Project
averaged GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
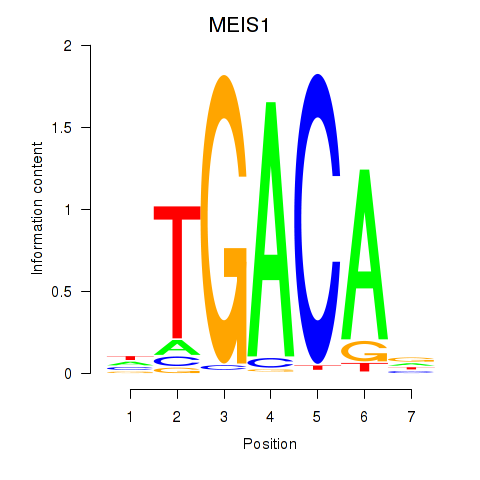
Results for MEIS1
Z-value: 1.04
Transcription factors associated with MEIS1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MEIS1
|
ENSG00000143995.15 | Meis homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MEIS1 | hg19_v2_chr2_+_66662690_66662711 | -0.13 | 4.9e-02 | Click! |
Activity profile of MEIS1 motif
Sorted Z-values of MEIS1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_183892635 | 11.03 |
ENST00000427072.1
ENST00000411763.2 ENST00000292807.5 ENST00000448139.1 ENST00000455925.1 |
AP2M1
|
adaptor-related protein complex 2, mu 1 subunit |
| chr17_-_29648761 | 10.80 |
ENST00000247270.3
ENST00000462804.2 |
EVI2A
|
ecotropic viral integration site 2A |
| chr10_+_54074033 | 10.01 |
ENST00000373970.3
|
DKK1
|
dickkopf WNT signaling pathway inhibitor 1 |
| chr2_+_201170770 | 9.27 |
ENST00000409988.3
ENST00000409385.1 |
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr1_-_113247543 | 9.22 |
ENST00000414971.1
ENST00000534717.1 |
RHOC
|
ras homolog family member C |
| chr3_+_159570722 | 9.11 |
ENST00000482804.1
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr1_+_26605618 | 8.72 |
ENST00000270792.5
|
SH3BGRL3
|
SH3 domain binding glutamic acid-rich protein like 3 |
| chr16_+_75600247 | 8.67 |
ENST00000037243.2
ENST00000565057.1 ENST00000563744.1 |
GABARAPL2
|
GABA(A) receptor-associated protein-like 2 |
| chr11_+_35198243 | 8.51 |
ENST00000528455.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr4_+_88896819 | 7.91 |
ENST00000237623.7
ENST00000395080.3 ENST00000508233.1 ENST00000360804.4 |
SPP1
|
secreted phosphoprotein 1 |
| chr2_+_201936707 | 7.89 |
ENST00000433898.1
ENST00000454214.1 |
NDUFB3
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3, 12kDa |
| chr5_-_175843569 | 7.81 |
ENST00000310418.4
ENST00000345807.2 |
CLTB
|
clathrin, light chain B |
| chr12_-_56694142 | 7.77 |
ENST00000550655.1
ENST00000548567.1 ENST00000551430.2 ENST00000351328.3 |
CS
|
citrate synthase |
| chr8_-_120651020 | 7.41 |
ENST00000522826.1
ENST00000520066.1 ENST00000259486.6 ENST00000075322.6 |
ENPP2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr3_-_49066811 | 6.87 |
ENST00000442157.1
ENST00000326739.4 |
IMPDH2
|
IMP (inosine 5'-monophosphate) dehydrogenase 2 |
| chr1_+_169079823 | 6.81 |
ENST00000367813.3
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr20_+_44036900 | 6.63 |
ENST00000443296.1
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chrX_+_118602363 | 6.62 |
ENST00000317881.8
|
SLC25A5
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 |
| chr17_+_49243176 | 6.57 |
ENST00000513177.1
|
NME1-NME2
|
NME1-NME2 readthrough |
| chr1_+_87797351 | 6.44 |
ENST00000370542.1
|
LMO4
|
LIM domain only 4 |
| chr2_-_175870085 | 6.38 |
ENST00000409156.3
|
CHN1
|
chimerin 1 |
| chr8_-_54755459 | 6.28 |
ENST00000524234.1
ENST00000521275.1 ENST00000396774.2 |
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H |
| chr8_-_54755789 | 6.27 |
ENST00000359530.2
|
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H |
| chr1_+_155657737 | 6.26 |
ENST00000471642.2
ENST00000471214.1 |
DAP3
|
death associated protein 3 |
| chr11_-_122929699 | 6.17 |
ENST00000526686.1
|
HSPA8
|
heat shock 70kDa protein 8 |
| chr3_-_127541194 | 6.13 |
ENST00000453507.2
|
MGLL
|
monoglyceride lipase |
| chrX_-_13835461 | 6.10 |
ENST00000316715.4
ENST00000356942.5 |
GPM6B
|
glycoprotein M6B |
| chr22_-_43355858 | 6.04 |
ENST00000402229.1
ENST00000407585.1 ENST00000453079.1 |
PACSIN2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr12_-_118498958 | 5.91 |
ENST00000315436.3
|
WSB2
|
WD repeat and SOCS box containing 2 |
| chr7_+_98923505 | 5.89 |
ENST00000432884.2
ENST00000262942.5 |
ARPC1A
|
actin related protein 2/3 complex, subunit 1A, 41kDa |
| chr1_+_165796753 | 5.78 |
ENST00000367879.4
|
UCK2
|
uridine-cytidine kinase 2 |
| chr7_+_149571045 | 5.73 |
ENST00000479613.1
ENST00000606024.1 ENST00000464662.1 ENST00000425642.2 |
ATP6V0E2
|
ATPase, H+ transporting V0 subunit e2 |
| chr3_+_158787041 | 5.60 |
ENST00000471575.1
ENST00000476809.1 ENST00000485419.1 |
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chrX_+_51636629 | 5.60 |
ENST00000375722.1
ENST00000326587.7 ENST00000375695.2 |
MAGED1
|
melanoma antigen family D, 1 |
| chr2_-_170681324 | 5.58 |
ENST00000409340.1
|
METTL5
|
methyltransferase like 5 |
| chr11_-_64052111 | 5.57 |
ENST00000394532.3
ENST00000394531.3 ENST00000309032.3 |
BAD
|
BCL2-associated agonist of cell death |
| chr18_-_21891460 | 5.57 |
ENST00000357041.4
|
OSBPL1A
|
oxysterol binding protein-like 1A |
| chr16_+_31044413 | 5.54 |
ENST00000394998.1
|
STX4
|
syntaxin 4 |
| chr5_+_49962495 | 5.51 |
ENST00000515175.1
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr16_+_4674814 | 5.49 |
ENST00000415496.1
ENST00000587747.1 ENST00000399577.5 ENST00000588994.1 ENST00000586183.1 |
MGRN1
|
mahogunin ring finger 1, E3 ubiquitin protein ligase |
| chr1_+_156163880 | 5.49 |
ENST00000359511.4
ENST00000423538.2 |
SLC25A44
|
solute carrier family 25, member 44 |
| chr17_-_79481666 | 5.46 |
ENST00000575659.1
|
ACTG1
|
actin, gamma 1 |
| chr19_-_47353547 | 5.42 |
ENST00000601498.1
|
AP2S1
|
adaptor-related protein complex 2, sigma 1 subunit |
| chr2_+_172544294 | 5.41 |
ENST00000358002.6
ENST00000435234.1 ENST00000443458.1 ENST00000412370.1 |
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2 |
| chr1_+_10271674 | 5.41 |
ENST00000377086.1
|
KIF1B
|
kinesin family member 1B |
| chr16_-_67493110 | 5.36 |
ENST00000602876.1
|
ATP6V0D1
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d1 |
| chr1_+_171810606 | 5.32 |
ENST00000358155.4
ENST00000367733.2 ENST00000355305.5 ENST00000367731.1 |
DNM3
|
dynamin 3 |
| chr16_+_31044812 | 5.30 |
ENST00000313843.3
|
STX4
|
syntaxin 4 |
| chr8_+_11666649 | 5.26 |
ENST00000528643.1
ENST00000525777.1 |
FDFT1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr6_+_63921351 | 5.24 |
ENST00000370659.1
|
FKBP1C
|
FK506 binding protein 1C |
| chr12_-_76462713 | 5.24 |
ENST00000552056.1
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr12_-_118810688 | 5.23 |
ENST00000542532.1
ENST00000392533.3 |
TAOK3
|
TAO kinase 3 |
| chr3_-_47934234 | 5.23 |
ENST00000420772.2
|
MAP4
|
microtubule-associated protein 4 |
| chr2_+_201170596 | 5.10 |
ENST00000439084.1
ENST00000409718.1 |
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr5_+_92919043 | 4.90 |
ENST00000327111.3
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr6_+_39760783 | 4.90 |
ENST00000398904.2
ENST00000538976.1 |
DAAM2
|
dishevelled associated activator of morphogenesis 2 |
| chr12_-_123756781 | 4.83 |
ENST00000544658.1
|
CDK2AP1
|
cyclin-dependent kinase 2 associated protein 1 |
| chr15_-_34394008 | 4.83 |
ENST00000527822.1
ENST00000528949.1 |
EMC7
|
ER membrane protein complex subunit 7 |
| chr16_+_14927538 | 4.79 |
ENST00000287667.7
|
NOMO1
|
NODAL modulator 1 |
| chrX_-_92928557 | 4.74 |
ENST00000373079.3
ENST00000475430.2 |
NAP1L3
|
nucleosome assembly protein 1-like 3 |
| chr5_-_175843524 | 4.70 |
ENST00000502877.1
|
CLTB
|
clathrin, light chain B |
| chr17_+_1944790 | 4.70 |
ENST00000575162.1
|
DPH1
|
diphthamide biosynthesis 1 |
| chr12_+_79439405 | 4.64 |
ENST00000552744.1
|
SYT1
|
synaptotagmin I |
| chr9_-_124922021 | 4.61 |
ENST00000537618.1
ENST00000373768.3 |
NDUFA8
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 8, 19kDa |
| chr1_-_155211017 | 4.59 |
ENST00000536770.1
ENST00000368373.3 |
GBA
|
glucosidase, beta, acid |
| chr11_+_35198118 | 4.59 |
ENST00000525211.1
ENST00000526000.1 ENST00000279452.6 ENST00000527889.1 |
CD44
|
CD44 molecule (Indian blood group) |
| chr13_-_67802549 | 4.59 |
ENST00000328454.5
ENST00000377865.2 |
PCDH9
|
protocadherin 9 |
| chr1_-_154909329 | 4.58 |
ENST00000368467.3
|
PMVK
|
phosphomevalonate kinase |
| chr8_-_102216925 | 4.58 |
ENST00000517844.1
|
ZNF706
|
zinc finger protein 706 |
| chr17_+_38137050 | 4.52 |
ENST00000264639.4
|
PSMD3
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 3 |
| chr14_+_90722839 | 4.50 |
ENST00000261303.8
ENST00000553835.1 |
PSMC1
|
proteasome (prosome, macropain) 26S subunit, ATPase, 1 |
| chr11_+_35211429 | 4.47 |
ENST00000525688.1
ENST00000278385.6 ENST00000533222.1 |
CD44
|
CD44 molecule (Indian blood group) |
| chr22_-_39096661 | 4.45 |
ENST00000216039.5
|
JOSD1
|
Josephin domain containing 1 |
| chr3_-_58419537 | 4.42 |
ENST00000474765.1
ENST00000485460.1 ENST00000302746.6 ENST00000383714.4 |
PDHB
|
pyruvate dehydrogenase (lipoamide) beta |
| chr14_-_58893832 | 4.39 |
ENST00000556007.2
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chrX_+_155110956 | 4.38 |
ENST00000286448.6
ENST00000262640.6 ENST00000460621.1 |
VAMP7
|
vesicle-associated membrane protein 7 |
| chr14_+_104029278 | 4.37 |
ENST00000472726.2
ENST00000409074.2 ENST00000440963.1 ENST00000556253.2 ENST00000247618.4 |
RP11-73M18.2
APOPT1
|
Kinesin light chain 1 apoptogenic 1, mitochondrial |
| chr17_-_76899275 | 4.35 |
ENST00000322630.2
ENST00000586713.1 |
DDC8
|
Protein DDC8 homolog |
| chr17_+_38137073 | 4.30 |
ENST00000541736.1
|
PSMD3
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 3 |
| chr14_+_58666824 | 4.29 |
ENST00000254286.4
|
ACTR10
|
actin-related protein 10 homolog (S. cerevisiae) |
| chr8_+_61429728 | 4.28 |
ENST00000529579.1
|
RAB2A
|
RAB2A, member RAS oncogene family |
| chr2_-_2334888 | 4.27 |
ENST00000428368.2
ENST00000399161.2 |
MYT1L
|
myelin transcription factor 1-like |
| chr6_-_46293378 | 4.25 |
ENST00000330430.6
|
RCAN2
|
regulator of calcineurin 2 |
| chr11_+_35211511 | 4.24 |
ENST00000524922.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr6_-_75960024 | 4.23 |
ENST00000370081.2
|
COX7A2
|
cytochrome c oxidase subunit VIIa polypeptide 2 (liver) |
| chr6_-_33385854 | 4.23 |
ENST00000488478.1
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr9_+_71820057 | 4.23 |
ENST00000539225.1
|
TJP2
|
tight junction protein 2 |
| chr18_+_3449695 | 4.21 |
ENST00000343820.5
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr4_-_102268628 | 4.21 |
ENST00000323055.6
ENST00000512215.1 ENST00000394854.3 |
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr15_-_34394119 | 4.21 |
ENST00000256545.4
|
EMC7
|
ER membrane protein complex subunit 7 |
| chr5_+_122110691 | 4.20 |
ENST00000379516.2
ENST00000505934.1 ENST00000514949.1 |
SNX2
|
sorting nexin 2 |
| chr9_+_71819927 | 4.19 |
ENST00000535702.1
|
TJP2
|
tight junction protein 2 |
| chr2_+_219081817 | 4.13 |
ENST00000315717.5
ENST00000420104.1 ENST00000295685.10 |
ARPC2
|
actin related protein 2/3 complex, subunit 2, 34kDa |
| chr8_-_120685608 | 4.13 |
ENST00000427067.2
|
ENPP2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr17_-_53809473 | 4.10 |
ENST00000575734.1
|
TMEM100
|
transmembrane protein 100 |
| chr1_-_161087802 | 4.09 |
ENST00000368010.3
|
PFDN2
|
prefoldin subunit 2 |
| chr4_+_41614909 | 4.01 |
ENST00000509454.1
ENST00000396595.3 ENST00000381753.4 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr22_+_41258250 | 3.97 |
ENST00000544094.1
|
XPNPEP3
|
X-prolyl aminopeptidase (aminopeptidase P) 3, putative |
| chr14_+_24540731 | 3.97 |
ENST00000558859.1
ENST00000559197.1 ENST00000560828.1 ENST00000216775.2 ENST00000560884.1 |
CPNE6
|
copine VI (neuronal) |
| chr2_+_102456277 | 3.96 |
ENST00000421882.1
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chrX_+_47082408 | 3.95 |
ENST00000518022.1
ENST00000276052.6 |
CDK16
|
cyclin-dependent kinase 16 |
| chr5_+_140864649 | 3.94 |
ENST00000306593.1
|
PCDHGC4
|
protocadherin gamma subfamily C, 4 |
| chr12_+_12870055 | 3.94 |
ENST00000228872.4
|
CDKN1B
|
cyclin-dependent kinase inhibitor 1B (p27, Kip1) |
| chr14_+_24540046 | 3.92 |
ENST00000397016.2
ENST00000537691.1 ENST00000560356.1 ENST00000558450.1 |
CPNE6
|
copine VI (neuronal) |
| chrX_+_70503037 | 3.91 |
ENST00000535149.1
|
NONO
|
non-POU domain containing, octamer-binding |
| chr5_+_125758865 | 3.91 |
ENST00000542322.1
ENST00000544396.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr8_-_141931287 | 3.89 |
ENST00000517887.1
|
PTK2
|
protein tyrosine kinase 2 |
| chr12_+_19358228 | 3.88 |
ENST00000424268.1
ENST00000543806.1 |
PLEKHA5
|
pleckstrin homology domain containing, family A member 5 |
| chr11_-_31832862 | 3.88 |
ENST00000379115.4
|
PAX6
|
paired box 6 |
| chr8_-_18744528 | 3.87 |
ENST00000523619.1
|
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chr15_+_80364901 | 3.82 |
ENST00000560228.1
ENST00000559835.1 ENST00000559775.1 ENST00000558688.1 ENST00000560392.1 ENST00000560976.1 ENST00000558272.1 ENST00000558390.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr5_+_125758813 | 3.75 |
ENST00000285689.3
ENST00000515200.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr20_+_4666882 | 3.75 |
ENST00000379440.4
ENST00000430350.2 |
PRNP
|
prion protein |
| chr11_-_31832785 | 3.74 |
ENST00000241001.8
|
PAX6
|
paired box 6 |
| chr5_+_32585605 | 3.72 |
ENST00000265073.4
ENST00000515355.1 ENST00000502897.1 ENST00000510442.1 |
SUB1
|
SUB1 homolog (S. cerevisiae) |
| chr3_+_35722487 | 3.71 |
ENST00000441454.1
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr7_+_95401851 | 3.70 |
ENST00000447467.2
|
DYNC1I1
|
dynein, cytoplasmic 1, intermediate chain 1 |
| chr7_+_94285637 | 3.69 |
ENST00000482108.1
ENST00000488574.1 |
PEG10
|
paternally expressed 10 |
| chr6_+_30297306 | 3.69 |
ENST00000420746.1
ENST00000513556.1 |
TRIM39
TRIM39-RPP21
|
tripartite motif containing 39 TRIM39-RPP21 readthrough |
| chr11_+_71938925 | 3.66 |
ENST00000538751.1
|
INPPL1
|
inositol polyphosphate phosphatase-like 1 |
| chr6_-_33385655 | 3.66 |
ENST00000440279.3
ENST00000607266.1 |
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr17_-_685493 | 3.62 |
ENST00000536578.1
ENST00000301328.5 ENST00000576419.1 |
GLOD4
|
glyoxalase domain containing 4 |
| chr3_+_184033135 | 3.61 |
ENST00000424196.1
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr15_-_37390482 | 3.61 |
ENST00000559085.1
ENST00000397624.3 |
MEIS2
|
Meis homeobox 2 |
| chr3_-_12705600 | 3.60 |
ENST00000542177.1
ENST00000442415.2 ENST00000251849.4 |
RAF1
|
v-raf-1 murine leukemia viral oncogene homolog 1 |
| chr5_-_150466692 | 3.60 |
ENST00000315050.7
ENST00000523338.1 ENST00000522100.1 |
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr6_+_111195973 | 3.58 |
ENST00000368885.3
ENST00000368882.3 ENST00000451850.2 ENST00000368877.5 |
AMD1
|
adenosylmethionine decarboxylase 1 |
| chr1_-_43638168 | 3.56 |
ENST00000431635.2
|
EBNA1BP2
|
EBNA1 binding protein 2 |
| chr12_-_123756687 | 3.56 |
ENST00000261692.2
|
CDK2AP1
|
cyclin-dependent kinase 2 associated protein 1 |
| chr15_-_37392086 | 3.53 |
ENST00000561208.1
|
MEIS2
|
Meis homeobox 2 |
| chr12_-_76953573 | 3.53 |
ENST00000549646.1
ENST00000550628.1 ENST00000553139.1 ENST00000261183.3 ENST00000393250.4 |
OSBPL8
|
oxysterol binding protein-like 8 |
| chr1_-_193075180 | 3.50 |
ENST00000367440.3
|
GLRX2
|
glutaredoxin 2 |
| chr17_+_66245341 | 3.49 |
ENST00000577985.1
|
AMZ2
|
archaelysin family metallopeptidase 2 |
| chrX_+_150148976 | 3.48 |
ENST00000419110.1
|
HMGB3
|
high mobility group box 3 |
| chr4_-_102268484 | 3.47 |
ENST00000394853.4
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr3_-_127541679 | 3.46 |
ENST00000265052.5
|
MGLL
|
monoglyceride lipase |
| chr5_+_140810132 | 3.45 |
ENST00000252085.3
|
PCDHGA12
|
protocadherin gamma subfamily A, 12 |
| chr11_+_4116054 | 3.45 |
ENST00000423050.2
|
RRM1
|
ribonucleotide reductase M1 |
| chr3_+_130569429 | 3.44 |
ENST00000505330.1
ENST00000504381.1 ENST00000507488.2 ENST00000393221.4 |
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr20_+_36405665 | 3.41 |
ENST00000373469.1
|
CTNNBL1
|
catenin, beta like 1 |
| chr2_-_220119280 | 3.40 |
ENST00000392088.2
|
TUBA4A
|
tubulin, alpha 4a |
| chr14_+_24701819 | 3.39 |
ENST00000560139.1
ENST00000559910.1 |
GMPR2
|
guanosine monophosphate reductase 2 |
| chr3_-_58613323 | 3.36 |
ENST00000474531.1
ENST00000465970.1 |
FAM107A
|
family with sequence similarity 107, member A |
| chr11_+_65686952 | 3.34 |
ENST00000527119.1
|
DRAP1
|
DR1-associated protein 1 (negative cofactor 2 alpha) |
| chr14_+_90722886 | 3.32 |
ENST00000543772.2
|
PSMC1
|
proteasome (prosome, macropain) 26S subunit, ATPase, 1 |
| chr16_+_4674787 | 3.32 |
ENST00000262370.7
|
MGRN1
|
mahogunin ring finger 1, E3 ubiquitin protein ligase |
| chr22_+_42017987 | 3.31 |
ENST00000405506.1
|
XRCC6
|
X-ray repair complementing defective repair in Chinese hamster cells 6 |
| chr12_+_28410128 | 3.31 |
ENST00000381259.1
ENST00000381256.1 |
CCDC91
|
coiled-coil domain containing 91 |
| chr1_+_15736359 | 3.27 |
ENST00000375980.4
|
EFHD2
|
EF-hand domain family, member D2 |
| chr3_+_130569592 | 3.25 |
ENST00000533801.2
|
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr16_-_67514982 | 3.24 |
ENST00000565835.1
ENST00000540149.1 ENST00000290949.3 |
ATP6V0D1
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d1 |
| chr17_-_39684550 | 3.23 |
ENST00000455635.1
ENST00000361566.3 |
KRT19
|
keratin 19 |
| chrX_-_47004437 | 3.22 |
ENST00000276062.8
|
NDUFB11
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 11, 17.3kDa |
| chr16_-_66968055 | 3.19 |
ENST00000568572.1
|
FAM96B
|
family with sequence similarity 96, member B |
| chr11_-_31832581 | 3.16 |
ENST00000379111.2
|
PAX6
|
paired box 6 |
| chr17_-_685559 | 3.13 |
ENST00000301329.6
|
GLOD4
|
glyoxalase domain containing 4 |
| chr9_+_87285539 | 3.11 |
ENST00000359847.3
|
NTRK2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr18_+_3449821 | 3.11 |
ENST00000407501.2
ENST00000405385.3 ENST00000546979.1 |
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr14_-_102552659 | 3.09 |
ENST00000441629.2
|
HSP90AA1
|
heat shock protein 90kDa alpha (cytosolic), class A member 1 |
| chr20_+_33104199 | 3.09 |
ENST00000357156.2
ENST00000417166.2 ENST00000300469.9 ENST00000374846.3 |
DYNLRB1
|
dynein, light chain, roadblock-type 1 |
| chr16_-_18573396 | 3.09 |
ENST00000543392.1
ENST00000381474.3 ENST00000330537.6 |
NOMO2
|
NODAL modulator 2 |
| chr2_-_98280383 | 3.08 |
ENST00000289228.5
|
ACTR1B
|
ARP1 actin-related protein 1 homolog B, centractin beta (yeast) |
| chr6_-_84418860 | 3.07 |
ENST00000521743.1
|
SNAP91
|
synaptosomal-associated protein, 91kDa |
| chr4_-_100871506 | 3.06 |
ENST00000296417.5
|
H2AFZ
|
H2A histone family, member Z |
| chr12_+_113344755 | 3.05 |
ENST00000550883.1
|
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr17_-_73389854 | 3.05 |
ENST00000578961.1
ENST00000392564.1 ENST00000582582.1 |
GRB2
|
growth factor receptor-bound protein 2 |
| chr14_-_71107921 | 3.03 |
ENST00000553982.1
ENST00000500016.1 |
CTD-2540L5.5
CTD-2540L5.6
|
CTD-2540L5.5 CTD-2540L5.6 |
| chr1_-_21948906 | 3.02 |
ENST00000374761.2
ENST00000599760.1 |
RAP1GAP
|
RAP1 GTPase activating protein |
| chr1_+_40506392 | 3.02 |
ENST00000414893.1
ENST00000414281.1 ENST00000420216.1 ENST00000372792.2 ENST00000372798.1 ENST00000340450.3 ENST00000372805.3 ENST00000435719.1 ENST00000427843.1 ENST00000417287.1 ENST00000424977.1 ENST00000446031.1 |
CAP1
|
CAP, adenylate cyclase-associated protein 1 (yeast) |
| chr11_+_74660278 | 3.02 |
ENST00000263672.6
ENST00000530257.1 ENST00000526361.1 ENST00000532972.1 |
SPCS2
|
signal peptidase complex subunit 2 homolog (S. cerevisiae) |
| chr1_-_155211065 | 3.02 |
ENST00000427500.3
|
GBA
|
glucosidase, beta, acid |
| chr6_-_33385823 | 3.01 |
ENST00000494751.1
ENST00000374496.3 |
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr1_-_201476274 | 3.00 |
ENST00000340006.2
|
CSRP1
|
cysteine and glycine-rich protein 1 |
| chr11_+_4116005 | 3.00 |
ENST00000300738.5
|
RRM1
|
ribonucleotide reductase M1 |
| chr7_-_102213030 | 2.99 |
ENST00000511313.1
ENST00000513438.1 ENST00000513506.1 |
POLR2J3
|
polymerase (RNA) II (DNA directed) polypeptide J3 |
| chr12_+_75874460 | 2.98 |
ENST00000266659.3
|
GLIPR1
|
GLI pathogenesis-related 1 |
| chr10_+_114709999 | 2.97 |
ENST00000355995.4
ENST00000545257.1 ENST00000543371.1 ENST00000536810.1 ENST00000355717.4 ENST00000538897.1 ENST00000534894.1 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr8_-_103876965 | 2.96 |
ENST00000337198.5
|
AZIN1
|
antizyme inhibitor 1 |
| chr7_-_102119342 | 2.96 |
ENST00000393794.3
ENST00000292614.5 |
POLR2J
|
polymerase (RNA) II (DNA directed) polypeptide J, 13.3kDa |
| chr6_-_84418841 | 2.95 |
ENST00000369694.2
ENST00000195649.6 |
SNAP91
|
synaptosomal-associated protein, 91kDa |
| chr6_-_41909191 | 2.94 |
ENST00000512426.1
ENST00000372987.4 |
CCND3
|
cyclin D3 |
| chr2_-_56150910 | 2.93 |
ENST00000424836.2
ENST00000438672.1 ENST00000440439.1 ENST00000429909.1 ENST00000424207.1 ENST00000452337.1 ENST00000355426.3 ENST00000439193.1 ENST00000421664.1 |
EFEMP1
|
EGF containing fibulin-like extracellular matrix protein 1 |
| chr18_+_3451584 | 2.93 |
ENST00000551541.1
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr16_+_56691838 | 2.92 |
ENST00000394501.2
|
MT1F
|
metallothionein 1F |
| chr1_+_244214577 | 2.91 |
ENST00000358704.4
|
ZBTB18
|
zinc finger and BTB domain containing 18 |
| chr16_+_56691606 | 2.91 |
ENST00000334350.6
|
MT1F
|
metallothionein 1F |
| chr17_-_73150599 | 2.91 |
ENST00000392566.2
ENST00000581874.1 |
HN1
|
hematological and neurological expressed 1 |
| chr12_+_8234807 | 2.91 |
ENST00000339754.5
|
NECAP1
|
NECAP endocytosis associated 1 |
| chr11_-_36310958 | 2.91 |
ENST00000532705.1
ENST00000263401.5 ENST00000452374.2 |
COMMD9
|
COMM domain containing 9 |
| chr7_-_137028534 | 2.90 |
ENST00000348225.2
|
PTN
|
pleiotrophin |
| chr8_-_13372395 | 2.90 |
ENST00000276297.4
ENST00000511869.1 |
DLC1
|
deleted in liver cancer 1 |
| chr16_+_56691911 | 2.89 |
ENST00000568475.1
|
MT1F
|
metallothionein 1F |
| chr9_-_136004782 | 2.88 |
ENST00000393157.3
|
RALGDS
|
ral guanine nucleotide dissociation stimulator |
| chr14_+_23340822 | 2.88 |
ENST00000359591.4
|
LRP10
|
low density lipoprotein receptor-related protein 10 |
| chr7_-_137028498 | 2.88 |
ENST00000393083.2
|
PTN
|
pleiotrophin |
| chr5_-_176057365 | 2.85 |
ENST00000310112.3
|
SNCB
|
synuclein, beta |
| chr19_-_43702231 | 2.83 |
ENST00000597374.1
ENST00000599371.1 |
PSG4
|
pregnancy specific beta-1-glycoprotein 4 |
| chr10_-_75012360 | 2.83 |
ENST00000416782.2
ENST00000372945.3 ENST00000372940.3 |
MRPS16
|
mitochondrial ribosomal protein S16 |
| chr2_+_201173667 | 2.80 |
ENST00000409755.3
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr17_-_73389737 | 2.79 |
ENST00000392563.1
|
GRB2
|
growth factor receptor-bound protein 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of MEIS1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 10.8 | GO:0043311 | positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 3.3 | 10.0 | GO:0009996 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) negative regulation of cell fate specification(GO:0009996) Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 2.7 | 13.6 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 2.6 | 7.9 | GO:2000861 | estrogen secretion(GO:0035937) estradiol secretion(GO:0035938) regulation of estrogen secretion(GO:2000861) regulation of estradiol secretion(GO:2000864) |
| 2.6 | 20.5 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 2.6 | 7.7 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 2.0 | 11.9 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 1.9 | 5.8 | GO:1904395 | retinal rod cell differentiation(GO:0060221) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) negative regulation of neuromuscular junction development(GO:1904397) |
| 1.9 | 5.7 | GO:1902938 | regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 1.9 | 7.6 | GO:1901804 | beta-glucoside metabolic process(GO:1901804) beta-glucoside catabolic process(GO:1901805) positive regulation of neuronal action potential(GO:1904457) |
| 1.8 | 5.4 | GO:1904647 | response to rotenone(GO:1904647) |
| 1.8 | 5.3 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 1.7 | 6.8 | GO:1903288 | protein transport into plasma membrane raft(GO:0044861) positive regulation of potassium ion import(GO:1903288) |
| 1.5 | 4.6 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 1.5 | 4.6 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 1.5 | 4.4 | GO:1903595 | positive regulation of histamine secretion by mast cell(GO:1903595) |
| 1.4 | 5.8 | GO:0006238 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 1.4 | 5.6 | GO:0052330 | induction of programmed cell death(GO:0012502) positive regulation of apoptotic process in other organism(GO:0044533) positive regulation by symbiont of host programmed cell death(GO:0052042) positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052330) positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052501) positive regulation of type B pancreatic cell development(GO:2000078) |
| 1.4 | 1.4 | GO:0009146 | purine nucleoside triphosphate catabolic process(GO:0009146) |
| 1.4 | 9.6 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 1.3 | 8.1 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 1.3 | 1.3 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 1.3 | 6.6 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 1.2 | 6.2 | GO:1902904 | negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 1.2 | 3.7 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 1.2 | 3.6 | GO:0052250 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 1.2 | 13.1 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 1.2 | 3.6 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 1.1 | 6.9 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 1.1 | 6.7 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 1.1 | 7.8 | GO:0016129 | phytosteroid metabolic process(GO:0016128) phytosteroid biosynthetic process(GO:0016129) |
| 1.1 | 3.3 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 1.1 | 3.3 | GO:1903526 | negative regulation of membrane tubulation(GO:1903526) |
| 1.0 | 3.0 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 1.0 | 4.0 | GO:0001971 | negative regulation of activation of membrane attack complex(GO:0001971) |
| 1.0 | 3.0 | GO:1903697 | negative regulation of microvillus assembly(GO:1903697) |
| 1.0 | 9.1 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 1.0 | 4.0 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 1.0 | 11.7 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 1.0 | 3.8 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.9 | 4.7 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.9 | 3.8 | GO:0090202 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.9 | 16.9 | GO:0032802 | low-density lipoprotein particle receptor catabolic process(GO:0032802) |
| 0.9 | 5.2 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.9 | 8.7 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.8 | 28.7 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.8 | 14.0 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.8 | 4.9 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.8 | 2.4 | GO:0046136 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.8 | 4.0 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.8 | 5.6 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.8 | 2.4 | GO:0046680 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) response to DDT(GO:0046680) histone H3-S10 phosphorylation involved in chromosome condensation(GO:2000775) |
| 0.8 | 2.4 | GO:0098923 | NMDA glutamate receptor clustering(GO:0097114) retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.8 | 3.1 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.8 | 6.8 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.8 | 6.8 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.7 | 3.0 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.7 | 2.2 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.7 | 10.8 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.7 | 2.1 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 0.7 | 2.1 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.7 | 2.1 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.7 | 2.1 | GO:0031627 | telomeric loop formation(GO:0031627) negative regulation of protein localization to chromosome, telomeric region(GO:1904815) |
| 0.7 | 2.8 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.7 | 6.1 | GO:0051610 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.7 | 2.7 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.7 | 2.7 | GO:0021815 | modulation of microtubule cytoskeleton involved in cerebral cortex radial glia guided migration(GO:0021815) |
| 0.7 | 1.3 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.7 | 2.0 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.6 | 1.9 | GO:0000965 | mitochondrial RNA 3'-end processing(GO:0000965) |
| 0.6 | 1.9 | GO:0051097 | negative regulation of helicase activity(GO:0051097) oligodendrocyte apoptotic process(GO:0097252) |
| 0.6 | 1.9 | GO:1904338 | regulation of dopaminergic neuron differentiation(GO:1904338) |
| 0.6 | 1.9 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 0.6 | 3.1 | GO:0099540 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 0.6 | 6.0 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.6 | 1.8 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.6 | 4.7 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.6 | 0.6 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.6 | 2.9 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.6 | 5.1 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.6 | 4.5 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.6 | 2.2 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.6 | 5.6 | GO:0042659 | regulation of cell fate specification(GO:0042659) |
| 0.6 | 2.2 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.6 | 3.3 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.5 | 5.5 | GO:0042262 | DNA protection(GO:0042262) |
| 0.5 | 1.6 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.5 | 12.5 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.5 | 1.1 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.5 | 4.3 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.5 | 2.7 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.5 | 2.6 | GO:2000639 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.5 | 2.6 | GO:1904798 | positive regulation of core promoter binding(GO:1904798) |
| 0.5 | 1.5 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.5 | 1.5 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.5 | 4.6 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.5 | 3.5 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.5 | 1.5 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.5 | 3.9 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.5 | 6.2 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.5 | 2.4 | GO:0030070 | insulin processing(GO:0030070) |
| 0.5 | 4.3 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.5 | 1.9 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.5 | 1.4 | GO:0051295 | establishment of meiotic spindle localization(GO:0051295) |
| 0.5 | 2.3 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.5 | 1.9 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.5 | 1.9 | GO:0001545 | primary ovarian follicle growth(GO:0001545) |
| 0.5 | 4.6 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.5 | 1.4 | GO:1901420 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) negative regulation of response to alcohol(GO:1901420) |
| 0.4 | 2.7 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.4 | 5.3 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.4 | 2.6 | GO:1902952 | positive regulation of dendritic spine maintenance(GO:1902952) regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.4 | 0.4 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.4 | 1.7 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.4 | 1.2 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.4 | 3.2 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.4 | 2.0 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.4 | 4.6 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.4 | 0.8 | GO:0071109 | superior temporal gyrus development(GO:0071109) |
| 0.4 | 1.5 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.4 | 1.9 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.4 | 2.3 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.4 | 8.3 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.4 | 1.1 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.4 | 2.2 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.4 | 1.8 | GO:1903273 | regulation of sodium ion export(GO:1903273) positive regulation of sodium ion export(GO:1903275) regulation of sodium ion export from cell(GO:1903276) positive regulation of sodium ion export from cell(GO:1903278) |
| 0.3 | 2.8 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.3 | 1.4 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.3 | 6.1 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.3 | 3.6 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.3 | 8.2 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.3 | 5.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.3 | 1.3 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.3 | 1.9 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.3 | 6.9 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.3 | 7.8 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.3 | 0.9 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.3 | 12.1 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.3 | 0.3 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.3 | 3.7 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) |
| 0.3 | 2.5 | GO:1990668 | vesicle fusion with endoplasmic reticulum-Golgi intermediate compartment (ERGIC) membrane(GO:1990668) |
| 0.3 | 6.4 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.3 | 4.9 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.3 | 3.3 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.3 | 15.2 | GO:0072583 | clathrin-mediated endocytosis(GO:0072583) |
| 0.3 | 17.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.3 | 1.8 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.3 | 4.4 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.3 | 0.6 | GO:0044111 | development involved in symbiotic interaction(GO:0044111) |
| 0.3 | 1.4 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.3 | 2.2 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.3 | 7.8 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.3 | 1.9 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.3 | 1.1 | GO:0061289 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.3 | 0.8 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.3 | 1.1 | GO:0061528 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.3 | 1.5 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.3 | 1.5 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.2 | 0.7 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.2 | 2.7 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.2 | 1.5 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.2 | 19.8 | GO:0002479 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent(GO:0002479) |
| 0.2 | 1.4 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.2 | 2.4 | GO:0030952 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) establishment or maintenance of cytoskeleton polarity(GO:0030952) |
| 0.2 | 3.7 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.2 | 0.9 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.2 | 1.8 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.2 | 1.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.2 | 2.2 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.2 | 1.5 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.2 | 3.5 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.2 | 0.7 | GO:0014876 | response to injury involved in regulation of muscle adaptation(GO:0014876) |
| 0.2 | 4.8 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.2 | 5.9 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.2 | 1.1 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.2 | 1.5 | GO:0030091 | protein repair(GO:0030091) |
| 0.2 | 0.6 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.2 | 1.5 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.2 | 1.5 | GO:2000795 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.2 | 0.9 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.2 | 0.6 | GO:0018406 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.2 | 3.2 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.2 | 1.2 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.2 | 3.9 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 | 1.2 | GO:0021859 | pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) |
| 0.2 | 0.6 | GO:0021831 | embryonic olfactory bulb interneuron precursor migration(GO:0021831) chemorepulsion involved in embryonic olfactory bulb interneuron precursor migration(GO:0021834) apoptotic process involved in luteolysis(GO:0061364) |
| 0.2 | 0.8 | GO:0010909 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.2 | 3.7 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.2 | 2.0 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.2 | 1.2 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.2 | 0.6 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.2 | 0.4 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) |
| 0.2 | 0.6 | GO:0036015 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) interleukin-5-mediated signaling pathway(GO:0038043) |
| 0.2 | 1.0 | GO:0015722 | canalicular bile acid transport(GO:0015722) glycerol transport(GO:0015793) pyrimidine nucleobase transport(GO:0015855) urea transmembrane transport(GO:0071918) purine nucleobase transmembrane transport(GO:1904823) |
| 0.2 | 1.0 | GO:2000630 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) positive regulation of miRNA metabolic process(GO:2000630) |
| 0.2 | 0.8 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.2 | 3.7 | GO:2000059 | negative regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000059) |
| 0.2 | 1.9 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.2 | 4.8 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.2 | 4.4 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.2 | 2.5 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.2 | 0.8 | GO:0030920 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.2 | 0.4 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.2 | 4.7 | GO:0072663 | protein targeting to peroxisome(GO:0006625) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.2 | 4.8 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.2 | 1.8 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.2 | 3.9 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.2 | 1.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.2 | 2.5 | GO:1900103 | positive regulation of endoplasmic reticulum unfolded protein response(GO:1900103) |
| 0.2 | 0.7 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.2 | 1.2 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.2 | 1.2 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.2 | 1.0 | GO:0043316 | cytotoxic T cell degranulation(GO:0043316) positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.2 | 2.3 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.2 | 11.8 | GO:0007632 | visual behavior(GO:0007632) |
| 0.2 | 0.5 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.2 | 5.2 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.2 | 1.0 | GO:0070458 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.2 | 4.3 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.2 | 2.7 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.2 | 1.3 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.2 | 5.0 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.2 | 0.5 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.2 | 0.6 | GO:0086097 | renin-angiotensin regulation of aldosterone production(GO:0002018) phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.2 | 1.7 | GO:1903204 | cellular response to hydroperoxide(GO:0071447) negative regulation of oxidative stress-induced neuron death(GO:1903204) |
| 0.2 | 6.4 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.2 | 3.0 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.2 | 1.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.2 | 1.5 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.1 | 0.4 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.1 | 1.0 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 0.6 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.1 | 9.5 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.1 | 1.2 | GO:0048262 | determination of dorsal/ventral asymmetry(GO:0048262) |
| 0.1 | 2.6 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 10.6 | GO:0070125 | mitochondrial translational elongation(GO:0070125) mitochondrial translational termination(GO:0070126) |
| 0.1 | 3.7 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 2.1 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.1 | 0.7 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.1 | 0.4 | GO:1902988 | neurofibrillary tangle assembly(GO:1902988) |
| 0.1 | 0.5 | GO:0002278 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) |
| 0.1 | 3.6 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 7.7 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.1 | 0.4 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.1 | 1.1 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 1.6 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.1 | 0.9 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.1 | 1.4 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.1 | 0.9 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.1 | 1.5 | GO:0035413 | positive regulation of catenin import into nucleus(GO:0035413) |
| 0.1 | 0.7 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 1.4 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.1 | 1.4 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 2.1 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.1 | 5.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 2.0 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.1 | 1.1 | GO:0010529 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.1 | 0.2 | GO:2000328 | regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.1 | 2.4 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.1 | 0.4 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.1 | 1.3 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.1 | 2.1 | GO:0039694 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.1 | 0.6 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.1 | 4.1 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.1 | 0.6 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.1 | 1.2 | GO:0035307 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.1 | 0.2 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.1 | 0.9 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.1 | 2.9 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.1 | 0.5 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.1 | 5.9 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 2.7 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.1 | 0.6 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 2.5 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.1 | 0.9 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 4.3 | GO:0051017 | actin filament bundle assembly(GO:0051017) actin filament bundle organization(GO:0061572) |
| 0.1 | 2.9 | GO:0042417 | dopamine metabolic process(GO:0042417) |
| 0.1 | 0.1 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.1 | 1.7 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 3.0 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.1 | 0.5 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.1 | 0.2 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.1 | 4.1 | GO:0051495 | positive regulation of cytoskeleton organization(GO:0051495) |
| 0.1 | 1.7 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.1 | 0.5 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.1 | 0.6 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.1 | 0.7 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.1 | 2.7 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.1 | 2.0 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.1 | 2.2 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.1 | 3.4 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.1 | 0.9 | GO:0060004 | reflex(GO:0060004) |
| 0.1 | 5.1 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.1 | 0.5 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.1 | 0.7 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.1 | 0.9 | GO:0001780 | neutrophil homeostasis(GO:0001780) |
| 0.1 | 0.2 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.1 | 1.1 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.1 | 1.7 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.1 | 0.4 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.1 | 1.6 | GO:0019370 | leukotriene biosynthetic process(GO:0019370) |
| 0.1 | 4.6 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.1 | 0.6 | GO:0071158 | positive regulation of cell cycle arrest(GO:0071158) |
| 0.1 | 1.2 | GO:0061045 | negative regulation of wound healing(GO:0061045) |
| 0.1 | 1.0 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.1 | 1.0 | GO:2000615 | regulation of histone H3-K9 acetylation(GO:2000615) |
| 0.1 | 2.5 | GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061418) |
| 0.1 | 1.9 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.1 | 0.5 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.1 | 0.3 | GO:0035897 | proteolysis in other organism(GO:0035897) |
| 0.1 | 1.5 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.1 | 0.9 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.1 | 1.2 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.1 | 2.2 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.1 | 0.2 | GO:0002774 | Fc receptor mediated inhibitory signaling pathway(GO:0002774) |
| 0.1 | 0.1 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.2 | GO:0032971 | diaphragm contraction(GO:0002086) involuntary skeletal muscle contraction(GO:0003011) regulation of muscle filament sliding(GO:0032971) regulation of muscle filament sliding speed(GO:0032972) |
| 0.0 | 1.2 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 1.4 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 2.0 | GO:1901016 | regulation of potassium ion transmembrane transporter activity(GO:1901016) |
| 0.0 | 1.6 | GO:0043620 | regulation of DNA-templated transcription in response to stress(GO:0043620) |
| 0.0 | 0.6 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 | 1.4 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.3 | GO:0043306 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 0.5 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 2.4 | GO:0055092 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.0 | 0.6 | GO:0071498 | cellular response to fluid shear stress(GO:0071498) |
| 0.0 | 1.3 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.0 | 0.7 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.1 | GO:0051685 | maintenance of ER location(GO:0051685) |
| 0.0 | 0.5 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.0 | 0.7 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.9 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.6 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.4 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.0 | 0.8 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.0 | 5.4 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 1.2 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.0 | 0.3 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.0 | 0.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.8 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 1.0 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.2 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.0 | 0.3 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 1.0 | GO:0071363 | cellular response to growth factor stimulus(GO:0071363) |
| 0.0 | 1.1 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.0 | 0.0 | GO:0034199 | activation of protein kinase A activity(GO:0034199) gastric motility(GO:0035482) gastric emptying(GO:0035483) |
| 0.0 | 2.3 | GO:0006672 | ceramide metabolic process(GO:0006672) |
| 0.0 | 0.9 | GO:0006458 | 'de novo' protein folding(GO:0006458) |
| 0.0 | 0.2 | GO:0006333 | chromatin assembly or disassembly(GO:0006333) |
| 0.0 | 1.0 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 3.2 | GO:0010389 | regulation of G2/M transition of mitotic cell cycle(GO:0010389) |
| 0.0 | 1.5 | GO:0048010 | vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
| 0.0 | 0.8 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.0 | 7.9 | GO:0000398 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 0.2 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.8 | GO:1904667 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051436) regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051439) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.0 | 3.6 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.4 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.0 | 0.7 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.0 | GO:1901725 | regulation of histone deacetylase activity(GO:1901725) |
| 0.0 | 1.9 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 0.2 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.6 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.2 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.1 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.0 | 2.6 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.0 | 0.3 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.2 | GO:0048003 | antigen processing and presentation via MHC class Ib(GO:0002475) antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.7 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 1.3 | GO:0048839 | inner ear development(GO:0048839) |
| 0.0 | 0.1 | GO:0060346 | bone trabecula formation(GO:0060346) |
| 0.0 | 0.6 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.0 | 0.3 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) |
| 0.0 | 0.2 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.1 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.2 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 2.0 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 21.8 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 2.5 | 12.5 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 2.3 | 6.8 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 1.7 | 6.8 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 1.6 | 9.8 | GO:0071817 | MMXD complex(GO:0071817) |
| 1.6 | 6.4 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 1.4 | 14.0 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 1.3 | 3.9 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 1.2 | 4.6 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 1.2 | 17.3 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 1.2 | 9.2 | GO:0000322 | storage vacuole(GO:0000322) |
| 1.2 | 10.4 | GO:0005955 | calcineurin complex(GO:0005955) |
| 1.1 | 5.3 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 1.1 | 16.9 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 1.0 | 9.0 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 1.0 | 14.0 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 1.0 | 5.9 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 1.0 | 5.8 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.9 | 6.2 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.7 | 5.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.7 | 7.8 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.7 | 2.1 | GO:0019034 | viral replication complex(GO:0019034) |
| 0.7 | 4.2 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.7 | 3.3 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.6 | 4.9 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.6 | 9.8 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.6 | 7.4 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.6 | 17.0 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.6 | 6.7 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.5 | 1.6 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.5 | 2.2 | GO:1990075 | kinesin II complex(GO:0016939) periciliary membrane compartment(GO:1990075) |
| 0.5 | 3.2 | GO:1990357 | terminal web(GO:1990357) |
| 0.5 | 2.7 | GO:0033503 | HULC complex(GO:0033503) |
| 0.5 | 3.1 | GO:0001740 | Barr body(GO:0001740) |
| 0.5 | 2.0 | GO:0070695 | FHF complex(GO:0070695) |
| 0.5 | 2.0 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.5 | 3.0 | GO:0000835 | ER ubiquitin ligase complex(GO:0000835) |
| 0.5 | 2.4 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.5 | 1.4 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.4 | 16.8 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.4 | 7.5 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.4 | 4.4 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.4 | 3.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.4 | 3.0 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.4 | 8.6 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.4 | 1.7 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.4 | 5.4 | GO:0097433 | dense body(GO:0097433) |
| 0.4 | 1.2 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.4 | 2.8 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.4 | 1.5 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.4 | 2.7 | GO:0070522 | ERCC4-ERCC1 complex(GO:0070522) |
| 0.4 | 3.8 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.4 | 2.3 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.4 | 7.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.4 | 2.6 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.4 | 11.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.3 | 5.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.3 | 9.1 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.3 | 4.6 | GO:0044754 | autolysosome(GO:0044754) |
| 0.3 | 10.9 | GO:0032420 | stereocilium(GO:0032420) |
| 0.3 | 4.4 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.3 | 2.5 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.3 | 1.2 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.3 | 1.5 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.3 | 2.7 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.3 | 6.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.3 | 1.1 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.3 | 2.1 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.3 | 3.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.3 | 15.7 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.3 | 1.5 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.2 | 2.7 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.2 | 2.6 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.2 | 1.9 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.2 | 6.4 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.2 | 1.2 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.2 | 2.6 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.2 | 2.8 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.2 | 1.9 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.2 | 2.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.2 | 2.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.2 | 1.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.2 | 1.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.2 | 1.1 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.2 | 0.9 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.2 | 0.8 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.2 | 0.6 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.2 | 2.1 | GO:0070187 | telosome(GO:0070187) |
| 0.2 | 0.8 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.2 | 0.6 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.2 | 5.5 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.2 | 3.1 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.2 | 2.3 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.2 | 2.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.2 | 0.7 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.2 | 2.0 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.2 | 0.9 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.2 | 7.0 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.2 | 2.0 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.2 | 3.3 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.2 | 3.2 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.2 | 3.6 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 12.3 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 1.5 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 0.9 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.1 | 0.6 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 1.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 1.3 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.1 | 1.3 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 5.8 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.1 | 4.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 1.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 11.0 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.1 | 19.5 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 2.5 | GO:0031968 | organelle outer membrane(GO:0031968) |
| 0.1 | 3.7 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 6.7 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 5.4 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 2.4 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 25.3 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.1 | 1.0 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 1.0 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 0.8 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.1 | 0.8 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 1.0 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.1 | 4.5 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.1 | 13.4 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 0.2 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.1 | 8.3 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 7.5 | GO:0016020 | membrane(GO:0016020) |
| 0.1 | 1.8 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 8.1 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 13.3 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.1 | 1.4 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 0.5 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.1 | 5.1 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 1.4 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 12.9 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 0.7 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.1 | 0.4 | GO:0097179 | protease inhibitor complex(GO:0097179) |
| 0.1 | 0.9 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 1.0 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 0.5 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.1 | 0.6 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 0.9 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.1 | 2.3 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 8.7 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.1 | 1.2 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 2.0 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 0.9 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.1 | 1.0 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 1.1 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 0.3 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.1 | 3.2 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.1 | 6.1 | GO:0005903 | brush border(GO:0005903) |
| 0.1 | 6.7 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 2.6 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.1 | 3.2 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.1 | 4.2 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 2.8 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 2.5 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 3.9 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 1.0 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.1 | 1.9 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 31.8 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.5 | GO:0035631 | IkappaB kinase complex(GO:0008385) CD40 receptor complex(GO:0035631) |
| 0.0 | 5.5 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.2 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 1.6 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.5 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 4.0 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 3.3 | GO:0031252 | cell leading edge(GO:0031252) |
| 0.0 | 0.6 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.4 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.4 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.9 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 2.2 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 1.3 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.9 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.8 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 1.2 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.0 | 7.0 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.3 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 1.3 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.8 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.1 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.8 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.2 | GO:0030666 | endocytic vesicle membrane(GO:0030666) |
| 0.0 | 1.1 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 2.2 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.4 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 4.0 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.0 | 1.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.0 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 1.0 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 0.5 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 0.6 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.5 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 1.4 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.0 | GO:0043196 | varicosity(GO:0043196) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 11.5 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 2.7 | 10.8 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 1.9 | 7.6 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 1.7 | 6.9 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 1.7 | 6.6 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 1.6 | 6.4 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 1.6 | 7.8 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 1.4 | 5.8 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 1.4 | 8.6 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 1.4 | 11.1 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 1.3 | 5.3 | GO:0004310 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 1.2 | 14.8 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 1.1 | 3.4 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 1.1 | 4.4 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 1.0 | 10.4 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 1.0 | 5.9 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.9 | 24.4 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.9 | 3.7 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.9 | 4.6 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.9 | 20.1 | GO:0046961 | hydrogen-exporting ATPase activity(GO:0036442) proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.8 | 4.2 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.8 | 5.8 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.8 | 2.5 | GO:0004853 | uroporphyrinogen decarboxylase activity(GO:0004853) |
| 0.8 | 2.4 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.8 | 22.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.7 | 9.6 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.7 | 2.1 | GO:0004756 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.7 | 7.8 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.7 | 2.1 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.7 | 4.2 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.7 | 3.5 | GO:0030614 | oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) |
| 0.7 | 14.7 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.7 | 3.4 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.7 | 2.7 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.6 | 8.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.6 | 1.9 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.6 | 3.8 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.6 | 3.1 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.6 | 6.2 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.6 | 14.3 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.6 | 3.5 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.6 | 7.5 | GO:0005167 | neurotrophin TRK receptor binding(GO:0005167) neurotrophin TRKA receptor binding(GO:0005168) |
| 0.6 | 2.3 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.6 | 13.8 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.5 | 9.1 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.5 | 1.6 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.5 | 1.5 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.5 | 1.5 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.5 | 2.0 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.5 | 6.0 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.5 | 1.5 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.5 | 3.8 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.5 | 1.9 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 0.5 | 13.4 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.5 | 2.3 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.4 | 1.3 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.4 | 2.7 | GO:0051022 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.4 | 3.9 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.4 | 6.9 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.4 | 1.3 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.4 | 5.8 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.4 | 1.7 | GO:0052812 | phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.4 | 2.5 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.4 | 2.4 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.4 | 1.2 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.4 | 1.6 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.4 | 3.2 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.4 | 3.9 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.4 | 4.6 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.4 | 5.4 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.4 | 2.7 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.4 | 2.3 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.4 | 2.6 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.4 | 6.0 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.4 | 2.6 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.4 | 5.4 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.4 | 2.9 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.3 | 1.4 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.3 | 5.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.3 | 2.4 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.3 | 3.4 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.3 | 11.5 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.3 | 2.6 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.3 | 1.9 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.3 | 11.5 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.3 | 3.1 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.3 | 1.5 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.3 | 0.9 | GO:0000285 | 1-phosphatidylinositol-3-phosphate 5-kinase activity(GO:0000285) |
| 0.3 | 2.4 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.3 | 1.8 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.3 | 1.7 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.3 | 8.6 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.3 | 5.9 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.3 | 1.4 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.3 | 8.7 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.3 | 3.0 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.3 | 0.5 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.3 | 3.5 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.3 | 4.8 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.3 | 1.1 | GO:0032145 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.3 | 6.1 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.3 | 14.0 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.3 | 1.0 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.3 | 6.7 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.3 | 2.0 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.2 | 6.7 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.2 | 1.0 | GO:0015265 | glycerol channel activity(GO:0015254) urea channel activity(GO:0015265) |
| 0.2 | 0.7 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.2 | 1.4 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.2 | 2.9 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.2 | 0.5 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.2 | 3.7 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.2 | 3.2 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.2 | 18.0 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.2 | 1.5 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.2 | 5.4 | GO:1990939 | ATP-dependent microtubule motor activity(GO:1990939) |
| 0.2 | 0.6 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.2 | 1.1 | GO:0015639 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.2 | 0.9 | GO:0042019 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.2 | 0.6 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.2 | 5.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.2 | 0.6 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.2 | 1.6 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.2 | 2.2 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.2 | 0.6 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.2 | 0.6 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.2 | 0.6 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.2 | 0.8 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.2 | 0.6 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 0.2 | 2.3 | GO:0031386 | protein tag(GO:0031386) |
| 0.2 | 0.8 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.2 | 0.4 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.2 | 4.7 | GO:0005521 | lamin binding(GO:0005521) |
| 0.2 | 6.3 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.2 | 2.2 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.2 | 2.4 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.2 | 5.3 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.2 | 1.5 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.2 | 1.1 | GO:0001190 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.2 | 1.8 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.2 | 3.9 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.2 | 4.9 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.2 | 1.5 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.2 | 1.2 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.2 | 4.6 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.2 | 0.5 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.2 | 0.3 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.2 | 26.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.2 | 7.4 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.2 | 3.5 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.2 | 1.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.2 | 5.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.2 | 0.6 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.1 | 2.6 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.1 | 1.3 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.1 | 6.4 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 13.2 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.1 | 1.4 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 1.4 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 3.5 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 0.4 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.1 | 4.1 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 0.4 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.1 | 0.9 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.1 | 0.8 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 1.5 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 0.6 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 1.8 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 4.1 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 2.3 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 3.2 | GO:0001848 | complement binding(GO:0001848) |
| 0.1 | 5.9 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 7.1 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.1 | 3.0 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.1 | 2.5 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 5.1 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 2.4 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 2.1 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.1 | 4.0 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 1.9 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.1 | 0.9 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.1 | 1.0 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 2.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 1.9 | GO:0004549 | tRNA-specific ribonuclease activity(GO:0004549) |
| 0.1 | 0.6 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.1 | 0.9 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.1 | 1.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 3.9 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.1 | 3.7 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 1.1 | GO:0089720 | caspase binding(GO:0089720) |
| 0.1 | 1.1 | GO:0016273 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.1 | 2.5 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.1 | 10.1 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.1 | 0.8 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 0.9 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.1 | 0.9 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 1.0 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 22.8 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 0.6 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 1.0 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.1 | 2.0 | GO:0071949 | FAD binding(GO:0071949) |
| 0.1 | 1.9 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 2.0 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 0.8 | GO:0019864 | IgG binding(GO:0019864) |
| 0.1 | 0.5 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.1 | 2.2 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 3.6 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.1 | 11.0 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 0.8 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.1 | 0.7 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.1 | 1.3 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.1 | 2.2 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 1.2 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 0.2 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 0.2 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 1.5 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.1 | 1.8 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.9 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 2.3 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.5 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.0 | 0.8 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 4.6 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.3 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 1.8 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 2.3 | GO:0016651 | oxidoreductase activity, acting on NAD(P)H(GO:0016651) |
| 0.0 | 0.2 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.3 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.9 | GO:0008233 | peptidase activity(GO:0008233) |
| 0.0 | 0.3 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 8.7 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 2.6 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.1 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.0 | 2.3 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 0.3 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 1.9 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 2.7 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 6.8 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 5.9 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 5.1 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.2 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.3 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.1 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.0 | 0.5 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.8 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 3.3 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 1.0 | GO:0016791 | phosphatase activity(GO:0016791) |
| 0.0 | 1.4 | GO:0005319 | lipid transporter activity(GO:0005319) |
| 0.0 | 0.2 | GO:0030883 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.0 | 0.3 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.2 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.0 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 0.3 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 3.7 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 2.6 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.1 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 0.4 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.2 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.7 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.0 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 1.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.4 | GO:0050840 | extracellular matrix binding(GO:0050840) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 23.2 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.7 | 23.6 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.6 | 25.1 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.4 | 4.9 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.3 | 4.8 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.3 | 9.1 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.3 | 1.4 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.3 | 8.1 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.3 | 10.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.3 | 9.5 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.3 | 9.4 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.2 | 24.1 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.2 | 19.1 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.2 | 6.9 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.2 | 3.8 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.2 | 6.4 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.2 | 8.9 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.2 | 4.9 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.2 | 7.8 | PID MYC PATHWAY | C-MYC pathway |
| 0.2 | 8.0 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.2 | 18.4 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.2 | 1.6 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.2 | 9.7 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.2 | 5.2 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.1 | 4.6 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.1 | 4.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 6.5 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 1.0 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 6.5 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 1.7 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 4.5 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 1.4 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 1.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 3.9 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 1.9 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 4.4 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 8.8 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 4.9 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.1 | 4.6 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 5.3 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 2.5 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 0.6 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 2.6 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 2.1 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 2.1 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 0.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 0.4 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 1.1 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.8 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 2.2 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.8 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 2.2 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.6 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 1.8 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 1.8 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.1 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 1.2 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.7 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.7 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.5 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 1.3 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.4 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.3 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 0.5 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 1.4 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.9 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.8 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.5 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 26.8 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.9 | 16.6 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.8 | 14.7 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.8 | 21.9 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.6 | 1.9 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.6 | 19.0 | REACTOME BOTULINUM NEUROTOXICITY | Genes involved in Botulinum neurotoxicity |
| 0.5 | 12.5 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.5 | 9.3 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.5 | 16.0 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.5 | 8.5 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.5 | 5.6 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.4 | 41.7 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.4 | 12.8 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.4 | 12.8 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.4 | 6.9 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.4 | 4.8 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.4 | 3.6 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.3 | 22.3 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.3 | 13.6 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.3 | 3.0 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.3 | 6.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.3 | 4.4 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.3 | 10.7 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.3 | 3.6 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.2 | 12.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.2 | 9.9 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.2 | 3.2 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.2 | 10.0 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.2 | 1.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.2 | 6.4 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.2 | 3.1 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.2 | 3.3 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.2 | 8.1 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.2 | 2.6 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.2 | 2.2 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.2 | 0.2 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.2 | 3.5 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.2 | 4.8 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.2 | 8.9 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.2 | 2.8 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.2 | 10.8 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.2 | 2.0 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.2 | 2.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.2 | 3.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.2 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.2 | 3.8 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.2 | 5.8 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.2 | 7.5 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.2 | 3.9 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.2 | 6.2 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.2 | 3.6 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.2 | 3.1 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 15.4 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 4.7 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 0.9 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.1 | 11.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 0.3 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.1 | 6.6 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 1.3 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.1 | 2.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 4.7 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.1 | 3.0 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 0.7 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.1 | 4.7 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 4.1 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 1.0 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 6.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 2.6 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.1 | 3.4 | REACTOME GLUCAGON SIGNALING IN METABOLIC REGULATION | Genes involved in Glucagon signaling in metabolic regulation |
| 0.1 | 1.9 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.1 | 4.4 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 1.8 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 3.0 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.1 | 1.8 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 0.5 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.1 | 2.0 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 2.7 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 0.4 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 0.9 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.1 | 3.9 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 1.0 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 2.1 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 1.8 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 0.3 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.1 | 2.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 6.3 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 1.9 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.1 | 0.5 | REACTOME PROTEIN FOLDING | Genes involved in Protein folding |
| 0.1 | 2.9 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 1.2 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 1.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 1.1 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.5 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 2.2 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 1.0 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 1.1 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.6 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 2.0 | REACTOME RECRUITMENT OF MITOTIC CENTROSOME PROTEINS AND COMPLEXES | Genes involved in Recruitment of mitotic centrosome proteins and complexes |
| 0.0 | 1.3 | REACTOME HOST INTERACTIONS OF HIV FACTORS | Genes involved in Host Interactions of HIV factors |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 1.0 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.8 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.4 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.6 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.3 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.5 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.3 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.0 | 0.7 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 2.7 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.7 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.7 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.3 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 1.1 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.5 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 1.8 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |