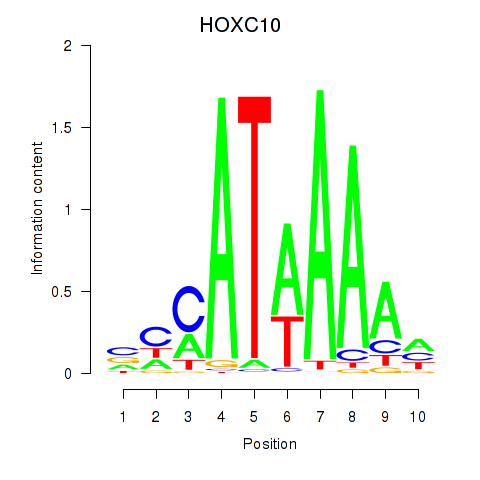
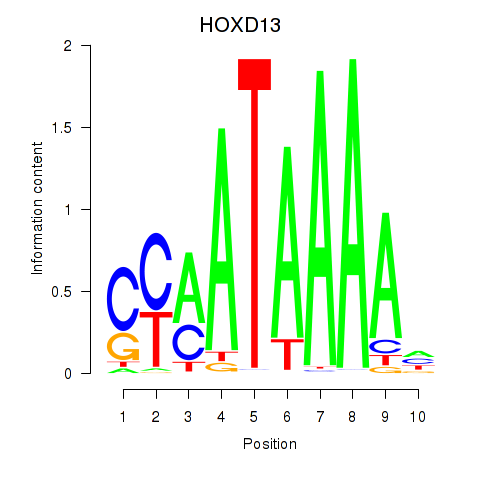
Project
averaged GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for HOXC10_HOXD13
Z-value: 1.93
Transcription factors associated with HOXC10_HOXD13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXC10
|
ENSG00000180818.4 | homeobox C10 |
|
HOXD13
|
ENSG00000128714.5 | homeobox D13 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXD13 | hg19_v2_chr2_+_176957619_176957619 | 0.39 | 1.5e-09 | Click! |
| HOXC10 | hg19_v2_chr12_+_54378923_54378970 | -0.15 | 2.8e-02 | Click! |
Activity profile of HOXC10_HOXD13 motif
Sorted Z-values of HOXC10_HOXD13 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXC10_HOXD13
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 16.4 | 196.4 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 9.5 | 28.5 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 6.3 | 18.9 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 5.3 | 15.9 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 4.7 | 23.7 | GO:0048478 | replication fork protection(GO:0048478) |
| 4.4 | 17.6 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 4.4 | 26.3 | GO:0051012 | microtubule sliding(GO:0051012) |
| 4.1 | 20.4 | GO:1904207 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 3.9 | 31.5 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 3.9 | 15.7 | GO:1904749 | regulation of protein localization to nucleolus(GO:1904749) |
| 3.3 | 9.9 | GO:1904395 | retinal rod cell differentiation(GO:0060221) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) negative regulation of neuromuscular junction development(GO:1904397) |
| 3.3 | 13.1 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 3.1 | 37.1 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 2.9 | 14.4 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 2.8 | 8.5 | GO:0086092 | regulation of the force of heart contraction by cardiac conduction(GO:0086092) regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) |
| 2.8 | 8.4 | GO:0021538 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 2.5 | 5.1 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 2.3 | 9.3 | GO:1902159 | regulation of inward rectifier potassium channel activity(GO:1901979) regulation of cyclic nucleotide-gated ion channel activity(GO:1902159) |
| 2.2 | 56.1 | GO:0001502 | cartilage condensation(GO:0001502) |
| 2.2 | 4.5 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 2.2 | 15.2 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 2.2 | 8.6 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 2.0 | 8.1 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 1.9 | 5.7 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 1.7 | 5.0 | GO:2000276 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 1.6 | 6.5 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 1.6 | 4.7 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 1.6 | 11.0 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 1.5 | 4.5 | GO:1990641 | response to iron ion starvation(GO:1990641) |
| 1.5 | 7.4 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 1.4 | 23.0 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 1.4 | 4.2 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 1.4 | 11.1 | GO:0019720 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 1.4 | 5.5 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 1.4 | 8.2 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 1.3 | 10.6 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 1.3 | 3.9 | GO:0002384 | hepatic immune response(GO:0002384) regulation of STAT protein import into nucleus(GO:2000364) positive regulation of STAT protein import into nucleus(GO:2000366) |
| 1.2 | 6.2 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 1.2 | 5.0 | GO:0007525 | somatic muscle development(GO:0007525) |
| 1.2 | 6.1 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 1.2 | 3.5 | GO:0014028 | mesoderm migration involved in gastrulation(GO:0007509) notochord formation(GO:0014028) |
| 1.1 | 5.7 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 1.1 | 28.7 | GO:0006825 | copper ion transport(GO:0006825) |
| 1.1 | 3.2 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 1.1 | 13.9 | GO:0050942 | positive regulation of pigment cell differentiation(GO:0050942) |
| 1.1 | 2.1 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 1.1 | 3.2 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 1.0 | 7.3 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 1.0 | 9.3 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 1.0 | 2.1 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 1.0 | 8.2 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 1.0 | 6.0 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.9 | 3.5 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.9 | 4.4 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.9 | 3.5 | GO:0060029 | convergent extension involved in organogenesis(GO:0060029) |
| 0.9 | 2.6 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.9 | 15.7 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.9 | 8.7 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.8 | 11.5 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.8 | 2.5 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.8 | 4.8 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.8 | 61.5 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.8 | 6.0 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.7 | 4.5 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.7 | 12.6 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.7 | 1.5 | GO:0048867 | stem cell fate determination(GO:0048867) |
| 0.7 | 2.1 | GO:0021779 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.7 | 2.1 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.7 | 3.5 | GO:0070778 | L-aspartate transport(GO:0070778) L-aspartate transmembrane transport(GO:0089712) |
| 0.7 | 11.6 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.7 | 30.5 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.7 | 2.0 | GO:0050917 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.7 | 5.4 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.7 | 4.0 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.6 | 5.8 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.6 | 3.8 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.6 | 34.0 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.6 | 10.8 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.6 | 8.9 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.6 | 1.8 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.6 | 2.3 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.6 | 9.7 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.6 | 5.7 | GO:0060371 | regulation of atrial cardiac muscle cell membrane depolarization(GO:0060371) |
| 0.6 | 2.8 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.6 | 10.1 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.6 | 3.9 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.5 | 2.2 | GO:0002434 | immune complex clearance(GO:0002434) immune complex clearance by monocytes and macrophages(GO:0002436) response to high density lipoprotein particle(GO:0055099) T cell extravasation(GO:0072683) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) |
| 0.5 | 2.2 | GO:1901205 | regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) negative regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901205) |
| 0.5 | 2.1 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.5 | 3.2 | GO:0036016 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.5 | 1.6 | GO:1900158 | positive regulation of osteoclast proliferation(GO:0090290) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.5 | 5.5 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.5 | 8.4 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.5 | 24.7 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.5 | 3.9 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.5 | 3.9 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.5 | 5.8 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.5 | 5.8 | GO:0042354 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.5 | 2.3 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.5 | 1.4 | GO:1904252 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) negative regulation of bile acid biosynthetic process(GO:0070858) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) negative regulation of bile acid metabolic process(GO:1904252) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.5 | 9.5 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.4 | 2.2 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.4 | 2.6 | GO:1903301 | fructose 2,6-bisphosphate metabolic process(GO:0006003) positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.4 | 1.7 | GO:0003335 | corneocyte development(GO:0003335) |
| 0.4 | 2.9 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.4 | 4.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.4 | 8.1 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.4 | 3.2 | GO:0071798 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.4 | 8.8 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.4 | 1.6 | GO:0061743 | motor learning(GO:0061743) |
| 0.4 | 3.8 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.4 | 1.5 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.4 | 34.9 | GO:0070268 | cornification(GO:0070268) |
| 0.4 | 2.3 | GO:0006540 | glutamate decarboxylation to succinate(GO:0006540) |
| 0.4 | 4.8 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.4 | 1.5 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.4 | 1.8 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.4 | 0.7 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) |
| 0.3 | 3.1 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.3 | 3.8 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.3 | 12.1 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.3 | 1.7 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.3 | 3.4 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.3 | 10.2 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.3 | 3.4 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.3 | 6.1 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.3 | 2.7 | GO:0048260 | positive regulation of receptor-mediated endocytosis(GO:0048260) |
| 0.3 | 1.3 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.3 | 3.0 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.3 | 4.9 | GO:0006183 | GTP biosynthetic process(GO:0006183) |
| 0.3 | 3.3 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.3 | 0.9 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.3 | 4.4 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.3 | 1.8 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.3 | 3.5 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.3 | 3.8 | GO:0046325 | negative regulation of glucose import(GO:0046325) |
| 0.3 | 10.9 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.3 | 1.1 | GO:0006408 | snRNA export from nucleus(GO:0006408) |
| 0.3 | 2.0 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.3 | 2.4 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.3 | 4.0 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.3 | 13.9 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.3 | 3.4 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.3 | 12.8 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.3 | 2.0 | GO:0001505 | regulation of neurotransmitter levels(GO:0001505) |
| 0.2 | 42.7 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.2 | 12.4 | GO:0007517 | muscle organ development(GO:0007517) |
| 0.2 | 4.9 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.2 | 6.0 | GO:0060271 | cilium morphogenesis(GO:0060271) |
| 0.2 | 3.7 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.2 | 14.9 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.2 | 3.9 | GO:0034616 | response to laminar fluid shear stress(GO:0034616) |
| 0.2 | 10.5 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.2 | 1.8 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.2 | 3.9 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) |
| 0.2 | 1.2 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.2 | 3.6 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.2 | 4.4 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.2 | 0.8 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.2 | 0.4 | GO:0072366 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) regulation of cellular ketone metabolic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072366) |
| 0.2 | 1.7 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.2 | 2.9 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.2 | 1.6 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.2 | 2.0 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.2 | 0.5 | GO:0090675 | intermicrovillar adhesion(GO:0090675) |
| 0.2 | 1.0 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.2 | 0.5 | GO:0098907 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) regulation of SA node cell action potential(GO:0098907) |
| 0.2 | 40.1 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.2 | 5.9 | GO:1900181 | negative regulation of protein localization to nucleus(GO:1900181) |
| 0.2 | 1.9 | GO:0051573 | negative regulation of histone H3-K9 methylation(GO:0051573) |
| 0.2 | 3.6 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 0.2 | 12.3 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.1 | 6.6 | GO:0009994 | oocyte differentiation(GO:0009994) |
| 0.1 | 0.6 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.1 | 1.8 | GO:0051775 | response to redox state(GO:0051775) |
| 0.1 | 0.8 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.1 | 3.4 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 3.0 | GO:0007183 | SMAD protein complex assembly(GO:0007183) |
| 0.1 | 3.4 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.1 | 1.2 | GO:0015705 | iodide transport(GO:0015705) |
| 0.1 | 1.2 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.1 | 2.9 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 0.8 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.1 | 7.3 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.1 | 5.5 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.1 | 3.9 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 1.4 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.1 | 1.0 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 4.3 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.1 | 6.3 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.1 | 1.8 | GO:0010623 | programmed cell death involved in cell development(GO:0010623) |
| 0.1 | 0.7 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.1 | 5.8 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.1 | 1.4 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.1 | 2.0 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 1.1 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.1 | 0.8 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 0.4 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 0.1 | 2.2 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 2.9 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.1 | 3.4 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.1 | 1.3 | GO:0033630 | positive regulation of cell adhesion mediated by integrin(GO:0033630) |
| 0.1 | 3.1 | GO:0035904 | aorta development(GO:0035904) |
| 0.1 | 2.3 | GO:0048738 | cardiac muscle tissue development(GO:0048738) |
| 0.1 | 12.0 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.1 | 11.9 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 0.5 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.1 | 3.1 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.1 | 1.8 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.1 | 0.2 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.1 | 0.8 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.1 | 5.1 | GO:0035722 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.1 | 0.4 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 0.9 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.1 | 1.4 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.1 | 6.0 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.1 | 1.9 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.1 | 0.5 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 2.1 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.1 | 1.8 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 1.4 | GO:0035137 | hindlimb morphogenesis(GO:0035137) |
| 0.0 | 0.3 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.0 | 1.4 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 2.9 | GO:1902108 | regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902108) |
| 0.0 | 0.3 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 2.0 | GO:0021954 | central nervous system neuron development(GO:0021954) |
| 0.0 | 1.3 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 0.9 | GO:0036258 | multivesicular body assembly(GO:0036258) |
| 0.0 | 1.3 | GO:0050890 | cognition(GO:0050890) |
| 0.0 | 0.6 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.3 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 2.3 | GO:0010833 | telomere maintenance via telomere lengthening(GO:0010833) |
| 0.0 | 1.7 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.6 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.1 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.0 | 0.3 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 | 0.9 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.0 | 0.6 | GO:0050771 | negative regulation of axonogenesis(GO:0050771) |
| 0.0 | 4.6 | GO:0016197 | endosomal transport(GO:0016197) |
| 0.0 | 0.5 | GO:0060338 | regulation of type I interferon-mediated signaling pathway(GO:0060338) |
| 0.0 | 2.4 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.0 | 1.5 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 1.2 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.9 | GO:0034620 | cellular response to unfolded protein(GO:0034620) |
| 0.0 | 1.1 | GO:0030838 | positive regulation of actin filament polymerization(GO:0030838) |
| 0.0 | 4.4 | GO:0030198 | extracellular matrix organization(GO:0030198) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 16.1 | 225.0 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 11.3 | 34.0 | GO:0044609 | DBIRD complex(GO:0044609) |
| 3.9 | 11.7 | GO:0033150 | perinuclear theca(GO:0033011) cytoskeletal calyx(GO:0033150) |
| 3.7 | 11.1 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 3.6 | 10.8 | GO:0097679 | other organism cytoplasm(GO:0097679) |
| 2.7 | 41.1 | GO:0030478 | actin cap(GO:0030478) |
| 2.6 | 15.9 | GO:1990037 | Lewy body core(GO:1990037) |
| 2.5 | 24.7 | GO:0098651 | basement membrane collagen trimer(GO:0098651) |
| 2.2 | 15.7 | GO:0033269 | internode region of axon(GO:0033269) |
| 2.1 | 26.8 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 1.2 | 4.7 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 1.0 | 3.9 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 1.0 | 3.9 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.9 | 15.7 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.8 | 35.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.8 | 13.9 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.8 | 10.6 | GO:0036038 | MKS complex(GO:0036038) |
| 0.7 | 12.0 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.7 | 10.5 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.6 | 11.0 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.6 | 5.7 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.6 | 6.3 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.6 | 28.1 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.5 | 4.8 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.5 | 93.5 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.5 | 24.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.5 | 6.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.5 | 1.4 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.4 | 5.4 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.4 | 2.2 | GO:1990425 | ryanodine receptor complex(GO:1990425) |
| 0.4 | 34.8 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.4 | 3.8 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.4 | 1.7 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.4 | 2.4 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.4 | 1.2 | GO:0000806 | Y chromosome(GO:0000806) |
| 0.3 | 3.1 | GO:0070083 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.3 | 5.1 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.3 | 7.4 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.3 | 3.9 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.3 | 2.3 | GO:0061200 | clathrin-sculpted gamma-aminobutyric acid transport vesicle(GO:0061200) clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane(GO:0061202) |
| 0.3 | 124.2 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.3 | 28.9 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.3 | 8.5 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.3 | 0.9 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.3 | 2.1 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.3 | 2.3 | GO:0016012 | dystroglycan complex(GO:0016011) sarcoglycan complex(GO:0016012) |
| 0.3 | 28.6 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.3 | 3.4 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.3 | 6.6 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.3 | 8.4 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.3 | 2.8 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.3 | 6.0 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.3 | 11.0 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.2 | 4.7 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.2 | 1.7 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.2 | 28.5 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.2 | 8.1 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.2 | 0.6 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.2 | 2.3 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.2 | 20.0 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.2 | 4.4 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.2 | 3.5 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.2 | 34.2 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.2 | 3.7 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.2 | 1.1 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 14.9 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.1 | 8.6 | GO:0031672 | A band(GO:0031672) |
| 0.1 | 21.7 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 6.0 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 22.5 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 1.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 14.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 0.5 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.1 | 0.4 | GO:0030936 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.1 | 3.5 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 0.7 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 15.0 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 3.2 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 1.3 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 1.1 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 3.6 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 2.2 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.1 | 1.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 2.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 2.4 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 1.0 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.1 | 4.4 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.8 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 1.3 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 44.4 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 2.1 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 3.2 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 4.0 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 5.0 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 4.5 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 2.1 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 38.5 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 2.2 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.5 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 1.5 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 7.4 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 1.4 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.3 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 3.4 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 0.2 | GO:0005640 | nuclear outer membrane(GO:0005640) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.3 | 81.7 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 6.2 | 30.9 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 4.0 | 16.2 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 3.2 | 29.1 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 3.2 | 9.6 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 3.1 | 226.6 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 2.5 | 9.9 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 2.4 | 23.7 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 2.2 | 8.7 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 2.2 | 6.5 | GO:0047223 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,3-N-acetylglucosaminyltransferase activity(GO:0047223) |
| 2.1 | 8.5 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 2.0 | 8.1 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 1.7 | 30.5 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 1.6 | 9.6 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 1.5 | 41.1 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 1.4 | 19.1 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 1.4 | 17.6 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 1.3 | 15.7 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 1.2 | 6.2 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 1.2 | 5.8 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 1.1 | 34.0 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 1.1 | 5.6 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 1.1 | 3.2 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 1.0 | 3.1 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 1.0 | 3.0 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 0.9 | 12.0 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.9 | 15.9 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.9 | 15.7 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.8 | 9.3 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.8 | 75.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.8 | 4.7 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.8 | 2.3 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.7 | 6.7 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.7 | 8.8 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.7 | 2.2 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.7 | 2.9 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.7 | 3.5 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.6 | 3.1 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.6 | 1.8 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.6 | 3.5 | GO:0032558 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.6 | 2.8 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.6 | 11.2 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.6 | 7.8 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.6 | 28.5 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.5 | 2.2 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.5 | 8.5 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.5 | 2.6 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.5 | 4.1 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.5 | 10.8 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.5 | 2.4 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.5 | 3.9 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.5 | 10.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.5 | 18.6 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.5 | 1.4 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.5 | 20.9 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.4 | 6.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.4 | 4.4 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.4 | 2.2 | GO:0004306 | ethanolamine-phosphate cytidylyltransferase activity(GO:0004306) |
| 0.4 | 3.9 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.4 | 3.0 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.4 | 42.8 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.4 | 3.8 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.4 | 2.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.4 | 16.3 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.4 | 5.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.4 | 18.9 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.4 | 2.7 | GO:0005497 | androgen binding(GO:0005497) |
| 0.4 | 35.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.3 | 7.6 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.3 | 8.2 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.3 | 1.3 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.3 | 3.7 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.3 | 4.2 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.3 | 2.3 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.3 | 6.3 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.3 | 3.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.3 | 6.5 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.3 | 5.8 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.3 | 3.0 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.3 | 2.9 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.3 | 9.9 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.3 | 5.4 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.3 | 1.4 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.3 | 1.4 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.3 | 6.0 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.3 | 12.8 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.2 | 1.2 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.2 | 1.9 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.2 | 4.9 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.2 | 3.9 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.2 | 5.5 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.2 | 0.9 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.2 | 5.5 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.2 | 1.7 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.2 | 5.9 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.2 | 22.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.2 | 1.4 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.2 | 8.6 | GO:0070888 | E-box binding(GO:0070888) |
| 0.2 | 9.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.2 | 0.7 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.2 | 5.7 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.2 | 5.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.2 | 2.3 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.2 | 2.9 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 12.6 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.1 | 3.8 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 0.9 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.1 | 33.0 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 0.7 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.1 | 3.4 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 13.9 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.1 | 6.0 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 1.0 | GO:0001093 | TFIIB-class transcription factor binding(GO:0001093) |
| 0.1 | 2.2 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 6.0 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 2.0 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 1.6 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.1 | 0.9 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.1 | 1.8 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 2.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 3.9 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 3.5 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 18.5 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 1.6 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.1 | 3.2 | GO:0038187 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.1 | 0.8 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.1 | 7.1 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.1 | 1.8 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 7.7 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.1 | 3.4 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 12.9 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 0.6 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 1.1 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 0.3 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.1 | 3.3 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.1 | 4.0 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 1.9 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 2.0 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 2.0 | GO:0000217 | DNA secondary structure binding(GO:0000217) |
| 0.1 | 1.6 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 3.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 1.4 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 1.9 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 1.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 20.8 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.1 | 2.0 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 0.8 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.7 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 0.5 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 0.8 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 16.8 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.1 | 6.3 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.1 | 3.5 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 1.2 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.1 | 7.1 | GO:0008528 | G-protein coupled peptide receptor activity(GO:0008528) |
| 0.1 | 3.2 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 8.3 | GO:0003712 | transcription cofactor activity(GO:0003712) |
| 0.1 | 2.2 | GO:0008200 | ion channel inhibitor activity(GO:0008200) |
| 0.1 | 0.3 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.1 | 4.5 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 0.6 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 4.5 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 7.2 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.1 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 3.4 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 1.8 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.5 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 3.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 8.0 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 1.7 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 6.1 | GO:0017171 | serine hydrolase activity(GO:0017171) |
| 0.0 | 2.4 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 10.0 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.5 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 4.0 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 1.4 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.0 | 0.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 1.6 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 1.7 | GO:0016782 | transferase activity, transferring sulfur-containing groups(GO:0016782) |
| 0.0 | 3.1 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.7 | GO:0005548 | phospholipid transporter activity(GO:0005548) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 262.1 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 1.4 | 58.5 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 1.3 | 62.0 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.8 | 13.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.8 | 3.2 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.7 | 84.8 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.5 | 30.9 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.5 | 6.0 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.4 | 12.1 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.3 | 5.9 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.3 | 58.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.3 | 7.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.2 | 17.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.2 | 5.4 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.2 | 8.5 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.2 | 2.5 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.2 | 6.2 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.2 | 9.7 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 4.0 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 3.3 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.1 | 1.4 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 3.4 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.1 | 6.2 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 1.4 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 4.4 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 12.0 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 2.9 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 20.1 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.1 | 0.9 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 14.3 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 2.0 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 4.2 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 2.6 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 25.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 1.1 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 4.3 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 2.1 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 1.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 4.8 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.1 | 1.8 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 3.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 1.5 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 2.7 | NABA CORE MATRISOME | Ensemble of genes encoding core extracellular matrix including ECM glycoproteins, collagens and proteoglycans |
| 0.1 | 2.3 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 3.9 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 9.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.2 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 1.1 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 1.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 1.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.8 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 1.2 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.9 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.6 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.3 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.5 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.3 | 196.4 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 1.5 | 79.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 1.4 | 24.6 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 1.1 | 27.5 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 1.1 | 72.0 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 1.0 | 24.6 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 1.0 | 62.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.9 | 30.7 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.7 | 7.8 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.6 | 6.2 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.6 | 11.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.5 | 9.8 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.4 | 8.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.4 | 3.4 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.4 | 6.0 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.3 | 29.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.3 | 6.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.3 | 10.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.3 | 11.4 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.3 | 11.3 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.3 | 3.5 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.3 | 3.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.3 | 12.8 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.3 | 8.1 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.2 | 48.5 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.2 | 4.8 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.2 | 3.9 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.2 | 5.4 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.2 | 13.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.2 | 3.8 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.2 | 0.7 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.2 | 3.9 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.2 | 2.9 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.2 | 3.1 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.2 | 2.3 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 1.8 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 1.9 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 6.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 3.2 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 1.3 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 2.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 6.3 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.1 | 7.8 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 0.8 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.1 | 10.3 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 2.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.8 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 6.9 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 4.8 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 3.4 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 3.7 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 1.7 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.1 | 1.4 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.1 | 1.1 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.1 | 6.3 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 4.4 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 3.6 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 0.8 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.1 | 1.4 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 2.0 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 0.9 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 2.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 6.2 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 3.0 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 1.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.5 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.7 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 1.2 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.5 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 1.2 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.5 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 1.1 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |