Project
averaged GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for HOXC8
Z-value: 1.53
Transcription factors associated with HOXC8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXC8
|
ENSG00000037965.4 | homeobox C8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXC8 | hg19_v2_chr12_+_54402790_54402832 | -0.34 | 1.8e-07 | Click! |
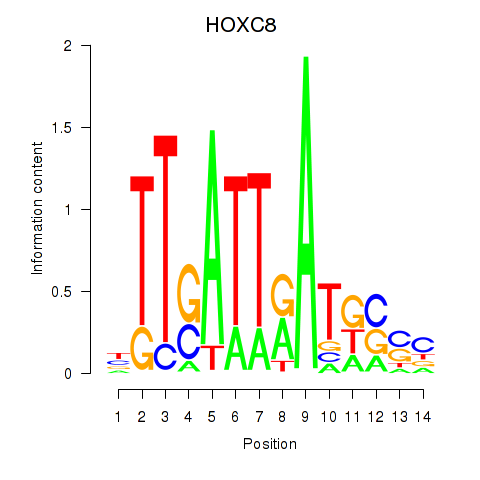
Activity profile of HOXC8 motif
Sorted Z-values of HOXC8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_-_109590174 | 24.44 |
ENST00000372054.1
|
GNG5P2
|
guanine nucleotide binding protein (G protein), gamma 5 pseudogene 2 |
| chr8_-_27695552 | 22.95 |
ENST00000522944.1
ENST00000301905.4 |
PBK
|
PDZ binding kinase |
| chrX_+_119737806 | 18.79 |
ENST00000371317.5
|
MCTS1
|
malignant T cell amplified sequence 1 |
| chr3_-_64009658 | 15.72 |
ENST00000394431.2
|
PSMD6
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 6 |
| chr18_+_657578 | 15.41 |
ENST00000323274.10
|
TYMS
|
thymidylate synthetase |
| chr16_-_46655538 | 15.26 |
ENST00000303383.3
|
SHCBP1
|
SHC SH2-domain binding protein 1 |
| chr5_+_159848854 | 14.55 |
ENST00000517480.1
ENST00000520452.1 ENST00000393964.1 |
PTTG1
|
pituitary tumor-transforming 1 |
| chr2_-_89292422 | 13.81 |
ENST00000495489.1
|
IGKV1-8
|
immunoglobulin kappa variable 1-8 |
| chr2_+_89890533 | 13.37 |
ENST00000429992.2
|
IGKV2D-40
|
immunoglobulin kappa variable 2D-40 |
| chr5_+_135385202 | 13.28 |
ENST00000514554.1
|
TGFBI
|
transforming growth factor, beta-induced, 68kDa |
| chr2_-_89513402 | 13.23 |
ENST00000498435.1
|
IGKV1-27
|
immunoglobulin kappa variable 1-27 |
| chr1_+_45205478 | 12.73 |
ENST00000452259.1
ENST00000372224.4 |
KIF2C
|
kinesin family member 2C |
| chr7_-_105752971 | 12.52 |
ENST00000011473.2
|
SYPL1
|
synaptophysin-like 1 |
| chr10_+_62538089 | 12.47 |
ENST00000519078.2
ENST00000395284.3 ENST00000316629.4 |
CDK1
|
cyclin-dependent kinase 1 |
| chr12_-_102455846 | 12.33 |
ENST00000545679.1
|
CCDC53
|
coiled-coil domain containing 53 |
| chr2_-_150444116 | 12.13 |
ENST00000428879.1
ENST00000422782.2 |
MMADHC
|
methylmalonic aciduria (cobalamin deficiency) cblD type, with homocystinuria |
| chr2_+_187371440 | 11.88 |
ENST00000445547.1
|
ZC3H15
|
zinc finger CCCH-type containing 15 |
| chr7_-_6523688 | 11.83 |
ENST00000490996.1
|
KDELR2
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2 |
| chr1_-_149900122 | 11.52 |
ENST00000271628.8
|
SF3B4
|
splicing factor 3b, subunit 4, 49kDa |
| chr15_-_59949693 | 11.46 |
ENST00000396063.1
ENST00000396064.3 ENST00000484743.1 ENST00000559706.1 ENST00000396060.2 |
GTF2A2
|
general transcription factor IIA, 2, 12kDa |
| chr9_+_134065506 | 11.34 |
ENST00000483497.2
|
NUP214
|
nucleoporin 214kDa |
| chr15_-_59949667 | 11.13 |
ENST00000396061.1
|
GTF2A2
|
general transcription factor IIA, 2, 12kDa |
| chr22_+_42017987 | 11.01 |
ENST00000405506.1
|
XRCC6
|
X-ray repair complementing defective repair in Chinese hamster cells 6 |
| chr1_-_225615599 | 10.91 |
ENST00000421383.1
ENST00000272163.4 |
LBR
|
lamin B receptor |
| chr12_-_54652060 | 10.83 |
ENST00000552562.1
|
CBX5
|
chromobox homolog 5 |
| chr1_+_45205498 | 10.78 |
ENST00000372218.4
|
KIF2C
|
kinesin family member 2C |
| chr5_+_172385732 | 10.77 |
ENST00000519974.1
ENST00000521476.1 |
RPL26L1
|
ribosomal protein L26-like 1 |
| chr2_+_90139056 | 10.61 |
ENST00000492446.1
|
IGKV1D-16
|
immunoglobulin kappa variable 1D-16 |
| chr2_-_89521942 | 10.55 |
ENST00000482769.1
|
IGKV2-28
|
immunoglobulin kappa variable 2-28 |
| chr10_+_62538248 | 10.30 |
ENST00000448257.2
|
CDK1
|
cyclin-dependent kinase 1 |
| chr4_+_79472673 | 10.23 |
ENST00000264908.6
|
ANXA3
|
annexin A3 |
| chr5_+_167913450 | 10.12 |
ENST00000231572.3
ENST00000538719.1 |
RARS
|
arginyl-tRNA synthetase |
| chr10_+_5135981 | 10.11 |
ENST00000380554.3
|
AKR1C3
|
aldo-keto reductase family 1, member C3 |
| chr15_+_59397275 | 9.93 |
ENST00000288207.2
|
CCNB2
|
cyclin B2 |
| chr2_-_89568263 | 9.84 |
ENST00000473726.1
|
IGKV1-33
|
immunoglobulin kappa variable 1-33 |
| chr11_+_35198118 | 9.80 |
ENST00000525211.1
ENST00000526000.1 ENST00000279452.6 ENST00000527889.1 |
CD44
|
CD44 molecule (Indian blood group) |
| chr1_+_226250379 | 9.67 |
ENST00000366815.3
ENST00000366814.3 |
H3F3A
|
H3 histone, family 3A |
| chr7_-_132766818 | 9.54 |
ENST00000262570.5
|
CHCHD3
|
coiled-coil-helix-coiled-coil-helix domain containing 3 |
| chr7_-_132766800 | 9.54 |
ENST00000542753.1
ENST00000448878.1 |
CHCHD3
|
coiled-coil-helix-coiled-coil-helix domain containing 3 |
| chr5_-_68665084 | 9.42 |
ENST00000509462.1
|
TAF9
|
TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 32kDa |
| chrX_-_38080077 | 9.33 |
ENST00000378533.3
ENST00000544439.1 ENST00000432886.2 ENST00000538295.1 |
SRPX
|
sushi-repeat containing protein, X-linked |
| chr1_-_146040968 | 9.32 |
ENST00000401010.3
|
NBPF11
|
neuroblastoma breakpoint family, member 11 |
| chr14_-_107083690 | 9.23 |
ENST00000455737.1
ENST00000390629.2 |
IGHV4-59
|
immunoglobulin heavy variable 4-59 |
| chr2_+_90108504 | 9.21 |
ENST00000390271.2
|
IGKV6D-41
|
immunoglobulin kappa variable 6D-41 (non-functional) |
| chr3_-_149095652 | 9.01 |
ENST00000305366.3
|
TM4SF1
|
transmembrane 4 L six family member 1 |
| chr6_+_63921399 | 9.00 |
ENST00000356170.3
|
FKBP1C
|
FK506 binding protein 1C |
| chr6_+_27114861 | 8.92 |
ENST00000377459.1
|
HIST1H2AH
|
histone cluster 1, H2ah |
| chr14_-_106642049 | 8.84 |
ENST00000390605.2
|
IGHV1-18
|
immunoglobulin heavy variable 1-18 |
| chr12_+_21525818 | 8.78 |
ENST00000240652.3
ENST00000542023.1 ENST00000537593.1 |
IAPP
|
islet amyloid polypeptide |
| chr10_-_58120996 | 8.59 |
ENST00000361148.6
ENST00000395405.1 ENST00000373944.3 |
ZWINT
|
ZW10 interacting kinetochore protein |
| chr13_+_34392185 | 8.49 |
ENST00000380071.3
|
RFC3
|
replication factor C (activator 1) 3, 38kDa |
| chr3_-_141719195 | 8.43 |
ENST00000397991.4
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr2_-_176046391 | 8.34 |
ENST00000392541.3
ENST00000409194.1 |
ATP5G3
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9) |
| chr16_-_24942411 | 8.32 |
ENST00000571843.1
|
ARHGAP17
|
Rho GTPase activating protein 17 |
| chr11_+_60223225 | 8.27 |
ENST00000524807.1
ENST00000345732.4 |
MS4A1
|
membrane-spanning 4-domains, subfamily A, member 1 |
| chrX_+_48433326 | 8.23 |
ENST00000376755.1
|
RBM3
|
RNA binding motif (RNP1, RRM) protein 3 |
| chr19_-_10697895 | 7.88 |
ENST00000591240.1
ENST00000589684.1 ENST00000591676.1 ENST00000250244.6 ENST00000590923.1 |
AP1M2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr16_-_24942273 | 7.85 |
ENST00000571406.1
|
ARHGAP17
|
Rho GTPase activating protein 17 |
| chr12_+_75874580 | 7.80 |
ENST00000456650.3
|
GLIPR1
|
GLI pathogenesis-related 1 |
| chr12_-_53320245 | 7.60 |
ENST00000552150.1
|
KRT8
|
keratin 8 |
| chr18_+_3252265 | 7.57 |
ENST00000580887.1
ENST00000536605.1 |
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr5_+_110074685 | 7.53 |
ENST00000355943.3
ENST00000447245.2 |
SLC25A46
|
solute carrier family 25, member 46 |
| chr2_-_70520539 | 7.50 |
ENST00000482975.2
ENST00000438261.1 |
SNRPG
|
small nuclear ribonucleoprotein polypeptide G |
| chr7_-_87856280 | 7.42 |
ENST00000490437.1
ENST00000431660.1 |
SRI
|
sorcin |
| chr2_+_192109911 | 7.40 |
ENST00000418908.1
ENST00000339514.4 ENST00000392318.3 |
MYO1B
|
myosin IB |
| chr6_-_27114577 | 7.29 |
ENST00000356950.1
ENST00000396891.4 |
HIST1H2BK
|
histone cluster 1, H2bk |
| chr14_-_75643296 | 7.29 |
ENST00000303575.4
|
TMED10
|
transmembrane emp24-like trafficking protein 10 (yeast) |
| chr22_+_23063100 | 7.28 |
ENST00000390309.2
|
IGLV3-19
|
immunoglobulin lambda variable 3-19 |
| chr13_+_53216565 | 7.27 |
ENST00000357495.2
|
HNRNPA1L2
|
heterogeneous nuclear ribonucleoprotein A1-like 2 |
| chr2_-_70520832 | 7.25 |
ENST00000454893.1
ENST00000272348.2 |
SNRPG
|
small nuclear ribonucleoprotein polypeptide G |
| chr2_+_216176540 | 7.23 |
ENST00000236959.9
|
ATIC
|
5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase |
| chr1_+_53480598 | 7.16 |
ENST00000430330.2
ENST00000408941.3 ENST00000478274.2 ENST00000484100.1 ENST00000435345.2 ENST00000488965.1 |
SCP2
|
sterol carrier protein 2 |
| chr3_-_64009102 | 7.12 |
ENST00000478185.1
ENST00000482510.1 ENST00000497323.1 ENST00000492933.1 ENST00000295901.4 |
PSMD6
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 6 |
| chr14_-_58894223 | 7.11 |
ENST00000555593.1
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr14_-_106733624 | 7.09 |
ENST00000390610.2
|
IGHV1-24
|
immunoglobulin heavy variable 1-24 |
| chr12_+_104609550 | 7.08 |
ENST00000525566.1
ENST00000429002.2 |
TXNRD1
|
thioredoxin reductase 1 |
| chr7_-_87856303 | 7.07 |
ENST00000394641.3
|
SRI
|
sorcin |
| chr8_-_101718991 | 7.04 |
ENST00000517990.1
|
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr12_-_102455902 | 7.03 |
ENST00000240079.6
|
CCDC53
|
coiled-coil domain containing 53 |
| chr6_-_159065741 | 7.00 |
ENST00000367085.3
ENST00000367089.3 |
DYNLT1
|
dynein, light chain, Tctex-type 1 |
| chr2_-_20251744 | 6.99 |
ENST00000175091.4
|
LAPTM4A
|
lysosomal protein transmembrane 4 alpha |
| chr2_-_89310012 | 6.94 |
ENST00000493819.1
|
IGKV1-9
|
immunoglobulin kappa variable 1-9 |
| chr17_-_8113542 | 6.91 |
ENST00000578549.1
ENST00000535053.1 ENST00000582368.1 |
AURKB
|
aurora kinase B |
| chr1_+_224544572 | 6.90 |
ENST00000366857.5
ENST00000366856.3 |
CNIH4
|
cornichon family AMPA receptor auxiliary protein 4 |
| chr4_+_169418195 | 6.90 |
ENST00000261509.6
ENST00000335742.7 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr8_-_101962777 | 6.87 |
ENST00000395951.3
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr2_-_24308051 | 6.84 |
ENST00000238721.4
ENST00000335934.4 |
TP53I3
|
tumor protein p53 inducible protein 3 |
| chr1_-_40042416 | 6.80 |
ENST00000372857.3
ENST00000372856.3 ENST00000531243.2 ENST00000451091.2 |
PABPC4
|
poly(A) binding protein, cytoplasmic 4 (inducible form) |
| chr6_+_57037089 | 6.78 |
ENST00000370693.5
|
BAG2
|
BCL2-associated athanogene 2 |
| chr14_+_89060739 | 6.72 |
ENST00000318308.6
|
ZC3H14
|
zinc finger CCCH-type containing 14 |
| chr14_-_106322288 | 6.69 |
ENST00000390559.2
|
IGHM
|
immunoglobulin heavy constant mu |
| chr4_+_83956237 | 6.69 |
ENST00000264389.2
|
COPS4
|
COP9 signalosome subunit 4 |
| chr15_+_66797627 | 6.68 |
ENST00000565627.1
ENST00000564179.1 |
ZWILCH
|
zwilch kinetochore protein |
| chr2_-_89247338 | 6.68 |
ENST00000496168.1
|
IGKV1-5
|
immunoglobulin kappa variable 1-5 |
| chr2_+_178257372 | 6.67 |
ENST00000264167.4
ENST00000409888.1 |
AGPS
|
alkylglycerone phosphate synthase |
| chr2_-_106054952 | 6.67 |
ENST00000336660.5
ENST00000393352.3 ENST00000607522.1 |
FHL2
|
four and a half LIM domains 2 |
| chr1_+_165864821 | 6.66 |
ENST00000470820.1
|
UCK2
|
uridine-cytidine kinase 2 |
| chr14_-_106926724 | 6.65 |
ENST00000434710.1
|
IGHV3-43
|
immunoglobulin heavy variable 3-43 |
| chr10_+_94352956 | 6.61 |
ENST00000260731.3
|
KIF11
|
kinesin family member 11 |
| chr14_+_89060749 | 6.54 |
ENST00000555900.1
ENST00000406216.3 ENST00000557737.1 |
ZC3H14
|
zinc finger CCCH-type containing 14 |
| chr14_-_106878083 | 6.52 |
ENST00000390619.2
|
IGHV4-39
|
immunoglobulin heavy variable 4-39 |
| chr11_+_60223312 | 6.51 |
ENST00000532491.1
ENST00000532073.1 ENST00000534668.1 ENST00000528313.1 ENST00000533306.1 |
MS4A1
|
membrane-spanning 4-domains, subfamily A, member 1 |
| chr1_+_33116765 | 6.49 |
ENST00000544435.1
ENST00000373485.1 ENST00000458695.2 ENST00000490500.1 ENST00000445722.2 |
RBBP4
|
retinoblastoma binding protein 4 |
| chr11_+_35198243 | 6.44 |
ENST00000528455.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr14_+_20937538 | 6.41 |
ENST00000361505.5
ENST00000553591.1 |
PNP
|
purine nucleoside phosphorylase |
| chr12_-_10875831 | 6.32 |
ENST00000279550.7
ENST00000228251.4 |
YBX3
|
Y box binding protein 3 |
| chr12_-_65146636 | 6.31 |
ENST00000418919.2
|
GNS
|
glucosamine (N-acetyl)-6-sulfatase |
| chr17_-_55038375 | 6.30 |
ENST00000240316.4
|
COIL
|
coilin |
| chr10_+_13652047 | 6.29 |
ENST00000601460.1
|
RP11-295P9.3
|
Uncharacterized protein |
| chr4_+_69962212 | 6.27 |
ENST00000508661.1
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr8_+_104310661 | 6.26 |
ENST00000522566.1
|
FZD6
|
frizzled family receptor 6 |
| chr17_-_40288449 | 6.21 |
ENST00000552162.1
ENST00000550504.1 |
RAB5C
|
RAB5C, member RAS oncogene family |
| chr19_+_49496705 | 6.17 |
ENST00000595090.1
|
RUVBL2
|
RuvB-like AAA ATPase 2 |
| chr12_+_96252706 | 6.16 |
ENST00000266735.5
ENST00000553192.1 ENST00000552085.1 |
SNRPF
|
small nuclear ribonucleoprotein polypeptide F |
| chr2_-_89459813 | 6.14 |
ENST00000390256.2
|
IGKV6-21
|
immunoglobulin kappa variable 6-21 (non-functional) |
| chr20_+_1099233 | 6.11 |
ENST00000246015.4
ENST00000335877.6 ENST00000438768.2 |
PSMF1
|
proteasome (prosome, macropain) inhibitor subunit 1 (PI31) |
| chrX_-_16887963 | 6.11 |
ENST00000380084.4
|
RBBP7
|
retinoblastoma binding protein 7 |
| chr14_-_106781017 | 6.09 |
ENST00000390612.2
|
IGHV4-28
|
immunoglobulin heavy variable 4-28 |
| chr2_+_102456277 | 6.06 |
ENST00000421882.1
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr14_-_106406090 | 6.04 |
ENST00000390593.2
|
IGHV6-1
|
immunoglobulin heavy variable 6-1 |
| chr15_-_59225758 | 5.98 |
ENST00000558486.1
ENST00000560682.1 ENST00000249736.7 ENST00000559880.1 ENST00000536328.1 |
SLTM
|
SAFB-like, transcription modulator |
| chr1_+_144173162 | 5.97 |
ENST00000356801.6
|
NBPF8
|
neuroblastoma breakpoint family, member 8 |
| chr3_+_151986709 | 5.97 |
ENST00000495875.2
ENST00000493459.1 ENST00000324210.5 ENST00000459747.1 |
MBNL1
|
muscleblind-like splicing regulator 1 |
| chr11_+_101983176 | 5.97 |
ENST00000524575.1
|
YAP1
|
Yes-associated protein 1 |
| chr5_-_68665296 | 5.96 |
ENST00000512152.1
ENST00000503245.1 ENST00000512561.1 ENST00000380822.4 |
TAF9
|
TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 32kDa |
| chr6_-_34393825 | 5.95 |
ENST00000605528.1
ENST00000326199.8 |
RPS10-NUDT3
RPS10
|
RPS10-NUDT3 readthrough ribosomal protein S10 |
| chr3_-_81811312 | 5.92 |
ENST00000429644.2
|
GBE1
|
glucan (1,4-alpha-), branching enzyme 1 |
| chr10_+_79793518 | 5.90 |
ENST00000440692.1
ENST00000435275.1 ENST00000372360.3 ENST00000360830.4 |
RPS24
|
ribosomal protein S24 |
| chr8_-_101719159 | 5.86 |
ENST00000520868.1
ENST00000522658.1 |
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr3_-_189840223 | 5.83 |
ENST00000427335.2
|
LEPREL1
|
leprecan-like 1 |
| chr12_+_117013656 | 5.83 |
ENST00000556529.1
|
MAP1LC3B2
|
microtubule-associated protein 1 light chain 3 beta 2 |
| chr5_+_34757309 | 5.83 |
ENST00000397449.1
|
RAI14
|
retinoic acid induced 14 |
| chr2_-_89399845 | 5.80 |
ENST00000479981.1
|
IGKV1-16
|
immunoglobulin kappa variable 1-16 |
| chr22_-_36924944 | 5.78 |
ENST00000405442.1
ENST00000402116.1 |
EIF3D
|
eukaryotic translation initiation factor 3, subunit D |
| chr2_+_90153696 | 5.78 |
ENST00000417279.2
|
IGKV3D-15
|
immunoglobulin kappa variable 3D-15 (gene/pseudogene) |
| chr9_-_32573130 | 5.78 |
ENST00000350021.2
ENST00000379847.3 |
NDUFB6
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 6, 17kDa |
| chr6_+_24667257 | 5.75 |
ENST00000537591.1
ENST00000230048.4 |
ACOT13
|
acyl-CoA thioesterase 13 |
| chr14_-_106478603 | 5.68 |
ENST00000390596.2
|
IGHV4-4
|
immunoglobulin heavy variable 4-4 |
| chr4_-_71532339 | 5.68 |
ENST00000254801.4
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr15_-_101835110 | 5.68 |
ENST00000560496.1
|
SNRPA1
|
small nuclear ribonucleoprotein polypeptide A' |
| chr17_-_38574169 | 5.65 |
ENST00000423485.1
|
TOP2A
|
topoisomerase (DNA) II alpha 170kDa |
| chr12_-_56122220 | 5.64 |
ENST00000552692.1
|
CD63
|
CD63 molecule |
| chr2_-_151344172 | 5.56 |
ENST00000375734.2
ENST00000263895.4 ENST00000454202.1 |
RND3
|
Rho family GTPase 3 |
| chr17_+_66509019 | 5.55 |
ENST00000585981.1
ENST00000589480.1 ENST00000585815.1 |
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr9_-_127624194 | 5.50 |
ENST00000373570.4
ENST00000348462.3 |
RPL35
|
ribosomal protein L35 |
| chr1_+_145301735 | 5.50 |
ENST00000605176.1
|
NBPF10
|
neuroblastoma breakpoint family, member 10 |
| chr19_-_14945933 | 5.48 |
ENST00000322301.3
|
OR7A5
|
olfactory receptor, family 7, subfamily A, member 5 |
| chr1_+_158900568 | 5.48 |
ENST00000458222.1
|
PYHIN1
|
pyrin and HIN domain family, member 1 |
| chr17_+_7155819 | 5.48 |
ENST00000570322.1
ENST00000576496.1 ENST00000574841.2 |
ELP5
|
elongator acetyltransferase complex subunit 5 |
| chr2_+_89952792 | 5.46 |
ENST00000390265.2
|
IGKV1D-33
|
immunoglobulin kappa variable 1D-33 |
| chr7_+_116502605 | 5.45 |
ENST00000458284.2
ENST00000490693.1 |
CAPZA2
|
capping protein (actin filament) muscle Z-line, alpha 2 |
| chr18_+_3252206 | 5.45 |
ENST00000578562.2
|
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr2_+_201994042 | 5.41 |
ENST00000417748.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr2_+_90060377 | 5.41 |
ENST00000436451.2
|
IGKV6D-21
|
immunoglobulin kappa variable 6D-21 (non-functional) |
| chr7_-_7679633 | 5.40 |
ENST00000401447.1
|
RPA3
|
replication protein A3, 14kDa |
| chr6_+_7541845 | 5.39 |
ENST00000418664.2
|
DSP
|
desmoplakin |
| chr10_-_5046042 | 5.38 |
ENST00000421196.3
ENST00000455190.1 |
AKR1C2
|
aldo-keto reductase family 1, member C2 |
| chr4_+_69962185 | 5.35 |
ENST00000305231.7
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr14_+_52456193 | 5.34 |
ENST00000261700.3
|
C14orf166
|
chromosome 14 open reading frame 166 |
| chr11_-_47664072 | 5.32 |
ENST00000542981.1
ENST00000530428.1 ENST00000302503.3 |
MTCH2
|
mitochondrial carrier 2 |
| chr10_-_95241951 | 5.31 |
ENST00000358334.5
ENST00000359263.4 ENST00000371488.3 |
MYOF
|
myoferlin |
| chr15_+_65843130 | 5.28 |
ENST00000569894.1
|
PTPLAD1
|
protein tyrosine phosphatase-like A domain containing 1 |
| chr12_+_75874460 | 5.24 |
ENST00000266659.3
|
GLIPR1
|
GLI pathogenesis-related 1 |
| chr12_+_48357401 | 5.24 |
ENST00000429772.2
ENST00000449758.2 |
TMEM106C
|
transmembrane protein 106C |
| chr1_-_211848899 | 5.24 |
ENST00000366998.3
ENST00000540251.1 ENST00000366999.4 |
NEK2
|
NIMA-related kinase 2 |
| chr14_+_56127989 | 5.23 |
ENST00000555573.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr7_+_107224364 | 5.23 |
ENST00000491150.1
|
BCAP29
|
B-cell receptor-associated protein 29 |
| chr6_-_8102279 | 5.21 |
ENST00000488226.2
|
EEF1E1
|
eukaryotic translation elongation factor 1 epsilon 1 |
| chr1_-_153518270 | 5.21 |
ENST00000354332.4
ENST00000368716.4 |
S100A4
|
S100 calcium binding protein A4 |
| chr11_+_35201826 | 5.20 |
ENST00000531873.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr2_+_219110149 | 5.17 |
ENST00000456575.1
|
ARPC2
|
actin related protein 2/3 complex, subunit 2, 34kDa |
| chr19_+_30414551 | 5.17 |
ENST00000360605.4
ENST00000570564.1 ENST00000574233.1 ENST00000585655.1 |
URI1
|
URI1, prefoldin-like chaperone |
| chr16_+_33605231 | 5.16 |
ENST00000570121.2
|
IGHV3OR16-12
|
immunoglobulin heavy variable 3/OR16-12 (non-functional) |
| chr2_-_169769787 | 5.15 |
ENST00000451987.1
|
SPC25
|
SPC25, NDC80 kinetochore complex component |
| chr1_-_146082633 | 5.15 |
ENST00000605317.1
ENST00000604938.1 ENST00000339388.5 |
NBPF11
|
neuroblastoma breakpoint family, member 11 |
| chr11_-_10828892 | 5.13 |
ENST00000525681.1
|
EIF4G2
|
eukaryotic translation initiation factor 4 gamma, 2 |
| chr16_+_33006369 | 5.13 |
ENST00000425181.3
|
IGHV3OR16-10
|
immunoglobulin heavy variable 3/OR16-10 (non-functional) |
| chr14_-_106552755 | 5.12 |
ENST00000390600.2
|
IGHV3-9
|
immunoglobulin heavy variable 3-9 |
| chr5_-_10761206 | 5.11 |
ENST00000432074.2
ENST00000230895.6 |
DAP
|
death-associated protein |
| chr12_+_113344755 | 5.07 |
ENST00000550883.1
|
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr7_+_116502527 | 5.07 |
ENST00000361183.3
|
CAPZA2
|
capping protein (actin filament) muscle Z-line, alpha 2 |
| chr8_+_30244580 | 5.06 |
ENST00000523115.1
ENST00000519647.1 |
RBPMS
|
RNA binding protein with multiple splicing |
| chr10_-_94257512 | 5.05 |
ENST00000371581.5
|
IDE
|
insulin-degrading enzyme |
| chr9_-_95056010 | 5.04 |
ENST00000443024.2
|
IARS
|
isoleucyl-tRNA synthetase |
| chr14_-_106539557 | 5.02 |
ENST00000390599.2
|
IGHV1-8
|
immunoglobulin heavy variable 1-8 |
| chr2_+_120125245 | 5.01 |
ENST00000393103.2
|
DBI
|
diazepam binding inhibitor (GABA receptor modulator, acyl-CoA binding protein) |
| chr2_-_89340242 | 5.00 |
ENST00000480492.1
|
IGKV1-12
|
immunoglobulin kappa variable 1-12 |
| chr8_+_55047763 | 4.99 |
ENST00000260102.4
ENST00000519831.1 |
MRPL15
|
mitochondrial ribosomal protein L15 |
| chr1_-_63988846 | 4.98 |
ENST00000283568.8
ENST00000371092.3 ENST00000271002.10 |
ITGB3BP
|
integrin beta 3 binding protein (beta3-endonexin) |
| chr2_-_89630186 | 4.98 |
ENST00000390264.2
|
IGKV2-40
|
immunoglobulin kappa variable 2-40 |
| chr7_+_117824210 | 4.97 |
ENST00000422760.1
ENST00000411938.1 |
NAA38
|
N(alpha)-acetyltransferase 38, NatC auxiliary subunit |
| chr5_-_96518907 | 4.96 |
ENST00000508447.1
ENST00000283109.3 |
RIOK2
|
RIO kinase 2 |
| chr4_+_83956312 | 4.95 |
ENST00000509317.1
ENST00000503682.1 ENST00000511653.1 |
COPS4
|
COP9 signalosome subunit 4 |
| chr2_+_73461410 | 4.94 |
ENST00000399032.2
ENST00000398422.2 ENST00000537131.1 ENST00000538797.1 |
CCT7
|
chaperonin containing TCP1, subunit 7 (eta) |
| chr2_+_73461364 | 4.94 |
ENST00000540468.1
ENST00000539919.1 ENST00000258091.5 |
CCT7
|
chaperonin containing TCP1, subunit 7 (eta) |
| chr2_+_89901292 | 4.92 |
ENST00000448155.2
|
IGKV1D-39
|
immunoglobulin kappa variable 1D-39 |
| chr1_-_145827015 | 4.89 |
ENST00000534502.1
ENST00000313835.9 ENST00000454423.3 |
GPR89A
|
G protein-coupled receptor 89A |
| chr14_-_58893832 | 4.87 |
ENST00000556007.2
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr15_-_22473353 | 4.87 |
ENST00000557788.2
|
IGHV4OR15-8
|
immunoglobulin heavy variable 4/OR15-8 (non-functional) |
| chr7_+_107531580 | 4.86 |
ENST00000537148.1
ENST00000440410.1 ENST00000437604.2 |
DLD
|
dihydrolipoamide dehydrogenase |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXC8
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.3 | 18.8 | GO:0002188 | translation reinitiation(GO:0002188) |
| 5.0 | 19.9 | GO:0009212 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) |
| 4.8 | 4.8 | GO:0009205 | purine nucleoside triphosphate metabolic process(GO:0009144) purine ribonucleoside triphosphate metabolic process(GO:0009205) |
| 3.6 | 21.5 | GO:0014038 | regulation of Schwann cell differentiation(GO:0014038) |
| 3.5 | 24.6 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 3.4 | 10.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 2.9 | 8.7 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 2.9 | 5.8 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 2.8 | 11.2 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 2.7 | 2.7 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) medium-chain fatty acid biosynthetic process(GO:0051792) |
| 2.7 | 11.0 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 2.7 | 21.4 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 2.6 | 12.8 | GO:0042100 | B cell proliferation(GO:0042100) |
| 2.5 | 7.5 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 2.4 | 9.7 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 2.4 | 23.5 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 2.3 | 6.9 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 2.2 | 6.7 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 2.1 | 6.4 | GO:0006738 | nicotinamide riboside catabolic process(GO:0006738) inosine metabolic process(GO:0046102) nicotinamide riboside metabolic process(GO:0046495) pyridine nucleoside metabolic process(GO:0070637) pyridine nucleoside catabolic process(GO:0070638) |
| 1.9 | 1.9 | GO:0035690 | cellular response to drug(GO:0035690) |
| 1.9 | 13.4 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 1.8 | 11.0 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 1.8 | 7.2 | GO:0046452 | dihydrofolate metabolic process(GO:0046452) |
| 1.7 | 8.5 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 1.7 | 5.1 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 1.7 | 5.1 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 1.7 | 5.0 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 1.7 | 6.7 | GO:0006238 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 1.7 | 21.5 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 1.7 | 8.3 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 1.6 | 4.8 | GO:1900195 | spindle assembly involved in female meiosis I(GO:0007057) positive regulation of oocyte maturation(GO:1900195) |
| 1.6 | 6.3 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 1.6 | 233.2 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 1.5 | 4.6 | GO:0090244 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) negative regulation of cell fate specification(GO:0009996) Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 1.5 | 4.5 | GO:0002514 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 1.5 | 13.6 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 1.5 | 5.8 | GO:0000912 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 1.4 | 4.3 | GO:0043311 | positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 1.4 | 7.2 | GO:1904977 | lymphatic endothelial cell migration(GO:1904977) |
| 1.4 | 12.9 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 1.4 | 7.2 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 1.4 | 8.4 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 1.4 | 5.5 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 1.3 | 6.7 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 1.3 | 9.3 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 1.3 | 7.7 | GO:0036111 | very long-chain fatty-acyl-CoA metabolic process(GO:0036111) |
| 1.3 | 12.6 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 1.2 | 9.9 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 1.2 | 6.1 | GO:0070370 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 1.2 | 4.9 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 1.2 | 4.8 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 1.2 | 9.5 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 1.2 | 7.0 | GO:0051708 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 1.2 | 9.3 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 1.1 | 1.1 | GO:0051673 | pore formation in membrane of other organism(GO:0035915) membrane disruption in other organism(GO:0051673) |
| 1.1 | 5.5 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 1.1 | 3.2 | GO:0070632 | spindle pole body duplication(GO:0030474) spindle pole body organization(GO:0051300) spindle pole body localization(GO:0070631) establishment of spindle pole body localization(GO:0070632) spindle pole body localization to nuclear envelope(GO:0071789) establishment of spindle pole body localization to nuclear envelope(GO:0071790) |
| 1.1 | 11.8 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 1.1 | 3.2 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 1.0 | 15.5 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 1.0 | 13.3 | GO:0035826 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 1.0 | 3.0 | GO:0097205 | glomerular filtration(GO:0003094) renal filtration(GO:0097205) |
| 1.0 | 3.9 | GO:0015965 | diadenosine polyphosphate biosynthetic process(GO:0015960) diadenosine tetraphosphate metabolic process(GO:0015965) diadenosine tetraphosphate biosynthetic process(GO:0015966) |
| 1.0 | 8.8 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 1.0 | 6.8 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 1.0 | 5.8 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 1.0 | 3.8 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.9 | 2.8 | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) |
| 0.9 | 4.7 | GO:1904016 | response to Thyroglobulin triiodothyronine(GO:1904016) |
| 0.9 | 3.7 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.9 | 5.5 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.9 | 3.6 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.9 | 3.5 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.8 | 5.0 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.8 | 4.2 | GO:0060337 | response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.8 | 10.0 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.8 | 12.5 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.8 | 2.5 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.8 | 2.4 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.8 | 8.8 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.8 | 3.2 | GO:0050712 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.8 | 3.2 | GO:0098905 | regulation of bundle of His cell action potential(GO:0098905) |
| 0.8 | 19.1 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.8 | 1.6 | GO:0001302 | replicative cell aging(GO:0001302) |
| 0.8 | 4.7 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.8 | 3.9 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.8 | 2.3 | GO:0097527 | necroptotic signaling pathway(GO:0097527) |
| 0.8 | 9.9 | GO:1904869 | regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.8 | 37.1 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.8 | 9.8 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.8 | 2.3 | GO:1900138 | negative regulation of phospholipase A2 activity(GO:1900138) |
| 0.8 | 36.8 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.7 | 3.7 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.7 | 6.0 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.7 | 3.0 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.7 | 17.7 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.7 | 3.7 | GO:1902775 | mitochondrial ribosome assembly(GO:0061668) mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.7 | 11.6 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.7 | 4.3 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.7 | 8.6 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.7 | 4.3 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.7 | 4.1 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.7 | 12.3 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.7 | 4.1 | GO:0030952 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) establishment or maintenance of cytoskeleton polarity(GO:0030952) |
| 0.7 | 4.7 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.7 | 4.0 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.7 | 10.0 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.6 | 2.6 | GO:0043321 | regulation of natural killer cell degranulation(GO:0043321) positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.6 | 11.8 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.6 | 1.9 | GO:0043605 | cellular amide catabolic process(GO:0043605) |
| 0.6 | 6.1 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.6 | 2.4 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.6 | 5.4 | GO:0003223 | ventricular compact myocardium morphogenesis(GO:0003223) cardiac muscle cell-cardiac muscle cell adhesion(GO:0086042) bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.6 | 11.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.6 | 3.0 | GO:1903243 | negative regulation of cardiac muscle adaptation(GO:0010616) transforming growth factor beta3 production(GO:0032907) regulation of transforming growth factor beta3 production(GO:0032910) positive regulation of transforming growth factor beta3 production(GO:0032916) negative regulation of lung blood pressure(GO:0061767) negative regulation of cardiac muscle hypertrophy in response to stress(GO:1903243) |
| 0.6 | 1.2 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.6 | 3.5 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.6 | 5.2 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.6 | 1.7 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.6 | 9.8 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.6 | 2.9 | GO:0006696 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.6 | 32.3 | GO:0007077 | mitotic nuclear envelope disassembly(GO:0007077) |
| 0.6 | 5.6 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.6 | 3.4 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.6 | 2.8 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.5 | 3.8 | GO:1902962 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.5 | 14.1 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.5 | 13.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.5 | 1.6 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.5 | 2.6 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.5 | 1.0 | GO:0000479 | endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.5 | 3.1 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.5 | 56.6 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.5 | 4.1 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.5 | 5.4 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.5 | 2.5 | GO:0007063 | regulation of sister chromatid cohesion(GO:0007063) |
| 0.5 | 2.9 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.5 | 1.0 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.5 | 8.3 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.5 | 2.9 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.5 | 2.9 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.5 | 6.2 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.5 | 13.2 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.5 | 77.6 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.5 | 1.9 | GO:1903803 | glutamine secretion(GO:0010585) L-glutamine import(GO:0036229) L-glutamine import into cell(GO:1903803) |
| 0.5 | 9.7 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.4 | 4.5 | GO:0006739 | NADP metabolic process(GO:0006739) |
| 0.4 | 4.5 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.4 | 1.3 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.4 | 1.8 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.4 | 5.2 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.4 | 1.3 | GO:0045659 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.4 | 1.7 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.4 | 4.2 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.4 | 1.7 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.4 | 5.4 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.4 | 8.6 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.4 | 2.4 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 0.4 | 0.8 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 0.4 | 7.3 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.4 | 6.8 | GO:0072697 | protein localization to cell cortex(GO:0072697) |
| 0.4 | 3.2 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.4 | 4.8 | GO:0051775 | response to redox state(GO:0051775) |
| 0.4 | 2.8 | GO:0034351 | regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.4 | 4.7 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.4 | 3.1 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.4 | 6.2 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.4 | 8.2 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.4 | 11.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.4 | 0.4 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.4 | 1.1 | GO:0070858 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) negative regulation of bile acid biosynthetic process(GO:0070858) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) negative regulation of bile acid metabolic process(GO:1904252) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.4 | 2.2 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.4 | 6.3 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.4 | 3.0 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.4 | 6.6 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.4 | 2.9 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.4 | 1.8 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.4 | 1.4 | GO:0030047 | actin modification(GO:0030047) |
| 0.4 | 2.5 | GO:1900363 | regulation of mRNA polyadenylation(GO:1900363) |
| 0.4 | 47.9 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.4 | 1.1 | GO:0061713 | neural crest cell migration involved in heart formation(GO:0003147) cell migration involved in heart formation(GO:0060974) anterior neural tube closure(GO:0061713) |
| 0.4 | 1.8 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.4 | 2.8 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.3 | 1.7 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.3 | 2.0 | GO:0006207 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) response to oleic acid(GO:0034201) |
| 0.3 | 3.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.3 | 3.4 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.3 | 2.7 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.3 | 1.0 | GO:0036152 | phosphatidylethanolamine acyl-chain remodeling(GO:0036152) |
| 0.3 | 9.6 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.3 | 2.6 | GO:0042262 | DNA protection(GO:0042262) |
| 0.3 | 3.6 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.3 | 2.6 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) |
| 0.3 | 6.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.3 | 14.0 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.3 | 0.6 | GO:0035587 | purinergic receptor signaling pathway(GO:0035587) purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.3 | 6.0 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.3 | 1.8 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.3 | 21.4 | GO:0034644 | cellular response to UV(GO:0034644) |
| 0.3 | 1.1 | GO:0010976 | positive regulation of neuron projection development(GO:0010976) |
| 0.3 | 1.1 | GO:1904640 | response to methionine(GO:1904640) |
| 0.3 | 10.5 | GO:1900739 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.3 | 1.4 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.3 | 2.7 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.3 | 1.3 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.3 | 0.8 | GO:0061316 | regulation of Wnt signaling pathway involved in heart development(GO:0003307) arterial endothelial cell fate commitment(GO:0060844) canonical Wnt signaling pathway involved in heart development(GO:0061316) canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) blood vessel lumenization(GO:0072554) positive regulation of ephrin receptor signaling pathway(GO:1901189) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.3 | 7.4 | GO:0014904 | myotube cell development(GO:0014904) |
| 0.3 | 6.9 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.3 | 1.8 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.3 | 3.4 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.3 | 4.1 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.3 | 0.8 | GO:0045575 | basophil activation(GO:0045575) |
| 0.2 | 2.7 | GO:0006295 | nucleotide-excision repair, preincision complex stabilization(GO:0006293) nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.2 | 1.5 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.2 | 2.2 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.2 | 4.6 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.2 | 3.9 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.2 | 1.7 | GO:1900165 | negative regulation of interleukin-6 secretion(GO:1900165) |
| 0.2 | 1.4 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.2 | 2.9 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.2 | 3.8 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.2 | 0.9 | GO:0044332 | Wnt signaling pathway involved in dorsal/ventral axis specification(GO:0044332) negative regulation of cholesterol efflux(GO:0090370) |
| 0.2 | 1.2 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.2 | 1.8 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.2 | 2.2 | GO:1902969 | mitotic DNA replication(GO:1902969) |
| 0.2 | 1.3 | GO:0002760 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.2 | 0.6 | GO:0043137 | DNA replication, Okazaki fragment processing(GO:0033567) DNA replication, removal of RNA primer(GO:0043137) |
| 0.2 | 6.2 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.2 | 1.9 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.2 | 1.0 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.2 | 1.4 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.2 | 5.0 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.2 | 1.6 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.2 | 3.3 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.2 | 7.4 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.2 | 12.8 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.2 | 0.4 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.2 | 4.7 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.2 | 5.0 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.2 | 0.7 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.2 | 1.1 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.2 | 1.5 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.2 | 0.5 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) |
| 0.2 | 0.9 | GO:0051944 | peptidyl-cysteine methylation(GO:0018125) positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.2 | 0.9 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.2 | 1.5 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.2 | 1.0 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.2 | 2.0 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.2 | 2.9 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.2 | 0.5 | GO:0051685 | maintenance of ER location(GO:0051685) |
| 0.2 | 2.7 | GO:0060850 | regulation of transcription involved in cell fate commitment(GO:0060850) |
| 0.2 | 3.4 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.2 | 5.7 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.2 | 1.3 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.2 | 8.3 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 0.2 | 1.5 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.2 | 0.6 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 0.2 | 1.8 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.2 | 0.8 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.2 | 3.0 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.2 | 1.6 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.2 | 6.8 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.2 | 0.5 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.2 | 1.5 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.1 | 8.2 | GO:0051452 | intracellular pH reduction(GO:0051452) |
| 0.1 | 0.6 | GO:1904796 | regulation of core promoter binding(GO:1904796) |
| 0.1 | 0.7 | GO:1900019 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.1 | 1.0 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.1 | 1.6 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.1 | 3.2 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.1 | 9.5 | GO:2000177 | regulation of neural precursor cell proliferation(GO:2000177) |
| 0.1 | 1.3 | GO:0048714 | positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.1 | 1.1 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.1 | 1.9 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.1 | 0.7 | GO:0045618 | positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.1 | 1.0 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.1 | 0.7 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) glucose 1-phosphate metabolic process(GO:0019255) |
| 0.1 | 1.1 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.1 | 1.6 | GO:0010658 | striated muscle cell apoptotic process(GO:0010658) cardiac muscle cell apoptotic process(GO:0010659) |
| 0.1 | 8.1 | GO:0031100 | organ regeneration(GO:0031100) |
| 0.1 | 1.4 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.1 | 2.8 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.1 | 0.7 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.1 | 7.6 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.1 | 0.9 | GO:0033625 | positive regulation of integrin activation(GO:0033625) |
| 0.1 | 2.5 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.1 | 3.5 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.1 | 1.7 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.1 | 0.8 | GO:0043497 | regulation of protein heterodimerization activity(GO:0043497) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.1 | 5.4 | GO:0050850 | positive regulation of calcium-mediated signaling(GO:0050850) |
| 0.1 | 1.1 | GO:1900376 | regulation of melanin biosynthetic process(GO:0048021) positive regulation of melanin biosynthetic process(GO:0048023) regulation of secondary metabolite biosynthetic process(GO:1900376) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.1 | 2.8 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 5.2 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.1 | 0.6 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.1 | 0.9 | GO:0006772 | thiamine metabolic process(GO:0006772) |
| 0.1 | 0.7 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.1 | 1.1 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.1 | 1.1 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.1 | 2.4 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.1 | 1.7 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.1 | 3.3 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 1.3 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.1 | 0.7 | GO:0009744 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.1 | 2.8 | GO:0048846 | axon extension involved in axon guidance(GO:0048846) neuron projection extension involved in neuron projection guidance(GO:1902284) |
| 0.1 | 6.5 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.1 | 2.9 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 2.8 | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.1 | 1.8 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.1 | 2.5 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.1 | 0.2 | GO:0035630 | bone mineralization involved in bone maturation(GO:0035630) regulation of bone mineralization involved in bone maturation(GO:1900157) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.1 | 4.0 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.1 | 0.9 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.1 | 11.7 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.1 | 0.6 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.1 | 0.5 | GO:0043585 | nose morphogenesis(GO:0043585) alveolar primary septum development(GO:0061143) |
| 0.1 | 0.3 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.1 | 0.3 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.1 | 0.9 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.1 | 0.2 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.1 | 0.5 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 2.3 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 0.6 | GO:0030578 | PML body organization(GO:0030578) |
| 0.1 | 0.3 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.1 | 0.3 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 | 8.2 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.1 | 1.0 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.1 | 0.9 | GO:0002158 | osteoclast proliferation(GO:0002158) pancreatic juice secretion(GO:0030157) |
| 0.1 | 0.8 | GO:0051546 | keratinocyte migration(GO:0051546) |
| 0.1 | 4.2 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.1 | 0.7 | GO:0051437 | positive regulation of ubiquitin-protein ligase activity involved in regulation of mitotic cell cycle transition(GO:0051437) |
| 0.1 | 0.8 | GO:0070828 | heterochromatin organization(GO:0070828) |
| 0.1 | 1.1 | GO:0010629 | negative regulation of gene expression(GO:0010629) |
| 0.1 | 0.6 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 0.9 | GO:0017004 | cytochrome complex assembly(GO:0017004) |
| 0.1 | 1.9 | GO:0030728 | ovulation(GO:0030728) |
| 0.1 | 0.2 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 0.1 | 2.0 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.1 | 7.4 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.1 | 1.2 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.1 | 1.2 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.1 | 0.9 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.1 | 4.2 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.1 | 0.8 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.1 | 1.4 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 1.9 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 1.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 1.7 | GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061418) |
| 0.1 | 1.1 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.1 | 1.4 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.1 | 0.7 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 0.4 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.1 | 0.1 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 0.5 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |
| 0.1 | 0.1 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.1 | 2.7 | GO:0045815 | positive regulation of gene expression, epigenetic(GO:0045815) |
| 0.1 | 1.6 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.1 | 1.8 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) |
| 0.1 | 0.8 | GO:0032461 | positive regulation of protein oligomerization(GO:0032461) |
| 0.1 | 1.3 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.1 | 0.3 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.1 | 0.3 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.1 | 0.2 | GO:0048696 | regulation of collateral sprouting in absence of injury(GO:0048696) negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.1 | 1.3 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 1.8 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.1 | 1.8 | GO:0042246 | tissue regeneration(GO:0042246) |
| 0.1 | 0.2 | GO:1902744 | negative regulation of lamellipodium organization(GO:1902744) negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.1 | 4.2 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.1 | 4.9 | GO:0019827 | stem cell population maintenance(GO:0019827) maintenance of cell number(GO:0098727) |
| 0.1 | 1.8 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.1 | 0.1 | GO:1902994 | regulation of phospholipid efflux(GO:1902994) positive regulation of phospholipid efflux(GO:1902995) |
| 0.1 | 0.2 | GO:0002027 | regulation of heart rate(GO:0002027) |
| 0.1 | 4.2 | GO:0002062 | chondrocyte differentiation(GO:0002062) |
| 0.0 | 1.5 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 3.2 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.0 | 0.2 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.0 | 0.2 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 2.9 | GO:1903311 | regulation of mRNA metabolic process(GO:1903311) |
| 0.0 | 1.9 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.5 | GO:0045008 | depyrimidination(GO:0045008) |
| 0.0 | 0.7 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.0 | 1.1 | GO:0018146 | keratan sulfate biosynthetic process(GO:0018146) |
| 0.0 | 0.9 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 3.6 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 0.8 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.9 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.0 | 0.4 | GO:0042180 | cellular ketone metabolic process(GO:0042180) |
| 0.0 | 0.2 | GO:0033630 | positive regulation of cell adhesion mediated by integrin(GO:0033630) |
| 0.0 | 0.9 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 1.6 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 | 0.5 | GO:1902895 | positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.0 | 0.4 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 2.1 | GO:0006401 | RNA catabolic process(GO:0006401) |
| 0.0 | 0.2 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.2 | GO:0043586 | tongue development(GO:0043586) |
| 0.0 | 2.5 | GO:0001837 | epithelial to mesenchymal transition(GO:0001837) |
| 0.0 | 2.1 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.0 | 0.1 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) detection of muscle stretch(GO:0035995) |
| 0.0 | 0.1 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 0.4 | GO:0042267 | natural killer cell mediated cytotoxicity(GO:0042267) |
| 0.0 | 1.1 | GO:0071158 | positive regulation of cell cycle arrest(GO:0071158) |
| 0.0 | 0.2 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.1 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) negative regulation of leukocyte adhesion to vascular endothelial cell(GO:1904995) |
| 0.0 | 0.3 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.9 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.0 | 0.8 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.4 | GO:0045332 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.0 | 0.6 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.1 | GO:0098502 | DNA dephosphorylation(GO:0098502) |
| 0.0 | 0.9 | GO:2001243 | negative regulation of intrinsic apoptotic signaling pathway(GO:2001243) |
| 0.0 | 1.1 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 0.3 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 0.5 | GO:0048477 | oogenesis(GO:0048477) |
| 0.0 | 0.2 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 1.4 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 0.1 | GO:1902603 | carnitine transmembrane transport(GO:1902603) |
| 0.0 | 0.1 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 0.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.8 | GO:0008360 | regulation of cell shape(GO:0008360) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.6 | 22.8 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 3.8 | 11.3 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 3.6 | 14.3 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 3.1 | 21.4 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 2.8 | 22.6 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 2.7 | 19.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 2.6 | 20.9 | GO:0005683 | U7 snRNP(GO:0005683) |
| 2.4 | 9.6 | GO:0031417 | NatC complex(GO:0031417) |
| 2.2 | 11.0 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 1.9 | 97.3 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 1.9 | 15.4 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 1.9 | 13.4 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 1.9 | 5.6 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 1.8 | 5.5 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 1.7 | 5.2 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 1.7 | 5.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 1.7 | 6.7 | GO:1990423 | RZZ complex(GO:1990423) |
| 1.6 | 4.9 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 1.6 | 4.8 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 1.6 | 4.8 | GO:0043224 | nuclear SCF ubiquitin ligase complex(GO:0043224) |
| 1.6 | 9.4 | GO:0001740 | Barr body(GO:0001740) |
| 1.5 | 6.2 | GO:0001940 | male pronucleus(GO:0001940) |
| 1.5 | 9.2 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 1.5 | 6.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 1.5 | 11.7 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 1.4 | 10.0 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 1.4 | 11.2 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 1.3 | 6.5 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 1.3 | 40.5 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 1.2 | 3.7 | GO:0034455 | t-UTP complex(GO:0034455) |
| 1.2 | 6.2 | GO:0071986 | Ragulator complex(GO:0071986) |
| 1.2 | 2.5 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 1.2 | 5.8 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 1.1 | 6.7 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 1.1 | 12.1 | GO:0000243 | commitment complex(GO:0000243) |
| 1.1 | 5.4 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 1.1 | 3.2 | GO:0070762 | nuclear pore transmembrane ring(GO:0070762) |
| 1.1 | 16.2 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 1.0 | 9.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 1.0 | 11.8 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 1.0 | 2.9 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 1.0 | 20.4 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 1.0 | 3.9 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.9 | 6.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.8 | 5.1 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.8 | 15.1 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.8 | 6.7 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.8 | 2.5 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.8 | 4.0 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.8 | 13.2 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.8 | 5.4 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.8 | 2.3 | GO:0000438 | core TFIIH complex portion of holo TFIIH complex(GO:0000438) |
| 0.8 | 3.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.8 | 9.9 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.8 | 12.9 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.7 | 2.9 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.7 | 54.1 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.7 | 7.9 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.7 | 4.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.7 | 10.5 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.7 | 4.1 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.7 | 4.0 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.6 | 3.8 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.6 | 7.0 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.6 | 3.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.6 | 13.2 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.6 | 6.0 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.6 | 6.0 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.6 | 2.4 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.6 | 5.3 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.6 | 4.6 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.6 | 5.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.6 | 2.2 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.6 | 8.3 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.5 | 3.2 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.5 | 4.3 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.5 | 16.5 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.5 | 4.5 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.5 | 6.5 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.5 | 9.8 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.5 | 3.9 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.5 | 13.1 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.5 | 1.9 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.5 | 2.8 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.5 | 9.4 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.5 | 8.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.5 | 3.2 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.5 | 4.1 | GO:0032039 | integrator complex(GO:0032039) |
| 0.5 | 7.8 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.4 | 4.4 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.4 | 15.6 | GO:0005844 | polysome(GO:0005844) |
| 0.4 | 1.7 | GO:0030891 | VCB complex(GO:0030891) |
| 0.4 | 10.4 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.4 | 5.8 | GO:0010369 | chromocenter(GO:0010369) |
| 0.4 | 4.9 | GO:0043219 | lateral loop(GO:0043219) |
| 0.4 | 22.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.4 | 11.5 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.4 | 4.3 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.4 | 5.4 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.4 | 12.7 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.4 | 3.0 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.4 | 1.5 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.4 | 1.8 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.3 | 3.4 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.3 | 17.0 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.3 | 48.8 | GO:0015934 | large ribosomal subunit(GO:0015934) |
| 0.3 | 4.6 | GO:0038201 | TORC2 complex(GO:0031932) TOR complex(GO:0038201) |
| 0.3 | 3.6 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.3 | 1.3 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.3 | 16.6 | GO:0000786 | nucleosome(GO:0000786) |
| 0.3 | 4.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.3 | 5.5 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.3 | 3.6 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.3 | 2.1 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.3 | 0.9 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.3 | 6.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.3 | 3.4 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.3 | 6.1 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.3 | 4.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.3 | 1.9 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.3 | 3.2 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.3 | 4.2 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.3 | 2.3 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.3 | 5.6 | GO:0031105 | septin complex(GO:0031105) |
| 0.3 | 2.8 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.3 | 2.8 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.3 | 13.9 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.3 | 17.5 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.2 | 3.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.2 | 1.6 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.2 | 1.6 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.2 | 2.3 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.2 | 20.9 | GO:0016459 | myosin complex(GO:0016459) |
| 0.2 | 5.9 | GO:0005761 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
| 0.2 | 13.8 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.2 | 2.2 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.2 | 1.3 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.2 | 5.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.2 | 6.7 | GO:0031430 | M band(GO:0031430) |
| 0.2 | 10.9 | GO:0002102 | podosome(GO:0002102) |
| 0.2 | 1.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.2 | 4.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.2 | 7.0 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.2 | 11.1 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.2 | 3.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.2 | 1.8 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.2 | 33.4 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.2 | 0.4 | GO:0033011 | perinuclear theca(GO:0033011) cytoskeletal calyx(GO:0033150) |
| 0.2 | 3.2 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.2 | 1.9 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.2 | 1.3 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.2 | 5.4 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.2 | 7.9 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.2 | 25.9 | GO:0101002 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.2 | 0.7 | GO:0002177 | manchette(GO:0002177) |
| 0.2 | 5.6 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.2 | 0.7 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.2 | 2.4 | GO:0000785 | chromatin(GO:0000785) |
| 0.2 | 0.8 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.2 | 0.9 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.1 | 1.8 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 2.0 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 8.0 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 1.5 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 6.8 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 0.5 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 1.3 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 0.7 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.1 | 2.5 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.1 | 0.9 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 2.5 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 11.4 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 8.5 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 3.9 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.1 | 5.7 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 3.0 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.1 | 0.4 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 2.4 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.1 | 3.0 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 2.9 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 1.5 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.1 | 3.7 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 8.7 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 3.1 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 5.0 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 4.9 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.1 | 2.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 1.8 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 7.4 | GO:0000922 | spindle pole(GO:0000922) |
| 0.1 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.1 | 2.9 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.1 | 0.9 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.1 | 0.9 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 0.4 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.1 | 0.6 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 1.0 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 0.5 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 20.7 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 0.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 0.4 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 4.7 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.1 | 3.2 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 0.4 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.1 | 5.7 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 0.5 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.0 | 3.3 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.5 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 1.1 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.0 | 5.1 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 48.6 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 6.3 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.2 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.9 | GO:0030666 | endocytic vesicle membrane(GO:0030666) |
| 0.0 | 1.0 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.9 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 3.9 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 13.6 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 1.2 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.7 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.9 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.1 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 1.3 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 1.4 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.4 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.3 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.3 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.8 | 19.2 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 3.6 | 10.9 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 3.4 | 10.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 3.1 | 9.3 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 3.0 | 11.8 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 2.8 | 8.3 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 2.4 | 7.2 | GO:0070538 | oleic acid binding(GO:0070538) |
| 2.4 | 7.1 | GO:0098625 | methylselenol reductase activity(GO:0098625) methylseleninic acid reductase activity(GO:0098626) |
| 2.3 | 23.4 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 2.3 | 6.8 | GO:0017130 | poly(C) RNA binding(GO:0017130) |
| 2.1 | 6.4 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 2.0 | 6.0 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 1.9 | 5.6 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 1.8 | 5.4 | GO:0047086 | phenanthrene 9,10-monooxygenase activity(GO:0018636) ketosteroid monooxygenase activity(GO:0047086) trans-1,2-dihydrobenzene-1,2-diol dehydrogenase activity(GO:0047115) |
| 1.8 | 97.3 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 1.8 | 5.3 | GO:0000035 | acyl binding(GO:0000035) phosphopantetheine binding(GO:0031177) |
| 1.7 | 12.2 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 1.7 | 10.2 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 1.7 | 5.1 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 1.7 | 5.0 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 1.6 | 6.3 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 1.5 | 10.2 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 1.4 | 5.8 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 1.3 | 11.7 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 1.3 | 7.7 | GO:0033989 | 3alpha,7alpha,12alpha-trihydroxy-5beta-cholest-24-enoyl-CoA hydratase activity(GO:0033989) 17-beta-hydroxysteroid dehydrogenase (NAD+) activity(GO:0044594) |
| 1.3 | 17.6 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 1.3 | 5.0 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 1.2 | 12.3 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 1.2 | 3.5 | GO:0070260 | tyrosyl-RNA phosphodiesterase activity(GO:0036317) 5'-tyrosyl-DNA phosphodiesterase activity(GO:0070260) |
| 1.2 | 4.7 | GO:0016936 | galactoside binding(GO:0016936) |
| 1.2 | 12.9 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 1.2 | 11.6 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 1.1 | 6.8 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 1.1 | 4.5 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 1.1 | 208.4 | GO:0003823 | antigen binding(GO:0003823) |
| 1.1 | 25.8 | GO:0008143 | poly(A) binding(GO:0008143) |
| 1.1 | 11.0 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 1.1 | 15.4 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 1.1 | 4.4 | GO:0008466 | glycogenin glucosyltransferase activity(GO:0008466) |
| 1.1 | 4.3 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 1.1 | 22.8 | GO:0035173 | histone kinase activity(GO:0035173) |
| 1.1 | 4.3 | GO:0043515 | kinetochore binding(GO:0043515) |
| 1.0 | 11.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 1.0 | 4.1 | GO:0030760 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 1.0 | 3.0 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 1.0 | 6.7 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.9 | 8.2 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.9 | 4.5 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 0.9 | 3.6 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.9 | 6.2 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.9 | 2.6 | GO:0035529 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) NADH pyrophosphatase activity(GO:0035529) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 0.8 | 22.6 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.8 | 2.5 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.8 | 2.5 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.8 | 2.5 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.8 | 2.5 | GO:0016781 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.8 | 8.1 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.8 | 3.2 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.8 | 4.7 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.8 | 4.6 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.8 | 3.8 | GO:0070363 | mitochondrial light strand promoter sense binding(GO:0070363) |
| 0.8 | 23.7 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.8 | 6.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.7 | 22.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.7 | 2.9 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.7 | 2.2 | GO:0080101 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.7 | 4.3 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.7 | 2.9 | GO:0051996 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.7 | 2.8 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.7 | 7.7 | GO:0001224 | RNA polymerase II transcription cofactor binding(GO:0001224) |
| 0.7 | 2.0 | GO:0004088 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.7 | 6.8 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.7 | 3.3 | GO:0008263 | pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.6 | 3.9 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.6 | 2.6 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.6 | 3.9 | GO:0044388 | small protein activating enzyme binding(GO:0044388) |
| 0.6 | 3.2 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.6 | 10.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.6 | 5.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.6 | 6.2 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.6 | 3.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.6 | 4.9 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.6 | 6.0 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.6 | 5.4 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.6 | 2.4 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.6 | 3.0 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.6 | 4.7 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.6 | 5.8 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.6 | 2.3 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.6 | 6.8 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.6 | 2.3 | GO:0061134 | peptidase regulator activity(GO:0061134) |
| 0.6 | 2.2 | GO:0048763 | HLH domain binding(GO:0043398) calcium-induced calcium release activity(GO:0048763) |
| 0.6 | 2.8 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.5 | 11.7 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.5 | 2.6 | GO:0034597 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.5 | 3.7 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.5 | 3.7 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.5 | 4.6 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.5 | 5.0 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.5 | 5.4 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.5 | 9.8 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.5 | 3.4 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) L-lactate dehydrogenase activity(GO:0004459) |
| 0.5 | 1.4 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.5 | 7.6 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.5 | 4.7 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.5 | 21.0 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.5 | 8.7 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.5 | 33.5 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.4 | 2.2 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.4 | 1.8 | GO:0030305 | beta-glucuronidase activity(GO:0004566) heparanase activity(GO:0030305) |
| 0.4 | 9.2 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.4 | 1.3 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.4 | 2.5 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.4 | 2.4 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.4 | 1.2 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.4 | 3.9 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.4 | 1.2 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) |
| 0.4 | 10.1 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.4 | 2.7 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.4 | 1.1 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.4 | 1.1 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.4 | 14.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.4 | 1.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.4 | 9.6 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.4 | 9.5 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.3 | 11.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.3 | 4.8 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.3 | 1.0 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.3 | 2.6 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.3 | 1.3 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.3 | 5.9 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.3 | 3.2 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.3 | 7.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.3 | 2.9 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.3 | 67.5 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.3 | 0.9 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.3 | 2.8 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.3 | 7.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.3 | 4.9 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.3 | 4.7 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.3 | 5.9 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.3 | 6.1 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.3 | 5.2 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.3 | 2.9 | GO:0050543 | icosanoid binding(GO:0050542) icosatetraenoic acid binding(GO:0050543) arachidonic acid binding(GO:0050544) fatty acid derivative binding(GO:1901567) |
| 0.3 | 0.9 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.3 | 4.0 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.3 | 3.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.3 | 2.7 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.3 | 1.9 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.3 | 0.8 | GO:0052871 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.3 | 2.4 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.3 | 3.2 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.3 | 1.9 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.3 | 0.8 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.3 | 1.1 | GO:0061714 | methotrexate binding(GO:0051870) folic acid receptor activity(GO:0061714) |
| 0.3 | 2.9 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.3 | 23.6 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.3 | 6.0 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.3 | 12.2 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.3 | 28.8 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.3 | 0.8 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.2 | 1.7 | GO:0089720 | caspase binding(GO:0089720) |
| 0.2 | 0.7 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.2 | 2.5 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.2 | 3.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.2 | 3.1 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.2 | 5.2 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.2 | 0.7 | GO:0003983 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.2 | 4.2 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.2 | 3.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.2 | 3.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.2 | 3.0 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.2 | 6.8 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.2 | 2.4 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.2 | 7.8 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.2 | 1.3 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.2 | 3.4 | GO:0005537 | mannose binding(GO:0005537) |
| 0.2 | 5.5 | GO:0071949 | FAD binding(GO:0071949) |
| 0.2 | 2.3 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.2 | 1.0 | GO:0070728 | leucine binding(GO:0070728) |
| 0.2 | 16.8 | GO:0005518 | collagen binding(GO:0005518) |
| 0.2 | 13.2 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.2 | 1.8 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.2 | 9.9 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.2 | 19.2 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.2 | 1.6 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.2 | 3.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.2 | 0.4 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.2 | 1.8 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.2 | 2.3 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.2 | 0.9 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.2 | 8.6 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.2 | 5.2 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.2 | 5.5 | GO:0008308 | voltage-gated anion channel activity(GO:0008308) |
| 0.2 | 0.7 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.2 | 1.4 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.2 | 2.3 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.2 | 0.5 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.2 | 0.7 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.2 | 1.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.2 | 1.6 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.2 | 3.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.2 | 2.5 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.2 | 1.6 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.2 | 1.1 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.1 | 0.6 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 0.9 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 3.6 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 2.8 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.1 | 2.6 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.1 | 4.1 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 4.4 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.1 | 0.5 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.1 | 5.9 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.1 | 0.5 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 0.9 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.1 | 0.8 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.1 | 0.6 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 17.1 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 1.8 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 1.0 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.1 | 0.7 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 3.5 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 2.5 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 1.8 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 2.5 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 3.7 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.1 | 1.5 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 1.4 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 2.0 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 0.6 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.1 | 2.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 2.0 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 10.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 0.2 | GO:0001181 | transcription factor activity, core RNA polymerase I binding(GO:0001181) |
| 0.1 | 3.8 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.1 | 1.4 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 0.2 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.1 | 0.5 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.1 | 1.1 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.1 | 24.8 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 0.1 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.1 | 2.9 | GO:0061650 | ubiquitin conjugating enzyme activity(GO:0061631) ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.1 | 1.1 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 3.0 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 2.4 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 0.3 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.1 | 3.1 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 0.2 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.1 | 0.8 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.1 | 3.1 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.1 | 4.5 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 0.6 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 1.8 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 0.8 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.1 | 0.1 | GO:0030226 | apolipoprotein receptor activity(GO:0030226) apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.1 | 0.6 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 4.3 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.4 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.7 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.1 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 0.6 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.2 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.6 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.2 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.3 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 3.8 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 1.7 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 2.6 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 16.1 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 3.4 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 0.3 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 1.8 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 1.6 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 1.3 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.3 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.1 | GO:0070697 | activin receptor binding(GO:0070697) |
| 0.0 | 1.9 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.3 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.0 | 0.3 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.0 | 0.8 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.1 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 3.4 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 0.1 | GO:0004771 | sterol esterase activity(GO:0004771) methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.0 | 0.6 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.2 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.4 | GO:0016279 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 0.5 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.1 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.3 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.9 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 6.6 | GO:0003712 | transcription cofactor activity(GO:0003712) |
| 0.0 | 0.5 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.1 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 1.0 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 2.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.1 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) polynucleotide phosphatase activity(GO:0098518) |
| 0.0 | 0.5 | GO:0008013 | beta-catenin binding(GO:0008013) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 22.8 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.7 | 46.3 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.6 | 21.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.5 | 21.5 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.5 | 17.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.5 | 10.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.4 | 12.9 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.4 | 3.7 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.4 | 14.0 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.4 | 10.8 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.3 | 16.7 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.3 | 39.0 | PID E2F PATHWAY | E2F transcription factor network |
| 0.3 | 17.5 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.3 | 15.2 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.3 | 11.3 | PID MYC PATHWAY | C-MYC pathway |
| 0.2 | 2.7 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.2 | 14.9 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.2 | 11.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.2 | 7.7 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.2 | 13.3 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.2 | 13.8 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.2 | 6.3 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.2 | 10.6 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 4.5 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 7.5 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.1 | 2.7 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.1 | 7.1 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 2.8 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 1.5 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.1 | 1.8 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 3.0 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 3.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 2.7 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 3.5 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 1.8 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.1 | 3.9 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 2.0 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 3.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.7 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 2.6 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 3.2 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 3.5 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.1 | 11.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 5.0 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.1 | 1.6 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 1.9 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.3 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.7 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 2.4 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.6 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.8 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.9 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.5 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.7 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 2.2 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 3.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.7 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.7 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.1 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 22.8 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 1.3 | 20.9 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 1.2 | 22.4 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 1.1 | 29.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.9 | 23.9 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.9 | 20.6 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.9 | 5.2 | REACTOME APC CDC20 MEDIATED DEGRADATION OF NEK2A | Genes involved in APC-Cdc20 mediated degradation of Nek2A |
| 0.9 | 18.0 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.8 | 13.9 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.8 | 11.0 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.8 | 82.2 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.7 | 10.4 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.7 | 42.3 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.7 | 24.3 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.6 | 16.1 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.6 | 22.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.6 | 4.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.6 | 44.5 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.6 | 6.4 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.5 | 18.4 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.5 | 19.9 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.5 | 9.9 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.5 | 15.1 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.5 | 15.2 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.4 | 7.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.4 | 41.1 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.4 | 8.3 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.4 | 11.2 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.4 | 9.7 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.4 | 6.3 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.3 | 6.3 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.3 | 5.6 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.3 | 12.7 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.3 | 8.6 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.3 | 9.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.3 | 10.2 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.3 | 18.8 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.3 | 6.2 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.3 | 3.2 | REACTOME GLOBAL GENOMIC NER GG NER | Genes involved in Global Genomic NER (GG-NER) |
| 0.3 | 3.8 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.3 | 4.6 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.3 | 5.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.3 | 5.0 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.3 | 6.7 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.3 | 6.0 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.3 | 2.8 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.3 | 5.1 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.2 | 4.7 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.2 | 6.8 | REACTOME INTERFERON SIGNALING | Genes involved in Interferon Signaling |
| 0.2 | 17.1 | REACTOME TRANS GOLGI NETWORK VESICLE BUDDING | Genes involved in trans-Golgi Network Vesicle Budding |
| 0.2 | 19.3 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.2 | 4.5 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.2 | 3.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.2 | 15.6 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.2 | 4.0 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.2 | 23.6 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.2 | 3.8 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.2 | 6.9 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.2 | 4.5 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.2 | 1.8 | REACTOME OPSINS | Genes involved in Opsins |
| 0.2 | 2.4 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.2 | 4.4 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.2 | 2.8 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.2 | 10.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.2 | 3.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 14.7 | REACTOME PROCESSING OF CAPPED INTRON CONTAINING PRE MRNA | Genes involved in Processing of Capped Intron-Containing Pre-mRNA |
| 0.1 | 7.2 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 1.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 4.2 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 1.5 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 2.2 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 5.4 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 3.4 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.1 | 0.9 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.1 | 2.8 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 15.0 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 2.9 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.1 | 4.2 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 2.8 | REACTOME ASSEMBLY OF THE PRE REPLICATIVE COMPLEX | Genes involved in Assembly of the pre-replicative complex |
| 0.1 | 1.9 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 1.7 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.1 | 3.0 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 3.0 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.1 | 10.3 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.1 | 2.5 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 2.0 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.1 | 0.7 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.1 | 0.8 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.1 | 13.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 0.4 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.1 | 2.0 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 1.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 1.6 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.1 | 3.7 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.1 | 2.0 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 1.6 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 1.7 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 4.7 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |
| 0.0 | 0.1 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 1.5 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.6 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 1.0 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.5 | REACTOME RESOLUTION OF AP SITES VIA THE MULTIPLE NUCLEOTIDE PATCH REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the multiple-nucleotide patch replacement pathway |
| 0.0 | 0.9 | REACTOME PI 3K CASCADE | Genes involved in PI-3K cascade |
| 0.0 | 1.0 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.4 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.4 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.2 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.1 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.2 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.2 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |