Project
averaged GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for IRX5
Z-value: 0.82
Transcription factors associated with IRX5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
IRX5
|
ENSG00000176842.10 | iroquois homeobox 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IRX5 | hg19_v2_chr16_+_54964740_54964789 | -0.14 | 3.4e-02 | Click! |
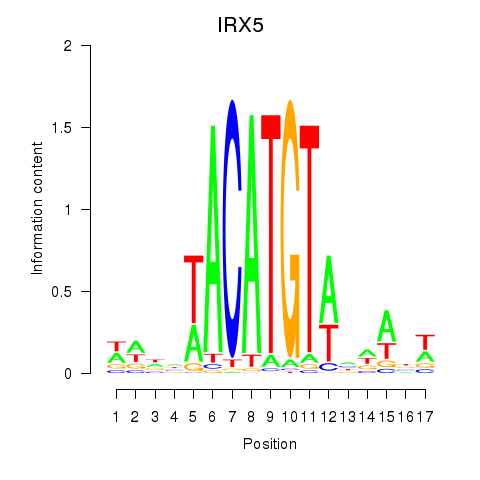
Activity profile of IRX5 motif
Sorted Z-values of IRX5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_73563735 | 13.73 |
ENST00000532192.1
|
RBM25
|
RNA binding motif protein 25 |
| chr14_+_56127989 | 11.98 |
ENST00000555573.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr1_-_197115818 | 10.89 |
ENST00000367409.4
ENST00000294732.7 |
ASPM
|
asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
| chr4_-_104119528 | 9.32 |
ENST00000380026.3
ENST00000503705.1 ENST00000265148.3 |
CENPE
|
centromere protein E, 312kDa |
| chrX_+_49294472 | 8.93 |
ENST00000361446.5
|
GAGE12B
|
G antigen 12B |
| chrX_-_52258669 | 7.88 |
ENST00000441417.1
|
XAGE1A
|
X antigen family, member 1A |
| chrX_+_52513455 | 7.38 |
ENST00000446098.1
|
XAGE1C
|
X antigen family, member 1C |
| chr14_+_35747825 | 7.03 |
ENST00000540871.1
|
PSMA6
|
proteasome (prosome, macropain) subunit, alpha type, 6 |
| chr1_+_220267429 | 6.70 |
ENST00000366922.1
ENST00000302637.5 |
IARS2
|
isoleucyl-tRNA synthetase 2, mitochondrial |
| chr3_+_180586536 | 6.22 |
ENST00000465551.1
|
FXR1
|
fragile X mental retardation, autosomal homolog 1 |
| chrX_+_12809463 | 6.10 |
ENST00000380663.3
ENST00000380668.5 ENST00000398491.2 ENST00000489404.1 |
PRPS2
|
phosphoribosyl pyrophosphate synthetase 2 |
| chr6_+_37012607 | 6.05 |
ENST00000423336.1
|
COX6A1P2
|
cytochrome c oxidase subunit VIa polypeptide 1 pseudogene 2 |
| chr5_+_169010638 | 5.69 |
ENST00000265295.4
ENST00000506574.1 ENST00000515224.1 ENST00000508247.1 ENST00000513941.1 |
SPDL1
|
spindle apparatus coiled-coil protein 1 |
| chr1_-_115124257 | 5.68 |
ENST00000369541.3
|
BCAS2
|
breast carcinoma amplified sequence 2 |
| chr16_-_58585513 | 5.57 |
ENST00000245138.4
ENST00000567285.1 |
CNOT1
|
CCR4-NOT transcription complex, subunit 1 |
| chr12_-_76462713 | 5.55 |
ENST00000552056.1
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr9_+_42704004 | 5.48 |
ENST00000457288.1
|
CBWD7
|
COBW domain containing 7 |
| chr7_-_87856280 | 5.36 |
ENST00000490437.1
ENST00000431660.1 |
SRI
|
sorcin |
| chr3_+_158787041 | 5.32 |
ENST00000471575.1
ENST00000476809.1 ENST00000485419.1 |
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr8_-_67976509 | 5.26 |
ENST00000518747.1
|
COPS5
|
COP9 signalosome subunit 5 |
| chr7_-_87856303 | 5.10 |
ENST00000394641.3
|
SRI
|
sorcin |
| chr4_+_113558612 | 5.09 |
ENST00000505034.1
ENST00000324052.6 |
LARP7
|
La ribonucleoprotein domain family, member 7 |
| chrX_+_52240504 | 4.88 |
ENST00000399805.2
|
XAGE1B
|
X antigen family, member 1B |
| chr1_-_241683001 | 4.87 |
ENST00000366560.3
|
FH
|
fumarate hydratase |
| chr11_+_114310102 | 4.79 |
ENST00000265881.5
|
REXO2
|
RNA exonuclease 2 |
| chr1_-_110284384 | 4.58 |
ENST00000540225.1
|
GSTM3
|
glutathione S-transferase mu 3 (brain) |
| chr5_+_96079240 | 4.55 |
ENST00000515663.1
|
CAST
|
calpastatin |
| chr8_-_101962777 | 4.44 |
ENST00000395951.3
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr2_-_32236002 | 4.39 |
ENST00000404530.1
|
MEMO1
|
mediator of cell motility 1 |
| chr11_+_114310237 | 4.34 |
ENST00000539119.1
|
REXO2
|
RNA exonuclease 2 |
| chr12_-_120315074 | 4.27 |
ENST00000261833.7
ENST00000392521.2 |
CIT
|
citron (rho-interacting, serine/threonine kinase 21) |
| chr15_-_63448973 | 4.06 |
ENST00000462430.1
|
RPS27L
|
ribosomal protein S27-like |
| chr4_+_17616253 | 3.95 |
ENST00000237380.7
|
MED28
|
mediator complex subunit 28 |
| chr15_-_64665911 | 3.85 |
ENST00000606793.1
ENST00000561349.1 ENST00000560278.1 |
CTD-2116N17.1
|
Uncharacterized protein |
| chr9_-_21202204 | 3.72 |
ENST00000239347.3
|
IFNA7
|
interferon, alpha 7 |
| chr4_+_122722466 | 3.72 |
ENST00000243498.5
ENST00000379663.3 ENST00000509800.1 |
EXOSC9
|
exosome component 9 |
| chr2_+_201173667 | 3.61 |
ENST00000409755.3
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr1_-_145826450 | 3.55 |
ENST00000462900.2
|
GPR89A
|
G protein-coupled receptor 89A |
| chr14_-_21737551 | 3.55 |
ENST00000554891.1
ENST00000555883.1 ENST00000553753.1 ENST00000555914.1 ENST00000557336.1 ENST00000555215.1 ENST00000556628.1 ENST00000555137.1 ENST00000556226.1 ENST00000555309.1 ENST00000556142.1 ENST00000554969.1 ENST00000554455.1 ENST00000556513.1 ENST00000557201.1 ENST00000420743.2 ENST00000557768.1 ENST00000553300.1 ENST00000554383.1 ENST00000554539.1 |
HNRNPC
|
heterogeneous nuclear ribonucleoprotein C (C1/C2) |
| chr3_+_52245458 | 3.53 |
ENST00000459884.1
|
ALAS1
|
aminolevulinate, delta-, synthase 1 |
| chr1_-_21377383 | 3.47 |
ENST00000374935.3
|
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3 |
| chr12_+_53848549 | 3.42 |
ENST00000439930.3
ENST00000548933.1 ENST00000562264.1 |
PCBP2
|
poly(rC) binding protein 2 |
| chr14_-_23623577 | 3.27 |
ENST00000422941.2
ENST00000453702.1 |
SLC7A8
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8 |
| chr3_-_81792780 | 3.26 |
ENST00000489715.1
|
GBE1
|
glucan (1,4-alpha-), branching enzyme 1 |
| chr14_+_51706886 | 3.21 |
ENST00000457354.2
|
TMX1
|
thioredoxin-related transmembrane protein 1 |
| chr2_+_160590469 | 3.14 |
ENST00000409591.1
|
MARCH7
|
membrane-associated ring finger (C3HC4) 7, E3 ubiquitin protein ligase |
| chr10_-_96829246 | 3.13 |
ENST00000371270.3
ENST00000535898.1 ENST00000539050.1 |
CYP2C8
|
cytochrome P450, family 2, subfamily C, polypeptide 8 |
| chr6_-_111804393 | 3.10 |
ENST00000368802.3
ENST00000368805.1 |
REV3L
|
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr7_+_77469439 | 3.09 |
ENST00000450574.1
ENST00000416283.2 ENST00000248550.7 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr1_+_53480598 | 3.08 |
ENST00000430330.2
ENST00000408941.3 ENST00000478274.2 ENST00000484100.1 ENST00000435345.2 ENST00000488965.1 |
SCP2
|
sterol carrier protein 2 |
| chr1_-_21377447 | 3.04 |
ENST00000374937.3
ENST00000264211.8 |
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3 |
| chr1_-_3566627 | 3.01 |
ENST00000419924.2
ENST00000270708.7 |
WRAP73
|
WD repeat containing, antisense to TP73 |
| chr4_-_143481822 | 2.94 |
ENST00000510812.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr15_+_41057818 | 2.89 |
ENST00000558467.1
|
GCHFR
|
GTP cyclohydrolase I feedback regulator |
| chr4_+_69962185 | 2.75 |
ENST00000305231.7
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr3_-_52713729 | 2.73 |
ENST00000296302.7
ENST00000356770.4 ENST00000337303.4 ENST00000409057.1 ENST00000410007.1 ENST00000409114.3 ENST00000409767.1 ENST00000423351.1 |
PBRM1
|
polybromo 1 |
| chr4_+_69962212 | 2.73 |
ENST00000508661.1
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr1_-_227505289 | 2.72 |
ENST00000366765.3
|
CDC42BPA
|
CDC42 binding protein kinase alpha (DMPK-like) |
| chr3_-_27498235 | 2.71 |
ENST00000295736.5
ENST00000428386.1 ENST00000428179.1 |
SLC4A7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr7_-_142232071 | 2.63 |
ENST00000390364.3
|
TRBV10-1
|
T cell receptor beta variable 10-1(gene/pseudogene) |
| chr2_+_58655461 | 2.58 |
ENST00000429095.1
ENST00000429664.1 ENST00000452840.1 |
AC007092.1
|
long intergenic non-protein coding RNA 1122 |
| chr12_-_100486668 | 2.57 |
ENST00000550544.1
ENST00000551980.1 ENST00000548045.1 ENST00000545232.2 ENST00000551973.1 |
UHRF1BP1L
|
UHRF1 binding protein 1-like |
| chr1_+_155579979 | 2.55 |
ENST00000452804.2
ENST00000538143.1 ENST00000245564.2 ENST00000368341.4 |
MSTO1
|
misato 1, mitochondrial distribution and morphology regulator |
| chr2_-_183106641 | 2.38 |
ENST00000346717.4
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr7_+_116502527 | 2.33 |
ENST00000361183.3
|
CAPZA2
|
capping protein (actin filament) muscle Z-line, alpha 2 |
| chr3_+_12392971 | 2.29 |
ENST00000287820.6
|
PPARG
|
peroxisome proliferator-activated receptor gamma |
| chr3_-_168865522 | 2.26 |
ENST00000464456.1
|
MECOM
|
MDS1 and EVI1 complex locus |
| chr1_-_3566590 | 2.23 |
ENST00000424367.1
ENST00000378322.3 |
WRAP73
|
WD repeat containing, antisense to TP73 |
| chr19_-_50266580 | 2.18 |
ENST00000246801.3
|
TSKS
|
testis-specific serine kinase substrate |
| chr7_-_16844611 | 2.09 |
ENST00000401412.1
ENST00000419304.2 |
AGR2
|
anterior gradient 2 |
| chr6_-_111804905 | 2.07 |
ENST00000358835.3
ENST00000435970.1 |
REV3L
|
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr2_-_88125471 | 2.03 |
ENST00000398146.3
|
RGPD2
|
RANBP2-like and GRIP domain containing 2 |
| chr4_+_75480629 | 1.99 |
ENST00000380846.3
|
AREGB
|
amphiregulin B |
| chr9_+_71819927 | 1.94 |
ENST00000535702.1
|
TJP2
|
tight junction protein 2 |
| chr19_-_22379753 | 1.94 |
ENST00000397121.2
|
ZNF676
|
zinc finger protein 676 |
| chr6_+_22569784 | 1.93 |
ENST00000510882.2
|
HDGFL1
|
hepatoma derived growth factor-like 1 |
| chrX_-_15353629 | 1.93 |
ENST00000333590.4
ENST00000428964.1 ENST00000542278.1 |
PIGA
|
phosphatidylinositol glycan anchor biosynthesis, class A |
| chr9_+_71820057 | 1.88 |
ENST00000539225.1
|
TJP2
|
tight junction protein 2 |
| chr21_-_34185944 | 1.82 |
ENST00000479548.1
|
C21orf62
|
chromosome 21 open reading frame 62 |
| chr15_-_44116873 | 1.78 |
ENST00000267812.3
|
MFAP1
|
microfibrillar-associated protein 1 |
| chr2_-_231989808 | 1.70 |
ENST00000258400.3
|
HTR2B
|
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chr4_+_76995855 | 1.69 |
ENST00000355810.4
ENST00000349321.3 |
ART3
|
ADP-ribosyltransferase 3 |
| chr7_-_41742697 | 1.58 |
ENST00000242208.4
|
INHBA
|
inhibin, beta A |
| chr16_+_31539197 | 1.58 |
ENST00000564707.1
|
AHSP
|
alpha hemoglobin stabilizing protein |
| chr6_+_42584847 | 1.55 |
ENST00000372883.3
|
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2 |
| chr1_-_62190793 | 1.54 |
ENST00000371177.2
ENST00000606498.1 |
TM2D1
|
TM2 domain containing 1 |
| chr17_+_75447326 | 1.53 |
ENST00000591088.1
|
SEPT9
|
septin 9 |
| chr10_-_35379524 | 1.48 |
ENST00000374751.3
ENST00000374742.1 ENST00000602371.1 |
CUL2
|
cullin 2 |
| chr19_+_35849362 | 1.46 |
ENST00000327809.4
|
FFAR3
|
free fatty acid receptor 3 |
| chr15_+_49170083 | 1.39 |
ENST00000530028.2
|
EID1
|
EP300 interacting inhibitor of differentiation 1 |
| chr14_+_96722539 | 1.39 |
ENST00000553356.1
|
BDKRB1
|
bradykinin receptor B1 |
| chr9_-_125667618 | 1.37 |
ENST00000423239.2
|
RC3H2
|
ring finger and CCCH-type domains 2 |
| chr10_-_14050522 | 1.34 |
ENST00000342409.2
|
FRMD4A
|
FERM domain containing 4A |
| chr1_+_145524891 | 1.31 |
ENST00000369304.3
|
ITGA10
|
integrin, alpha 10 |
| chrX_+_108779004 | 1.29 |
ENST00000218004.1
|
NXT2
|
nuclear transport factor 2-like export factor 2 |
| chr8_+_67976593 | 1.28 |
ENST00000262210.5
ENST00000412460.1 |
CSPP1
|
centrosome and spindle pole associated protein 1 |
| chr2_-_88427568 | 1.26 |
ENST00000393750.3
ENST00000295834.3 |
FABP1
|
fatty acid binding protein 1, liver |
| chr9_-_21166659 | 1.23 |
ENST00000380225.1
|
IFNA21
|
interferon, alpha 21 |
| chr18_+_616672 | 1.23 |
ENST00000338387.7
|
CLUL1
|
clusterin-like 1 (retinal) |
| chr3_-_46068969 | 1.22 |
ENST00000542109.1
ENST00000395946.2 |
XCR1
|
chemokine (C motif) receptor 1 |
| chr1_+_113010056 | 1.20 |
ENST00000369686.5
|
WNT2B
|
wingless-type MMTV integration site family, member 2B |
| chr2_+_173686303 | 1.18 |
ENST00000397087.3
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr21_-_34185989 | 1.17 |
ENST00000487113.1
ENST00000382373.4 |
C21orf62
|
chromosome 21 open reading frame 62 |
| chr15_-_56757329 | 1.14 |
ENST00000260453.3
|
MNS1
|
meiosis-specific nuclear structural 1 |
| chr3_+_173116225 | 1.07 |
ENST00000457714.1
|
NLGN1
|
neuroligin 1 |
| chr4_+_75310851 | 1.06 |
ENST00000395748.3
ENST00000264487.2 |
AREG
|
amphiregulin |
| chr9_+_105757590 | 1.05 |
ENST00000374798.3
ENST00000487798.1 |
CYLC2
|
cylicin, basic protein of sperm head cytoskeleton 2 |
| chr7_+_66386204 | 1.04 |
ENST00000341567.4
ENST00000607045.1 |
TMEM248
|
transmembrane protein 248 |
| chr1_+_196857144 | 1.03 |
ENST00000367416.2
ENST00000251424.4 ENST00000367418.2 |
CFHR4
|
complement factor H-related 4 |
| chr1_+_10509971 | 0.97 |
ENST00000320498.4
|
CORT
|
cortistatin |
| chr15_+_66585879 | 0.92 |
ENST00000319212.4
|
DIS3L
|
DIS3 mitotic control homolog (S. cerevisiae)-like |
| chr4_+_41361616 | 0.91 |
ENST00000513024.1
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr12_-_25102252 | 0.89 |
ENST00000261192.7
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr1_+_196743943 | 0.86 |
ENST00000471440.2
ENST00000391985.3 |
CFHR3
|
complement factor H-related 3 |
| chr17_+_3379284 | 0.84 |
ENST00000263080.2
|
ASPA
|
aspartoacylase |
| chr14_+_22386325 | 0.82 |
ENST00000390439.2
|
TRAV13-2
|
T cell receptor alpha variable 13-2 |
| chr22_-_24096562 | 0.73 |
ENST00000398465.3
|
VPREB3
|
pre-B lymphocyte 3 |
| chr16_+_87636474 | 0.71 |
ENST00000284262.2
|
JPH3
|
junctophilin 3 |
| chr14_+_22993296 | 0.68 |
ENST00000390517.1
|
TRAJ20
|
T cell receptor alpha joining 20 |
| chr21_-_34186006 | 0.68 |
ENST00000490358.1
|
C21orf62
|
chromosome 21 open reading frame 62 |
| chr4_+_70146217 | 0.61 |
ENST00000335568.5
ENST00000511240.1 |
UGT2B28
|
UDP glucuronosyltransferase 2 family, polypeptide B28 |
| chr16_+_31539183 | 0.61 |
ENST00000302312.4
|
AHSP
|
alpha hemoglobin stabilizing protein |
| chr10_+_96698406 | 0.58 |
ENST00000260682.6
|
CYP2C9
|
cytochrome P450, family 2, subfamily C, polypeptide 9 |
| chr3_-_72897545 | 0.57 |
ENST00000325599.8
|
SHQ1
|
SHQ1, H/ACA ribonucleoprotein assembly factor |
| chr10_+_118350468 | 0.56 |
ENST00000358834.4
ENST00000528052.1 ENST00000442761.1 |
PNLIPRP1
|
pancreatic lipase-related protein 1 |
| chr4_-_100356844 | 0.56 |
ENST00000437033.2
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr17_+_58018269 | 0.55 |
ENST00000591035.1
|
RP11-178C3.1
|
Uncharacterized protein |
| chrY_+_23698778 | 0.53 |
ENST00000303902.5
|
RBMY1A1
|
RNA binding motif protein, Y-linked, family 1, member A1 |
| chr1_+_67632083 | 0.52 |
ENST00000347310.5
ENST00000371002.1 |
IL23R
|
interleukin 23 receptor |
| chr8_+_27947746 | 0.51 |
ENST00000521015.1
ENST00000521570.1 |
ELP3
|
elongator acetyltransferase complex subunit 3 |
| chr5_-_64064508 | 0.49 |
ENST00000513458.4
|
SREK1IP1
|
SREK1-interacting protein 1 |
| chr6_-_154677900 | 0.48 |
ENST00000265198.4
ENST00000520261.1 |
IPCEF1
|
interaction protein for cytohesin exchange factors 1 |
| chr1_+_87012753 | 0.48 |
ENST00000370563.3
|
CLCA4
|
chloride channel accessory 4 |
| chr10_+_118350522 | 0.46 |
ENST00000530319.1
ENST00000527980.1 ENST00000471549.1 ENST00000534537.1 |
PNLIPRP1
|
pancreatic lipase-related protein 1 |
| chr17_-_42327236 | 0.43 |
ENST00000399246.2
|
AC003102.1
|
AC003102.1 |
| chr2_+_191792376 | 0.42 |
ENST00000409428.1
ENST00000409215.1 |
GLS
|
glutaminase |
| chr17_-_67264947 | 0.40 |
ENST00000586811.1
|
ABCA5
|
ATP-binding cassette, sub-family A (ABC1), member 5 |
| chr3_+_186383741 | 0.39 |
ENST00000232003.4
|
HRG
|
histidine-rich glycoprotein |
| chr3_-_9994021 | 0.38 |
ENST00000411976.2
ENST00000412055.1 |
PRRT3
|
proline-rich transmembrane protein 3 |
| chr1_-_201438282 | 0.37 |
ENST00000367311.3
ENST00000367309.1 |
PHLDA3
|
pleckstrin homology-like domain, family A, member 3 |
| chr6_+_26217159 | 0.36 |
ENST00000303910.2
|
HIST1H2AE
|
histone cluster 1, H2ae |
| chr12_+_5541267 | 0.35 |
ENST00000423158.3
|
NTF3
|
neurotrophin 3 |
| chr1_-_114355083 | 0.34 |
ENST00000261441.5
|
RSBN1
|
round spermatid basic protein 1 |
| chr12_-_10282742 | 0.34 |
ENST00000298523.5
ENST00000396484.2 ENST00000310002.4 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr11_-_89653576 | 0.32 |
ENST00000420869.1
|
TRIM49D1
|
tripartite motif containing 49D1 |
| chr4_-_75695366 | 0.31 |
ENST00000512743.1
|
BTC
|
betacellulin |
| chr6_+_29429217 | 0.30 |
ENST00000396792.2
|
OR2H1
|
olfactory receptor, family 2, subfamily H, member 1 |
| chr1_+_196743912 | 0.29 |
ENST00000367425.4
|
CFHR3
|
complement factor H-related 3 |
| chr6_+_146920116 | 0.27 |
ENST00000367493.3
|
ADGB
|
androglobin |
| chr13_-_46716969 | 0.26 |
ENST00000435666.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chrX_-_138790348 | 0.25 |
ENST00000414978.1
ENST00000519895.1 |
MCF2
|
MCF.2 cell line derived transforming sequence |
| chr14_+_72052983 | 0.23 |
ENST00000358550.2
|
SIPA1L1
|
signal-induced proliferation-associated 1 like 1 |
| chr8_-_108510224 | 0.20 |
ENST00000517746.1
ENST00000297450.3 |
ANGPT1
|
angiopoietin 1 |
| chr16_-_20709066 | 0.18 |
ENST00000520010.1
|
ACSM1
|
acyl-CoA synthetase medium-chain family member 1 |
| chr8_+_125463048 | 0.17 |
ENST00000328599.3
|
TRMT12
|
tRNA methyltransferase 12 homolog (S. cerevisiae) |
| chr17_+_41561317 | 0.16 |
ENST00000540306.1
ENST00000262415.3 ENST00000605777.1 |
DHX8
|
DEAH (Asp-Glu-Ala-His) box polypeptide 8 |
| chr12_-_102224704 | 0.16 |
ENST00000299314.7
|
GNPTAB
|
N-acetylglucosamine-1-phosphate transferase, alpha and beta subunits |
| chr6_-_26216872 | 0.14 |
ENST00000244601.3
|
HIST1H2BG
|
histone cluster 1, H2bg |
| chr21_+_38792602 | 0.12 |
ENST00000398960.2
ENST00000398956.2 |
DYRK1A
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A |
| chr11_-_104905840 | 0.10 |
ENST00000526568.1
ENST00000393136.4 ENST00000531166.1 ENST00000534497.1 ENST00000527979.1 ENST00000446369.1 ENST00000353247.5 ENST00000528974.1 ENST00000533400.1 ENST00000525825.1 ENST00000436863.3 |
CASP1
|
caspase 1, apoptosis-related cysteine peptidase |
| chr13_+_24844819 | 0.09 |
ENST00000399949.2
|
SPATA13
|
spermatogenesis associated 13 |
| chr18_+_6729725 | 0.08 |
ENST00000400091.2
ENST00000583410.1 ENST00000584387.1 |
ARHGAP28
|
Rho GTPase activating protein 28 |
| chrX_-_33229636 | 0.08 |
ENST00000357033.4
|
DMD
|
dystrophin |
| chrX_+_99899180 | 0.07 |
ENST00000373004.3
|
SRPX2
|
sushi-repeat containing protein, X-linked 2 |
| chr1_-_116383738 | 0.05 |
ENST00000320238.3
|
NHLH2
|
nescient helix loop helix 2 |
| chr12_-_21757774 | 0.05 |
ENST00000261195.2
|
GYS2
|
glycogen synthase 2 (liver) |
| chr3_+_171561127 | 0.03 |
ENST00000334567.5
ENST00000450693.1 |
TMEM212
|
transmembrane protein 212 |
| chr11_+_22688150 | 0.02 |
ENST00000454584.2
|
GAS2
|
growth arrest-specific 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of IRX5
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 6.7 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 1.8 | 10.9 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 1.6 | 4.9 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 1.6 | 9.3 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 1.5 | 10.5 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 1.2 | 6.1 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 1.0 | 2.9 | GO:0043105 | regulation of GTP cyclohydrolase I activity(GO:0043095) negative regulation of GTP cyclohydrolase I activity(GO:0043105) |
| 0.9 | 3.7 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.9 | 4.6 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.8 | 0.8 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.8 | 5.6 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.6 | 3.1 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.6 | 2.3 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.5 | 1.6 | GO:0060278 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.5 | 5.7 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.5 | 1.4 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.4 | 3.1 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.4 | 3.8 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.4 | 1.5 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.4 | 1.1 | GO:0099543 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.3 | 7.3 | GO:1903894 | regulation of IRE1-mediated unfolded protein response(GO:1903894) |
| 0.3 | 1.7 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.3 | 4.6 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.3 | 6.8 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.3 | 9.7 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.3 | 4.4 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.3 | 4.0 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.3 | 3.5 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.3 | 1.5 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 0.2 | 5.0 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.2 | 5.5 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.2 | 6.2 | GO:2000637 | positive regulation of posttranscriptional gene silencing(GO:0060148) positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.2 | 3.4 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.2 | 12.6 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.2 | 1.2 | GO:0060492 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) positive regulation of branching involved in lung morphogenesis(GO:0061047) |
| 0.2 | 0.6 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.2 | 5.2 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.2 | 3.6 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.2 | 4.1 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.2 | 1.1 | GO:0042695 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.1 | 0.9 | GO:0009082 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.1 | 0.6 | GO:2000232 | regulation of rRNA processing(GO:2000232) |
| 0.1 | 2.7 | GO:0060117 | auditory receptor cell development(GO:0060117) |
| 0.1 | 2.6 | GO:0048311 | mitochondrion distribution(GO:0048311) |
| 0.1 | 2.4 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 2.2 | GO:0020027 | hemoglobin metabolic process(GO:0020027) |
| 0.1 | 0.5 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.1 | 1.3 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.1 | 3.1 | GO:0002643 | regulation of tolerance induction(GO:0002643) |
| 0.1 | 0.6 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.1 | 5.5 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 0.4 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.1 | 1.0 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.1 | 1.9 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.1 | 1.1 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.1 | 0.4 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.1 | 3.6 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.1 | 2.3 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.1 | 6.5 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.1 | 8.5 | GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061418) |
| 0.1 | 2.3 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.1 | 2.3 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.1 | 2.0 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.1 | 0.2 | GO:0018874 | benzoate metabolic process(GO:0018874) butyrate metabolic process(GO:0019605) |
| 0.1 | 3.3 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 1.7 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 12.0 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 1.3 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.2 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 3.2 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 1.4 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 2.9 | GO:0043647 | inositol phosphate metabolic process(GO:0043647) |
| 0.0 | 0.5 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.2 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.0 | 4.3 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.3 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 3.3 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 2.2 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.1 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.0 | 0.1 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 | 0.8 | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.0 | 1.0 | GO:0006629 | lipid metabolic process(GO:0006629) |
| 0.0 | 0.3 | GO:0035810 | positive regulation of urine volume(GO:0035810) |
| 0.0 | 0.4 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.0 | 1.2 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 3.3 | GO:0032886 | regulation of microtubule-based process(GO:0032886) |
| 0.0 | 1.4 | GO:0035065 | regulation of histone acetylation(GO:0035065) |
| 0.0 | 2.7 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 0.4 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.1 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 10.9 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 1.5 | 6.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 1.5 | 10.5 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 1.3 | 15.0 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.6 | 7.0 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.6 | 5.6 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.5 | 1.6 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.4 | 1.3 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.4 | 4.6 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.4 | 5.7 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.4 | 5.2 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.4 | 1.1 | GO:0033150 | perinuclear theca(GO:0033011) cytoskeletal calyx(GO:0033150) |
| 0.3 | 4.9 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.3 | 3.7 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.3 | 6.5 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.3 | 1.5 | GO:0030891 | VCB complex(GO:0030891) |
| 0.2 | 5.3 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.2 | 1.9 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.2 | 0.7 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.2 | 1.3 | GO:0045179 | apical cortex(GO:0045179) |
| 0.2 | 2.2 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.2 | 2.3 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 0.5 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.1 | 5.8 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 4.0 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 3.6 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 9.1 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 0.6 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 1.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 1.8 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 2.7 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.1 | 2.7 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 3.6 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.1 | 12.9 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 1.5 | GO:0031105 | septin complex(GO:0031105) |
| 0.1 | 0.6 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.1 | 4.1 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 7.0 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 3.1 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 4.3 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 11.5 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 6.9 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.3 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 2.8 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 3.4 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.1 | GO:0097179 | protease inhibitor complex(GO:0097179) |
| 0.0 | 0.4 | GO:0036019 | endolysosome(GO:0036019) |
| 0.0 | 1.1 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 2.1 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 8.2 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 1.1 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.6 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 6.0 | GO:0030529 | intracellular ribonucleoprotein complex(GO:0030529) |
| 0.0 | 2.3 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 2.5 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 15.0 | GO:0043515 | kinetochore binding(GO:0043515) |
| 2.2 | 6.7 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 1.2 | 3.5 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 1.0 | 3.1 | GO:0070538 | oleic acid binding(GO:0070538) |
| 1.0 | 6.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 1.0 | 2.9 | GO:0044549 | GTP cyclohydrolase binding(GO:0044549) |
| 0.8 | 4.6 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.7 | 3.7 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.6 | 2.9 | GO:0052828 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.6 | 3.4 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.6 | 6.2 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) RNA strand annealing activity(GO:0033592) |
| 0.5 | 1.6 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.5 | 5.3 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.5 | 19.0 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.5 | 1.4 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.4 | 2.2 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.4 | 5.0 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.3 | 2.4 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.3 | 1.7 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.3 | 1.5 | GO:0070728 | leucine binding(GO:0070728) |
| 0.3 | 6.5 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.3 | 2.3 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.3 | 3.6 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.3 | 10.6 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.3 | 5.2 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.3 | 4.6 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.3 | 1.3 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.3 | 4.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.2 | 7.0 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.2 | 1.3 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.2 | 12.0 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.2 | 2.3 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.2 | 1.9 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.2 | 6.1 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.2 | 3.2 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.2 | 2.7 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.9 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.1 | 1.7 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.1 | 0.6 | GO:0035276 | aldehyde oxidase activity(GO:0004031) ethanol binding(GO:0035276) |
| 0.1 | 0.5 | GO:0042020 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.1 | 0.8 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 3.6 | GO:0008308 | voltage-gated anion channel activity(GO:0008308) |
| 0.1 | 1.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.5 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.1 | 3.8 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 0.3 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.1 | 0.4 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 15.6 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.1 | 4.3 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.1 | 0.2 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 1.0 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 2.3 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.5 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.7 | GO:0015278 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 0.4 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.5 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 1.0 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 10.0 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 2.2 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 1.4 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 1.2 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 1.2 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 3.3 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.0 | 1.0 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.2 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.4 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 2.2 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 2.7 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 1.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.7 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 4.4 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 4.7 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 4.3 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 2.3 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 5.0 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 3.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.6 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 1.3 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 2.7 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 3.8 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 4.2 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.4 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 1.3 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.2 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.5 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.3 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.7 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.3 | 6.7 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.2 | 4.4 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.2 | 6.1 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.2 | 3.7 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.2 | 4.9 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.2 | 3.8 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.2 | 2.9 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.2 | 3.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.2 | 3.5 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.2 | 5.0 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.1 | 2.9 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 1.7 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 1.6 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 7.0 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.1 | 3.4 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 1.9 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 1.5 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 0.6 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 6.5 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 2.4 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 2.3 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 1.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 3.3 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 1.0 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.9 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 4.8 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.0 | 1.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 2.7 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 3.6 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 5.1 | REACTOME METABOLISM OF CARBOHYDRATES | Genes involved in Metabolism of carbohydrates |
| 0.0 | 2.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.3 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 1.2 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 2.5 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 1.3 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |