Project
averaged GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
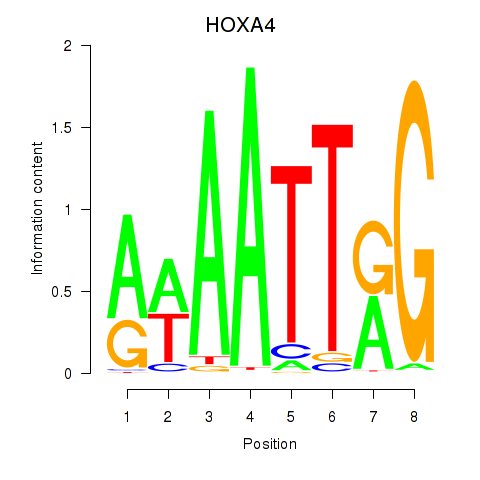
Results for HOXA4
Z-value: 0.63
Transcription factors associated with HOXA4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXA4
|
ENSG00000197576.9 | homeobox A4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXA4 | hg19_v2_chr7_-_27170352_27170418 | -0.19 | 4.8e-03 | Click! |
Activity profile of HOXA4 motif
Sorted Z-values of HOXA4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_50144381 | 8.94 |
ENST00000552370.1
|
TMBIM6
|
transmembrane BAX inhibitor motif containing 6 |
| chr15_+_75080883 | 8.64 |
ENST00000567571.1
|
CSK
|
c-src tyrosine kinase |
| chr5_+_49962495 | 7.69 |
ENST00000515175.1
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr13_-_46716969 | 7.05 |
ENST00000435666.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr3_-_52486841 | 6.84 |
ENST00000496590.1
|
TNNC1
|
troponin C type 1 (slow) |
| chr17_-_56082455 | 6.37 |
ENST00000578794.1
|
RP11-159D12.5
|
Uncharacterized protein |
| chr1_+_155658849 | 5.84 |
ENST00000368336.5
ENST00000343043.3 ENST00000421487.2 ENST00000535183.1 ENST00000465375.1 ENST00000470830.1 |
DAP3
|
death associated protein 3 |
| chr19_-_42806919 | 5.80 |
ENST00000595530.1
ENST00000538771.1 ENST00000601865.1 |
PAFAH1B3
|
platelet-activating factor acetylhydrolase 1b, catalytic subunit 3 (29kDa) |
| chr12_+_56521840 | 5.67 |
ENST00000394048.5
|
ESYT1
|
extended synaptotagmin-like protein 1 |
| chr6_+_63921399 | 5.28 |
ENST00000356170.3
|
FKBP1C
|
FK506 binding protein 1C |
| chr15_+_80351910 | 5.12 |
ENST00000261749.6
ENST00000561060.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr13_-_41593425 | 4.78 |
ENST00000239882.3
|
ELF1
|
E74-like factor 1 (ets domain transcription factor) |
| chr18_+_3247779 | 4.26 |
ENST00000578611.1
ENST00000583449.1 |
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr15_+_80351977 | 4.18 |
ENST00000559157.1
ENST00000561012.1 ENST00000564367.1 ENST00000558494.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr12_+_53848549 | 4.10 |
ENST00000439930.3
ENST00000548933.1 ENST00000562264.1 |
PCBP2
|
poly(rC) binding protein 2 |
| chr14_-_36988882 | 3.79 |
ENST00000498187.2
|
NKX2-1
|
NK2 homeobox 1 |
| chr6_+_63921351 | 3.73 |
ENST00000370659.1
|
FKBP1C
|
FK506 binding protein 1C |
| chr2_-_43453734 | 3.71 |
ENST00000282388.3
|
ZFP36L2
|
ZFP36 ring finger protein-like 2 |
| chr1_+_167298281 | 3.55 |
ENST00000367862.5
|
POU2F1
|
POU class 2 homeobox 1 |
| chr3_+_107241783 | 3.47 |
ENST00000415149.2
ENST00000402543.1 ENST00000325805.8 ENST00000427402.1 |
BBX
|
bobby sox homolog (Drosophila) |
| chr11_-_62313090 | 3.17 |
ENST00000528508.1
ENST00000533365.1 |
AHNAK
|
AHNAK nucleoprotein |
| chr6_-_55739542 | 3.14 |
ENST00000446683.2
|
BMP5
|
bone morphogenetic protein 5 |
| chr15_+_89631647 | 3.04 |
ENST00000569550.1
ENST00000565066.1 ENST00000565973.1 |
ABHD2
|
abhydrolase domain containing 2 |
| chr12_+_53848505 | 2.97 |
ENST00000552819.1
ENST00000455667.3 |
PCBP2
|
poly(rC) binding protein 2 |
| chr22_+_22930626 | 2.90 |
ENST00000390302.2
|
IGLV2-33
|
immunoglobulin lambda variable 2-33 (non-functional) |
| chr14_-_38064198 | 2.87 |
ENST00000250448.2
|
FOXA1
|
forkhead box A1 |
| chr7_-_140714430 | 2.84 |
ENST00000393008.3
|
MRPS33
|
mitochondrial ribosomal protein S33 |
| chr11_+_128563652 | 2.84 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr8_+_22424551 | 2.81 |
ENST00000523348.1
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr17_-_10452929 | 2.80 |
ENST00000532183.2
ENST00000397183.2 ENST00000420805.1 |
MYH2
|
myosin, heavy chain 2, skeletal muscle, adult |
| chr17_-_48785216 | 2.79 |
ENST00000285243.6
|
ANKRD40
|
ankyrin repeat domain 40 |
| chr7_-_29234802 | 2.72 |
ENST00000449801.1
ENST00000409850.1 |
CPVL
|
carboxypeptidase, vitellogenic-like |
| chr14_-_36989336 | 2.68 |
ENST00000522719.2
|
NKX2-1
|
NK2 homeobox 1 |
| chr8_-_95449155 | 2.57 |
ENST00000481490.2
|
FSBP
|
fibrinogen silencer binding protein |
| chr11_-_69633792 | 2.51 |
ENST00000334134.2
|
FGF3
|
fibroblast growth factor 3 |
| chr1_+_81771806 | 2.51 |
ENST00000370721.1
ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2
|
latrophilin 2 |
| chr12_-_57037284 | 2.47 |
ENST00000551570.1
|
ATP5B
|
ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide |
| chr10_+_11047259 | 2.35 |
ENST00000379261.4
ENST00000416382.2 |
CELF2
|
CUGBP, Elav-like family member 2 |
| chr4_-_41884620 | 2.23 |
ENST00000504870.1
|
LINC00682
|
long intergenic non-protein coding RNA 682 |
| chr11_-_27494279 | 2.19 |
ENST00000379214.4
|
LGR4
|
leucine-rich repeat containing G protein-coupled receptor 4 |
| chr1_+_214161272 | 2.04 |
ENST00000498508.2
ENST00000366958.4 |
PROX1
|
prospero homeobox 1 |
| chr10_-_101825151 | 2.02 |
ENST00000441382.1
|
CPN1
|
carboxypeptidase N, polypeptide 1 |
| chr17_-_77924627 | 1.95 |
ENST00000572862.1
ENST00000573782.1 ENST00000574427.1 ENST00000570373.1 ENST00000340848.7 ENST00000576768.1 |
TBC1D16
|
TBC1 domain family, member 16 |
| chr19_-_51220176 | 1.88 |
ENST00000359082.3
ENST00000293441.1 |
SHANK1
|
SH3 and multiple ankyrin repeat domains 1 |
| chr10_-_105845674 | 1.85 |
ENST00000353479.5
ENST00000369733.3 |
COL17A1
|
collagen, type XVII, alpha 1 |
| chr17_-_10421853 | 1.76 |
ENST00000226207.5
|
MYH1
|
myosin, heavy chain 1, skeletal muscle, adult |
| chr7_+_100273736 | 1.70 |
ENST00000412215.1
ENST00000393924.1 |
GNB2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chrX_-_119445306 | 1.68 |
ENST00000371369.4
ENST00000440464.1 ENST00000519908.1 |
TMEM255A
|
transmembrane protein 255A |
| chr12_-_86650077 | 1.66 |
ENST00000552808.2
ENST00000547225.1 |
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr2_-_145278475 | 1.66 |
ENST00000558170.2
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr2_+_87135076 | 1.62 |
ENST00000409776.2
|
RGPD1
|
RANBP2-like and GRIP domain containing 1 |
| chr11_+_112832133 | 1.58 |
ENST00000524665.1
|
NCAM1
|
neural cell adhesion molecule 1 |
| chr10_-_92681033 | 1.56 |
ENST00000371697.3
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle) |
| chr3_+_69134080 | 1.52 |
ENST00000273258.3
|
ARL6IP5
|
ADP-ribosylation-like factor 6 interacting protein 5 |
| chr5_+_49962772 | 1.51 |
ENST00000281631.5
ENST00000513738.1 ENST00000503665.1 ENST00000514067.2 ENST00000503046.1 |
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr11_-_13517565 | 1.42 |
ENST00000282091.1
ENST00000529816.1 |
PTH
|
parathyroid hormone |
| chrX_-_119445263 | 1.41 |
ENST00000309720.5
|
TMEM255A
|
transmembrane protein 255A |
| chr17_-_10372875 | 1.39 |
ENST00000255381.2
|
MYH4
|
myosin, heavy chain 4, skeletal muscle |
| chr3_+_88188254 | 1.38 |
ENST00000309495.5
|
ZNF654
|
zinc finger protein 654 |
| chr12_-_24103954 | 1.37 |
ENST00000441133.2
ENST00000545921.1 |
SOX5
|
SRY (sex determining region Y)-box 5 |
| chr1_-_42384343 | 1.37 |
ENST00000372584.1
|
HIVEP3
|
human immunodeficiency virus type I enhancer binding protein 3 |
| chr7_+_32996997 | 1.34 |
ENST00000242209.4
ENST00000538336.1 ENST00000538443.1 |
FKBP9
|
FK506 binding protein 9, 63 kDa |
| chr4_-_66536057 | 1.29 |
ENST00000273854.3
|
EPHA5
|
EPH receptor A5 |
| chr12_+_29376592 | 1.25 |
ENST00000182377.4
|
FAR2
|
fatty acyl CoA reductase 2 |
| chr15_+_89631381 | 1.24 |
ENST00000352732.5
|
ABHD2
|
abhydrolase domain containing 2 |
| chr12_-_91398796 | 1.23 |
ENST00000261172.3
ENST00000551767.1 |
EPYC
|
epiphycan |
| chr11_-_27494309 | 1.23 |
ENST00000389858.4
|
LGR4
|
leucine-rich repeat containing G protein-coupled receptor 4 |
| chr12_+_29376673 | 1.17 |
ENST00000547116.1
|
FAR2
|
fatty acyl CoA reductase 2 |
| chr6_-_55740352 | 1.12 |
ENST00000370830.3
|
BMP5
|
bone morphogenetic protein 5 |
| chr7_+_129015484 | 1.09 |
ENST00000490911.1
|
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chr3_-_57233966 | 1.08 |
ENST00000473921.1
ENST00000295934.3 |
HESX1
|
HESX homeobox 1 |
| chr20_+_32150140 | 1.08 |
ENST00000344201.3
ENST00000346541.3 ENST00000397800.1 ENST00000397798.2 ENST00000492345.1 |
CBFA2T2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2 |
| chr17_+_37894570 | 1.06 |
ENST00000394211.3
|
GRB7
|
growth factor receptor-bound protein 7 |
| chr17_-_39538550 | 1.06 |
ENST00000394001.1
|
KRT34
|
keratin 34 |
| chr3_+_69134124 | 1.02 |
ENST00000478935.1
|
ARL6IP5
|
ADP-ribosylation-like factor 6 interacting protein 5 |
| chr14_-_54423529 | 1.01 |
ENST00000245451.4
ENST00000559087.1 |
BMP4
|
bone morphogenetic protein 4 |
| chr2_+_128403439 | 0.99 |
ENST00000544369.1
|
GPR17
|
G protein-coupled receptor 17 |
| chr2_+_101437487 | 0.96 |
ENST00000427413.1
ENST00000542504.1 |
NPAS2
|
neuronal PAS domain protein 2 |
| chr11_+_128563948 | 0.94 |
ENST00000534087.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr17_-_10325261 | 0.93 |
ENST00000403437.2
|
MYH8
|
myosin, heavy chain 8, skeletal muscle, perinatal |
| chr4_-_66536196 | 0.88 |
ENST00000511294.1
|
EPHA5
|
EPH receptor A5 |
| chr2_+_54683419 | 0.87 |
ENST00000356805.4
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1 |
| chr12_-_86650045 | 0.87 |
ENST00000604798.1
|
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr2_-_190927447 | 0.81 |
ENST00000260950.4
|
MSTN
|
myostatin |
| chr7_-_14026063 | 0.73 |
ENST00000443608.1
ENST00000438956.1 |
ETV1
|
ets variant 1 |
| chr1_-_204329013 | 0.70 |
ENST00000272203.3
ENST00000414478.1 |
PLEKHA6
|
pleckstrin homology domain containing, family A member 6 |
| chr2_-_163100045 | 0.69 |
ENST00000188790.4
|
FAP
|
fibroblast activation protein, alpha |
| chr5_+_159343688 | 0.69 |
ENST00000306675.3
|
ADRA1B
|
adrenoceptor alpha 1B |
| chr3_+_121311966 | 0.65 |
ENST00000338040.4
|
FBXO40
|
F-box protein 40 |
| chr1_+_151739131 | 0.65 |
ENST00000400999.1
|
OAZ3
|
ornithine decarboxylase antizyme 3 |
| chr9_-_113342160 | 0.62 |
ENST00000401783.2
ENST00000374461.1 |
SVEP1
|
sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 |
| chr7_-_14026123 | 0.56 |
ENST00000420159.2
ENST00000399357.3 ENST00000403527.1 |
ETV1
|
ets variant 1 |
| chr14_+_39734482 | 0.55 |
ENST00000554392.1
ENST00000555716.1 ENST00000341749.3 ENST00000557038.1 |
CTAGE5
|
CTAGE family, member 5 |
| chr7_+_107110488 | 0.55 |
ENST00000304402.4
|
GPR22
|
G protein-coupled receptor 22 |
| chr2_-_163099885 | 0.53 |
ENST00000443424.1
|
FAP
|
fibroblast activation protein, alpha |
| chr6_+_152011628 | 0.52 |
ENST00000404742.1
ENST00000440973.1 |
ESR1
|
estrogen receptor 1 |
| chr2_+_128403720 | 0.52 |
ENST00000272644.3
|
GPR17
|
G protein-coupled receptor 17 |
| chr12_-_10959892 | 0.51 |
ENST00000240615.2
|
TAS2R8
|
taste receptor, type 2, member 8 |
| chr14_+_37131058 | 0.48 |
ENST00000361487.6
|
PAX9
|
paired box 9 |
| chr8_-_99954788 | 0.41 |
ENST00000523601.1
|
STK3
|
serine/threonine kinase 3 |
| chr1_+_160160346 | 0.38 |
ENST00000368078.3
|
CASQ1
|
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr6_+_50786414 | 0.37 |
ENST00000344788.3
ENST00000393655.3 ENST00000263046.4 |
TFAP2B
|
transcription factor AP-2 beta (activating enhancer binding protein 2 beta) |
| chr10_-_101690650 | 0.36 |
ENST00000543621.1
|
DNMBP
|
dynamin binding protein |
| chr10_-_33623310 | 0.33 |
ENST00000395995.1
ENST00000374823.5 ENST00000374821.5 ENST00000374816.3 |
NRP1
|
neuropilin 1 |
| chr11_+_120971882 | 0.24 |
ENST00000392793.1
|
TECTA
|
tectorin alpha |
| chr6_-_116447283 | 0.24 |
ENST00000452729.1
ENST00000243222.4 |
COL10A1
|
collagen, type X, alpha 1 |
| chr10_-_69455873 | 0.23 |
ENST00000433211.2
|
CTNNA3
|
catenin (cadherin-associated protein), alpha 3 |
| chr7_+_117251671 | 0.18 |
ENST00000468795.1
|
CFTR
|
cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7) |
| chr1_-_12677714 | 0.14 |
ENST00000376223.2
|
DHRS3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr12_+_53818855 | 0.13 |
ENST00000550839.1
|
AMHR2
|
anti-Mullerian hormone receptor, type II |
| chr17_+_73452545 | 0.07 |
ENST00000314256.7
|
KIAA0195
|
KIAA0195 |
| chr6_-_26235206 | 0.06 |
ENST00000244534.5
|
HIST1H1D
|
histone cluster 1, H1d |
| chr5_-_1882858 | 0.06 |
ENST00000511126.1
ENST00000231357.2 |
IRX4
|
iroquois homeobox 4 |
| chr2_-_69098566 | 0.05 |
ENST00000295379.1
|
BMP10
|
bone morphogenetic protein 10 |
| chr7_+_95115210 | 0.03 |
ENST00000428113.1
ENST00000325885.5 |
ASB4
|
ankyrin repeat and SOCS box containing 4 |
| chr1_+_160160283 | 0.02 |
ENST00000368079.3
|
CASQ1
|
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr8_-_67090825 | 0.01 |
ENST00000276571.3
|
CRH
|
corticotropin releasing hormone |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXA4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 8.6 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 1.7 | 6.8 | GO:0032972 | diaphragm contraction(GO:0002086) regulation of muscle filament sliding speed(GO:0032972) |
| 1.5 | 8.9 | GO:0060701 | negative regulation of mRNA cleavage(GO:0031438) negative regulation of immunoglobulin secretion(GO:0051025) negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 1.4 | 4.3 | GO:0071676 | negative regulation of mononuclear cell migration(GO:0071676) |
| 1.3 | 6.5 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.9 | 3.4 | GO:0060995 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.8 | 2.4 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.7 | 2.9 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.7 | 2.0 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.6 | 1.9 | GO:0050894 | determination of affect(GO:0050894) |
| 0.5 | 3.7 | GO:1904627 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.5 | 1.4 | GO:1900158 | positive regulation of osteoclast proliferation(GO:0090290) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.4 | 7.1 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.4 | 7.1 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.4 | 1.7 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.4 | 2.0 | GO:0030070 | insulin processing(GO:0030070) |
| 0.4 | 9.3 | GO:0072663 | protein targeting to peroxisome(GO:0006625) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.3 | 1.0 | GO:0072099 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm development(GO:0048389) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) regulation of transcription from RNA polymerase II promoter involved in kidney development(GO:0060994) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) pattern specification involved in mesonephros development(GO:0061227) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in kidney development(GO:0072098) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.3 | 2.5 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.3 | 2.5 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.3 | 1.2 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 0.2 | 2.8 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.2 | 1.4 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.2 | 0.9 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.2 | 10.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.2 | 0.8 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) negative regulation of skeletal muscle cell proliferation(GO:0014859) regulation of skeletal muscle tissue growth(GO:0048631) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.2 | 4.8 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.2 | 3.8 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.2 | 0.7 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.2 | 4.5 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.2 | 1.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 0.6 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 | 8.7 | GO:0070126 | mitochondrial translational elongation(GO:0070125) mitochondrial translational termination(GO:0070126) |
| 0.1 | 1.5 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.1 | 1.1 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.1 | 2.2 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.1 | 1.6 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.1 | 2.5 | GO:0055026 | negative regulation of cardiac muscle tissue development(GO:0055026) |
| 0.1 | 0.4 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.1 | 0.4 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.1 | 0.5 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) prostate gland morphogenetic growth(GO:0060737) |
| 0.1 | 0.3 | GO:0061552 | vestibulocochlear nerve structural organization(GO:0021649) neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) positive regulation of cytokine activity(GO:0060301) ganglion morphogenesis(GO:0061552) endothelial tip cell fate specification(GO:0097102) positive regulation of retinal ganglion cell axon guidance(GO:1902336) VEGF-activated neuropilin signaling pathway involved in axon guidance(GO:1902378) dorsal root ganglion morphogenesis(GO:1904835) otic placode development(GO:1905040) |
| 0.1 | 1.8 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 1.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 2.3 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 1.0 | GO:0051775 | response to redox state(GO:0051775) |
| 0.1 | 3.2 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.1 | 5.7 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.1 | 1.6 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.1 | 0.6 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 4.3 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 1.9 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.0 | 0.4 | GO:0072501 | cellular divalent inorganic anion homeostasis(GO:0072501) |
| 0.0 | 3.6 | GO:0042795 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 1.7 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.0 | 0.1 | GO:1990262 | regulation of anti-Mullerian hormone signaling pathway(GO:1902612) negative regulation of anti-Mullerian hormone signaling pathway(GO:1902613) anti-Mullerian hormone signaling pathway(GO:1990262) |
| 0.0 | 1.1 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 2.3 | GO:0033275 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 0.2 | GO:0086073 | cardiac muscle cell-cardiac muscle cell adhesion(GO:0086042) bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 0.0 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 0.0 | 0.5 | GO:0042481 | regulation of odontogenesis(GO:0042481) |
| 0.0 | 2.9 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.1 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.0 | GO:0042321 | negative regulation of epinephrine secretion(GO:0032811) negative regulation of circadian sleep/wake cycle, sleep(GO:0042321) positive regulation of corticotropin secretion(GO:0051461) |
| 0.0 | 0.1 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.3 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 1.6 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.0 | 5.8 | GO:0048232 | spermatogenesis(GO:0007283) male gamete generation(GO:0048232) |
| 0.0 | 4.6 | GO:0006936 | muscle contraction(GO:0006936) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 6.8 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.9 | 2.8 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.4 | 5.7 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.4 | 7.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.4 | 2.5 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.3 | 4.3 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.3 | 8.7 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.2 | 4.1 | GO:0032982 | myosin filament(GO:0032982) |
| 0.1 | 1.9 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 0.9 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.1 | 1.2 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 2.4 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 1.8 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 4.3 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.4 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.0 | 0.5 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 7.5 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 2.9 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.2 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 1.3 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.3 | GO:0002116 | semaphorin receptor complex(GO:0002116) sorting endosome(GO:0097443) |
| 0.0 | 0.2 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.6 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 11.6 | GO:0030055 | cell-substrate adherens junction(GO:0005924) focal adhesion(GO:0005925) cell-substrate junction(GO:0030055) |
| 0.0 | 3.3 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.1 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 3.7 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.5 | GO:0031674 | I band(GO:0031674) |
| 0.0 | 1.1 | GO:0005882 | intermediate filament(GO:0005882) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 8.9 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 1.2 | 7.1 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 1.2 | 5.8 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.8 | 2.4 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.6 | 1.9 | GO:0031877 | somatostatin receptor binding(GO:0031877) |
| 0.6 | 6.8 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.5 | 1.6 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.4 | 6.5 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.4 | 2.5 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.4 | 5.3 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.4 | 2.5 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.4 | 10.4 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.3 | 8.6 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.3 | 4.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.3 | 4.7 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.3 | 9.2 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.3 | 2.0 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.2 | 3.7 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.2 | 0.7 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.2 | 2.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.2 | 9.3 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.2 | 1.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.2 | 3.2 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.2 | 0.9 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.1 | 0.6 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 4.2 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.1 | 2.8 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 2.5 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 0.5 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.1 | 1.4 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.1 | 1.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 0.2 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.1 | 2.3 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 8.7 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 3.9 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 1.5 | GO:0004950 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) |
| 0.0 | 0.3 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.0 | 0.1 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.0 | 1.7 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.9 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 6.6 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 1.0 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.9 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 1.3 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 1.6 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 1.7 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.0 | 1.9 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.4 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 1.1 | GO:0001071 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.8 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 9.3 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 8.6 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.1 | 3.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 2.2 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 2.8 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 5.3 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 2.5 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 2.1 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.0 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 1.6 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 8.6 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.1 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.9 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 8.6 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 2.5 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.1 | 7.1 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 7.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 2.5 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 2.5 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 3.6 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.1 | 1.5 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.1 | 1.7 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.1 | 2.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 0.9 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 2.8 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 1.3 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.0 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 2.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.3 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.7 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 1.4 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |