2.0 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.1 | 0.9 | GO:1904627 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.1 | 2.6 | GO:0042711 | maternal behavior(GO:0042711) parental behavior(GO:0060746) |
| 0.1 | 2.3 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.1 | 0.6 | GO:1904379 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.1 | 4.3 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.1 | 1.1 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.1 | 3.9 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.1 | 1.5 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.1 | 0.8 | GO:0072364 | regulation of cellular ketone metabolic process by regulation of transcription from RNA polymerase II promoter(GO:0072364) |
| 0.1 | 1.4 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 1.9 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.1 | 0.5 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.1 | 4.4 | GO:0000226 | microtubule cytoskeleton organization(GO:0000226) |
| 0.1 | 0.7 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.1 | 7.5 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.1 | 6.6 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.1 | 8.6 | GO:0010812 | negative regulation of cell-substrate adhesion(GO:0010812) |
| 0.1 | 1.9 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.1 | 1.5 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.1 | 4.4 | GO:0014072 | response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.1 | 1.5 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.1 | 1.3 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 | 3.9 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.1 | 0.9 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 1.6 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 1.8 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.1 | 1.9 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.1 | 2.1 | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.1 | 0.9 | GO:0009125 | nucleoside monophosphate catabolic process(GO:0009125) |
| 0.1 | 3.2 | GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) |
| 0.1 | 1.1 | GO:0036035 | osteoclast development(GO:0036035) |
| 0.1 | 4.1 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.1 | 1.3 | GO:0097503 | sialylation(GO:0097503) |
| 0.0 | 1.9 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 1.6 | GO:0007099 | centriole replication(GO:0007099) |
| 0.0 | 2.2 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 1.2 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 1.1 | GO:1902402 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) signal transduction involved in cell cycle checkpoint(GO:0072395) signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in mitotic cell cycle checkpoint(GO:0072413) signal transduction involved in DNA damage checkpoint(GO:0072422) signal transduction involved in mitotic G1 DNA damage checkpoint(GO:0072431) intracellular signal transduction involved in G1 DNA damage checkpoint(GO:1902400) signal transduction involved in mitotic DNA damage checkpoint(GO:1902402) signal transduction involved in mitotic DNA integrity checkpoint(GO:1902403) |
| 0.0 | 0.4 | GO:0007625 | grooming behavior(GO:0007625) |
| 0.0 | 2.6 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 1.6 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.0 | 1.6 | GO:0007417 | central nervous system development(GO:0007417) |
| 0.0 | 0.7 | GO:0007399 | nervous system development(GO:0007399) |
| 0.0 | 5.8 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 0.1 | GO:0048213 | Golgi vesicle prefusion complex stabilization(GO:0048213) |
| 0.0 | 0.2 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
| 0.0 | 0.4 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 1.0 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) response to antibiotic(GO:0046677) negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.0 | 4.2 | GO:0007601 | visual perception(GO:0007601) sensory perception of light stimulus(GO:0050953) |
| 0.0 | 1.6 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 1.2 | GO:0040014 | regulation of multicellular organism growth(GO:0040014) |
| 0.0 | 2.0 | GO:0022604 | regulation of cell morphogenesis(GO:0022604) |
| 0.0 | 0.6 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.2 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.9 | GO:0007041 | lysosomal transport(GO:0007041) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 7.6 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 1.6 | 7.9 | GO:0045298 | tubulin complex(GO:0045298) |
| 1.4 | 16.3 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.8 | 7.1 | GO:1990812 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.8 | 3.1 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.7 | 9.0 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.6 | 5.8 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.6 | 7.1 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.5 | 4.3 | GO:0060203 | clathrin-sculpted glutamate transport vesicle(GO:0060199) clathrin-sculpted glutamate transport vesicle membrane(GO:0060203) |
| 0.5 | 5.9 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.5 | 4.3 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.5 | 4.3 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.4 | 2.5 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.4 | 2.4 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.3 | 2.1 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.3 | 8.4 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.3 | 4.2 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.3 | 1.1 | GO:0038038 | G-protein coupled receptor homodimeric complex(GO:0038038) |
| 0.3 | 0.8 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.2 | 1.5 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.2 | 12.8 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.2 | 0.7 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.2 | 1.6 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.2 | 1.6 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.2 | 1.5 | GO:0032039 | integrator complex(GO:0032039) |
| 0.2 | 1.7 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.2 | 5.1 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 1.5 | GO:0043235 | receptor complex(GO:0043235) |
| 0.1 | 1.8 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.1 | 0.9 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 3.5 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 10.2 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 1.7 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 0.8 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 4.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 7.4 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.1 | 1.5 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 1.8 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 5.6 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 1.2 | GO:0001741 | XY body(GO:0001741) |
| 0.1 | 5.8 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 2.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 2.5 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.1 | 1.2 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 7.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 13.3 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 6.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 14.2 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.1 | 2.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 3.9 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 6.2 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 2.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 3.1 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 5.0 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 4.6 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.6 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 1.9 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 0.8 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 12.1 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 2.9 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 1.6 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 1.6 | GO:0034774 | secretory granule lumen(GO:0034774) |
| 0.0 | 1.2 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 4.3 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 1.0 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.9 | GO:0070821 | tertiary granule membrane(GO:0070821) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 7.9 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 1.5 | 4.5 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 1.4 | 5.7 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 1.4 | 8.2 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 1.2 | 3.6 | GO:0047977 | hepoxilin-epoxide hydrolase activity(GO:0047977) |
| 1.2 | 16.3 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 1.1 | 4.3 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.8 | 4.5 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.6 | 1.9 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.6 | 2.5 | GO:0052590 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.6 | 1.8 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.6 | 6.6 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.5 | 1.5 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.5 | 1.5 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.5 | 2.3 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.4 | 9.0 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.4 | 5.5 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.4 | 7.1 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.4 | 1.7 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.4 | 4.4 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.4 | 2.4 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.3 | 3.5 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.3 | 8.6 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.3 | 6.6 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.3 | 1.8 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.3 | 5.5 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.3 | 1.0 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.2 | 3.4 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.2 | 5.1 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.2 | 2.1 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.2 | 3.1 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.2 | 0.7 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.2 | 1.8 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.2 | 0.7 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.2 | 1.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.2 | 4.4 | GO:0036041 | long-chain fatty acid binding(GO:0036041) |
| 0.2 | 0.9 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.2 | 4.5 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.2 | 1.3 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.2 | 4.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 0.4 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.1 | 1.4 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 3.7 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 1.1 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.1 | 5.3 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 1.9 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 2.4 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 1.6 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.8 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.1 | 0.9 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 1.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 4.3 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.1 | 2.3 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 3.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 5.9 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 1.5 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 2.3 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 7.1 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.1 | 0.6 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 0.9 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 2.6 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 0.9 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 6.7 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 2.8 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 5.2 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 2.6 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 0.4 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.8 | GO:0015278 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 1.0 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 7.7 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 4.1 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.5 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 6.7 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 8.9 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 1.3 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 2.5 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.7 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.9 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 3.1 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.9 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.3 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.7 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.7 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 13.8 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.5 | 10.4 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.3 | 16.3 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.3 | 12.8 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.2 | 2.4 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.2 | 7.6 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 7.9 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 8.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 8.2 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 7.3 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 5.8 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 5.1 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 2.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 12.9 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 2.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 7.1 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 5.0 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 1.5 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 1.6 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 2.3 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 3.2 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 3.6 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 2.4 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 1.6 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.7 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 1.9 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 2.0 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.8 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.6 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 6.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.4 | PID ENDOTHELIN PATHWAY | Endothelins |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 13.7 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.7 | 12.8 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.4 | 6.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.4 | 7.1 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.3 | 4.4 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.2 | 7.9 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 2.4 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.2 | 1.4 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.2 | 5.0 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 8.2 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 2.5 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.1 | 3.5 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 3.2 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 4.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 2.6 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 4.5 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 2.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 2.7 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 6.8 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 1.2 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 4.4 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.1 | 0.9 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 9.0 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 1.9 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 1.1 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 2.0 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 1.0 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 2.0 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 1.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.7 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 2.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.8 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 1.5 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 1.8 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.0 | 0.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.5 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.8 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.9 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |