Project
averaged GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
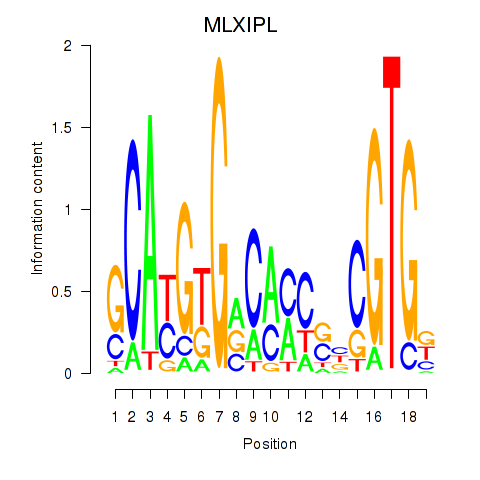
Results for MLXIPL
Z-value: 2.02
Transcription factors associated with MLXIPL
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MLXIPL
|
ENSG00000009950.11 | MLX interacting protein like |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MLXIPL | hg19_v2_chr7_-_73038867_73038878, hg19_v2_chr7_-_73038822_73038862 | 0.45 | 2.1e-12 | Click! |
Activity profile of MLXIPL motif
Sorted Z-values of MLXIPL motif
Network of associatons between targets according to the STRING database.
First level regulatory network of MLXIPL
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.3 | 18.8 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 5.4 | 5.4 | GO:0008355 | olfactory learning(GO:0008355) |
| 4.7 | 14.2 | GO:1902530 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 4.6 | 13.8 | GO:0030451 | regulation of complement activation, alternative pathway(GO:0030451) negative regulation of complement activation, alternative pathway(GO:0045957) |
| 4.5 | 13.6 | GO:0036079 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 3.8 | 15.2 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 3.7 | 3.7 | GO:0002688 | regulation of leukocyte chemotaxis(GO:0002688) |
| 3.6 | 3.6 | GO:0046425 | regulation of JAK-STAT cascade(GO:0046425) regulation of STAT cascade(GO:1904892) |
| 3.6 | 3.6 | GO:0036035 | osteoclast development(GO:0036035) |
| 3.4 | 10.3 | GO:1900073 | regulation of neuromuscular synaptic transmission(GO:1900073) positive regulation of neuromuscular synaptic transmission(GO:1900075) |
| 3.3 | 10.0 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 3.1 | 12.5 | GO:2001153 | regulation of renal water transport(GO:2001151) positive regulation of renal water transport(GO:2001153) |
| 3.1 | 9.4 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 3.1 | 9.2 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 2.9 | 8.8 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 2.9 | 26.3 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 2.9 | 28.7 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 2.9 | 42.8 | GO:0015671 | oxygen transport(GO:0015671) |
| 2.8 | 8.5 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 2.8 | 8.5 | GO:0097359 | UDP-glucosylation(GO:0097359) |
| 2.8 | 11.2 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 2.8 | 22.3 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 2.8 | 8.3 | GO:0100009 | circadian temperature homeostasis(GO:0060086) regulation of fever generation by regulation of prostaglandin secretion(GO:0071810) positive regulation of fever generation by positive regulation of prostaglandin secretion(GO:0071812) positive regulation of ERK1 and ERK2 cascade via TNFSF11-mediated signaling(GO:0071848) regulation of fever generation by prostaglandin secretion(GO:0100009) |
| 2.7 | 16.0 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 2.6 | 7.8 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 2.5 | 15.1 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 2.5 | 7.5 | GO:0048691 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 2.5 | 7.4 | GO:0019477 | L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 2.5 | 9.8 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 2.3 | 9.3 | GO:2000564 | CD8-positive, alpha-beta T cell proliferation(GO:0035740) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 2.2 | 11.2 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 2.2 | 6.6 | GO:0007439 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 2.2 | 22.1 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 2.2 | 6.6 | GO:1904933 | regulation of cell proliferation in midbrain(GO:1904933) |
| 2.1 | 6.4 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 2.1 | 15.0 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 2.1 | 17.1 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 2.1 | 6.4 | GO:0045659 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 2.1 | 14.8 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 2.1 | 6.3 | GO:0050870 | positive regulation of T cell activation(GO:0050870) |
| 2.1 | 4.2 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 2.1 | 14.7 | GO:0014050 | negative regulation of glutamate secretion(GO:0014050) |
| 2.1 | 6.3 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 2.1 | 10.4 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 2.1 | 12.5 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 2.1 | 8.3 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 2.0 | 6.1 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 2.0 | 6.0 | GO:2000417 | negative regulation of eosinophil migration(GO:2000417) |
| 2.0 | 7.8 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 2.0 | 5.9 | GO:2000410 | regulation of thymocyte migration(GO:2000410) |
| 1.9 | 15.5 | GO:0002223 | innate immune response activating cell surface receptor signaling pathway(GO:0002220) stimulatory C-type lectin receptor signaling pathway(GO:0002223) |
| 1.9 | 5.7 | GO:0002727 | natural killer cell cytokine production(GO:0002370) regulation of natural killer cell cytokine production(GO:0002727) |
| 1.9 | 5.7 | GO:0051295 | establishment of meiotic spindle localization(GO:0051295) |
| 1.9 | 7.5 | GO:0003356 | regulation of cilium movement(GO:0003352) regulation of cilium beat frequency(GO:0003356) |
| 1.8 | 7.2 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 1.8 | 7.2 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 1.8 | 30.4 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 1.8 | 14.1 | GO:0002774 | Fc receptor mediated inhibitory signaling pathway(GO:0002774) |
| 1.8 | 5.3 | GO:0090155 | glucosylceramide biosynthetic process(GO:0006679) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) |
| 1.7 | 1.7 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 1.7 | 13.7 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 1.7 | 18.6 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 1.7 | 21.8 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 1.7 | 16.5 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 1.6 | 11.3 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 1.6 | 107.7 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 1.6 | 4.8 | GO:0045210 | negative regulation of dendritic cell cytokine production(GO:0002731) FasL biosynthetic process(GO:0045210) |
| 1.6 | 17.6 | GO:0015886 | heme transport(GO:0015886) |
| 1.6 | 6.4 | GO:2000416 | regulation of eosinophil migration(GO:2000416) |
| 1.6 | 12.6 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 1.6 | 26.5 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 1.6 | 3.1 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) |
| 1.5 | 7.6 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 1.5 | 1.5 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 1.5 | 4.4 | GO:0001983 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 1.4 | 4.3 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 1.4 | 2.9 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 1.4 | 1.4 | GO:0015808 | L-alanine transport(GO:0015808) |
| 1.4 | 1.4 | GO:0045402 | interleukin-4 biosynthetic process(GO:0042097) regulation of interleukin-4 biosynthetic process(GO:0045402) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 1.4 | 4.3 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 1.4 | 2.8 | GO:0002765 | immune response-inhibiting signal transduction(GO:0002765) immune response-inhibiting cell surface receptor signaling pathway(GO:0002767) |
| 1.4 | 4.2 | GO:0042361 | menaquinone catabolic process(GO:0042361) vitamin K catabolic process(GO:0042377) |
| 1.4 | 4.2 | GO:1990641 | response to iron ion starvation(GO:1990641) |
| 1.4 | 6.9 | GO:0070370 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 1.4 | 5.5 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 1.4 | 1.4 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 1.3 | 4.0 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 1.3 | 9.2 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 1.3 | 6.5 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 1.3 | 6.5 | GO:0015824 | proline transport(GO:0015824) |
| 1.3 | 5.2 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 1.3 | 6.4 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 1.3 | 6.4 | GO:0086021 | SA node cell to atrial cardiac muscle cell communication by electrical coupling(GO:0086021) AV node cell to bundle of His cell communication by electrical coupling(GO:0086053) |
| 1.3 | 5.1 | GO:0003095 | pressure natriuresis(GO:0003095) |
| 1.3 | 10.2 | GO:0014827 | intestine smooth muscle contraction(GO:0014827) |
| 1.3 | 7.6 | GO:0015811 | L-cystine transport(GO:0015811) |
| 1.3 | 8.8 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 1.3 | 3.8 | GO:0055073 | cadmium ion homeostasis(GO:0055073) |
| 1.3 | 5.0 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 1.2 | 7.5 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 1.2 | 3.7 | GO:0097056 | seryl-tRNA aminoacylation(GO:0006434) selenocysteinyl-tRNA(Sec) biosynthetic process(GO:0097056) |
| 1.2 | 3.7 | GO:0031627 | telomeric loop formation(GO:0031627) |
| 1.2 | 7.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 1.2 | 3.6 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 1.2 | 4.8 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 1.2 | 3.6 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 1.2 | 7.1 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 1.2 | 7.1 | GO:2000834 | androgen secretion(GO:0035935) regulation of androgen secretion(GO:2000834) positive regulation of androgen secretion(GO:2000836) |
| 1.2 | 5.9 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 1.2 | 1.2 | GO:0019249 | lactate biosynthetic process(GO:0019249) |
| 1.2 | 8.2 | GO:0019532 | oxalate transport(GO:0019532) |
| 1.2 | 14.0 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 1.2 | 1.2 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) |
| 1.2 | 5.8 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 1.2 | 13.8 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 1.2 | 4.6 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 1.1 | 10.3 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 1.1 | 20.0 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 1.1 | 5.5 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 1.1 | 3.3 | GO:1990168 | protein K33-linked deubiquitination(GO:1990168) |
| 1.1 | 6.6 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 1.1 | 19.6 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 1.1 | 3.3 | GO:0003285 | septum secundum development(GO:0003285) |
| 1.1 | 1.1 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 1.1 | 1.1 | GO:0097267 | omega-hydroxylase P450 pathway(GO:0097267) |
| 1.1 | 7.5 | GO:0008218 | bioluminescence(GO:0008218) |
| 1.1 | 1.1 | GO:0003408 | optic cup formation involved in camera-type eye development(GO:0003408) |
| 1.1 | 3.2 | GO:0051710 | regulation of cytolysis in other organism(GO:0051710) |
| 1.1 | 4.2 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 1.1 | 14.8 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 1.0 | 3.1 | GO:0010165 | response to X-ray(GO:0010165) |
| 1.0 | 4.1 | GO:0010936 | positive regulation of tolerance induction(GO:0002645) negative regulation of macrophage cytokine production(GO:0010936) |
| 1.0 | 3.0 | GO:1905075 | proepicardium development(GO:0003342) septum transversum development(GO:0003343) occluding junction disassembly(GO:1905071) regulation of occluding junction disassembly(GO:1905073) positive regulation of occluding junction disassembly(GO:1905075) |
| 1.0 | 5.1 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 1.0 | 3.0 | GO:0070508 | sterol import(GO:0035376) cholesterol import(GO:0070508) |
| 1.0 | 6.0 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 1.0 | 2.0 | GO:0001575 | globoside metabolic process(GO:0001575) |
| 1.0 | 3.0 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 1.0 | 3.0 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 1.0 | 3.0 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 1.0 | 3.0 | GO:0071140 | resolution of recombination intermediates(GO:0071139) resolution of mitotic recombination intermediates(GO:0071140) |
| 1.0 | 8.9 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 1.0 | 11.8 | GO:0055089 | fatty acid homeostasis(GO:0055089) |
| 1.0 | 2.0 | GO:0032328 | alanine transport(GO:0032328) |
| 1.0 | 4.9 | GO:0033567 | DNA replication, Okazaki fragment processing(GO:0033567) |
| 1.0 | 3.9 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 1.0 | 15.4 | GO:0034382 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) |
| 1.0 | 2.9 | GO:0060084 | acetylcholine secretion, neurotransmission(GO:0014055) regulation of acetylcholine secretion, neurotransmission(GO:0014056) synaptic transmission involved in micturition(GO:0060084) |
| 1.0 | 14.4 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 1.0 | 3.8 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 1.0 | 1.9 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 1.0 | 4.8 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 1.0 | 2.9 | GO:0036118 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.9 | 2.8 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.9 | 2.8 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.9 | 4.7 | GO:1901091 | regulation of protein tetramerization(GO:1901090) negative regulation of protein tetramerization(GO:1901091) regulation of protein homotetramerization(GO:1901093) negative regulation of protein homotetramerization(GO:1901094) |
| 0.9 | 11.2 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 0.9 | 7.5 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.9 | 5.6 | GO:0052551 | response to defense-related nitric oxide production by other organism involved in symbiotic interaction(GO:0052551) response to defense-related host nitric oxide production(GO:0052565) |
| 0.9 | 4.6 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.9 | 37.0 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.9 | 18.5 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.9 | 5.3 | GO:1901376 | mycotoxin metabolic process(GO:0043385) aflatoxin metabolic process(GO:0046222) organic heteropentacyclic compound metabolic process(GO:1901376) |
| 0.9 | 8.7 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.9 | 7.0 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.9 | 6.1 | GO:0015705 | iodide transport(GO:0015705) |
| 0.9 | 6.0 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.9 | 2.6 | GO:0048075 | positive regulation of eye pigmentation(GO:0048075) |
| 0.8 | 3.4 | GO:0060423 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) |
| 0.8 | 10.9 | GO:1901072 | glucosamine-containing compound catabolic process(GO:1901072) |
| 0.8 | 1.7 | GO:0003257 | positive regulation of transcription from RNA polymerase II promoter involved in myocardial precursor cell differentiation(GO:0003257) positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) |
| 0.8 | 4.2 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.8 | 6.6 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.8 | 3.3 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.8 | 2.4 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.8 | 1.6 | GO:0045994 | positive regulation of translational initiation by iron(GO:0045994) |
| 0.8 | 2.4 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.8 | 3.2 | GO:0010816 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.8 | 4.8 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.8 | 2.4 | GO:0098937 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) |
| 0.8 | 1.6 | GO:0002329 | pre-B cell differentiation(GO:0002329) pre-B cell allelic exclusion(GO:0002331) |
| 0.8 | 4.7 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.8 | 3.9 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.8 | 7.0 | GO:0060633 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) |
| 0.8 | 3.9 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.8 | 3.1 | GO:0032954 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.8 | 3.8 | GO:1900242 | regulation of synaptic vesicle endocytosis(GO:1900242) positive regulation of synaptic vesicle endocytosis(GO:1900244) positive regulation of synaptic vesicle recycling(GO:1903423) |
| 0.8 | 3.1 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.8 | 4.6 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.8 | 0.8 | GO:0048073 | regulation of eye pigmentation(GO:0048073) |
| 0.8 | 5.3 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.8 | 2.3 | GO:0033212 | iron assimilation(GO:0033212) iron assimilation by chelation and transport(GO:0033214) positive regulation of bone mineralization involved in bone maturation(GO:1900159) negative regulation of tumor necrosis factor (ligand) superfamily member 11 production(GO:2000308) |
| 0.8 | 3.8 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.8 | 6.8 | GO:0060700 | regulation of ribonuclease activity(GO:0060700) |
| 0.8 | 7.5 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.7 | 3.7 | GO:2000035 | positive regulation of somatic stem cell population maintenance(GO:1904674) regulation of stem cell division(GO:2000035) |
| 0.7 | 1.5 | GO:0046618 | drug export(GO:0046618) |
| 0.7 | 7.4 | GO:0018209 | peptidyl-serine modification(GO:0018209) |
| 0.7 | 5.9 | GO:0010918 | positive regulation of mitochondrial membrane potential(GO:0010918) |
| 0.7 | 2.2 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.7 | 3.6 | GO:0072313 | metanephric glomerular epithelium development(GO:0072244) metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 0.7 | 2.2 | GO:0046125 | thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.7 | 9.4 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.7 | 3.6 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.7 | 1.4 | GO:0035356 | cellular triglyceride homeostasis(GO:0035356) |
| 0.7 | 6.4 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.7 | 2.8 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.7 | 4.1 | GO:0030397 | mitotic nuclear envelope disassembly(GO:0007077) membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.7 | 2.7 | GO:0035625 | negative regulation of epinephrine secretion(GO:0032811) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 0.7 | 0.7 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.7 | 61.5 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.7 | 4.0 | GO:0043045 | DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.7 | 3.3 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.7 | 2.7 | GO:0097421 | liver regeneration(GO:0097421) |
| 0.7 | 6.0 | GO:0044116 | growth involved in symbiotic interaction(GO:0044110) growth of symbiont involved in interaction with host(GO:0044116) |
| 0.7 | 9.9 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.7 | 3.3 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.7 | 2.0 | GO:0070781 | response to biotin(GO:0070781) |
| 0.7 | 1.3 | GO:0036022 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.7 | 2.0 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.7 | 2.0 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.6 | 7.8 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.6 | 1.9 | GO:1903925 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.6 | 1.9 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.6 | 1.9 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.6 | 11.5 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.6 | 4.5 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.6 | 3.8 | GO:0072364 | regulation of cellular ketone metabolic process by regulation of transcription from RNA polymerase II promoter(GO:0072364) |
| 0.6 | 2.6 | GO:0005986 | sucrose biosynthetic process(GO:0005986) |
| 0.6 | 7.0 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.6 | 5.0 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.6 | 6.3 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.6 | 5.0 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.6 | 5.0 | GO:0035771 | interleukin-4-mediated signaling pathway(GO:0035771) |
| 0.6 | 3.1 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 0.6 | 2.5 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.6 | 1.8 | GO:0035564 | regulation of kidney size(GO:0035564) regulation of pronephros size(GO:0035565) renal glucose absorption(GO:0035623) |
| 0.6 | 1.8 | GO:0038178 | complement component C5a signaling pathway(GO:0038178) |
| 0.6 | 9.0 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.6 | 2.4 | GO:0021784 | postganglionic parasympathetic fiber development(GO:0021784) |
| 0.6 | 1.8 | GO:0002644 | negative regulation of tolerance induction(GO:0002644) |
| 0.6 | 2.4 | GO:0006956 | complement activation(GO:0006956) |
| 0.6 | 1.8 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.6 | 1.8 | GO:1902809 | skeletal muscle fiber differentiation(GO:0098528) regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.6 | 4.8 | GO:0034384 | high-density lipoprotein particle clearance(GO:0034384) |
| 0.6 | 4.8 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.6 | 1.8 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.6 | 1.8 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.6 | 1.8 | GO:0015888 | thiamine transport(GO:0015888) thiamine transmembrane transport(GO:0071934) |
| 0.6 | 2.9 | GO:0046968 | peptide antigen transport(GO:0046968) |
| 0.6 | 5.3 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.6 | 22.3 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.6 | 2.3 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.6 | 0.6 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.6 | 2.9 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.6 | 2.3 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.6 | 2.3 | GO:0072299 | posterior mesonephric tubule development(GO:0072166) negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) |
| 0.6 | 14.3 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.6 | 2.3 | GO:1902159 | regulation of inward rectifier potassium channel activity(GO:1901979) regulation of cyclic nucleotide-gated ion channel activity(GO:1902159) |
| 0.6 | 5.6 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.6 | 2.3 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.6 | 5.0 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.6 | 5.0 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.6 | 2.2 | GO:0072402 | response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) |
| 0.6 | 42.9 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.6 | 3.3 | GO:0050666 | regulation of sulfur amino acid metabolic process(GO:0031335) regulation of homocysteine metabolic process(GO:0050666) |
| 0.6 | 5.6 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.6 | 6.1 | GO:0032364 | oxygen homeostasis(GO:0032364) |
| 0.6 | 13.9 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.6 | 9.4 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.5 | 2.2 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.5 | 2.2 | GO:0030856 | regulation of epithelial cell differentiation(GO:0030856) |
| 0.5 | 2.7 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.5 | 2.7 | GO:0036289 | peptidyl-serine autophosphorylation(GO:0036289) |
| 0.5 | 16.9 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.5 | 1.6 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.5 | 1.6 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.5 | 7.5 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.5 | 1.6 | GO:0045212 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.5 | 2.1 | GO:2000195 | negative regulation of female gonad development(GO:2000195) |
| 0.5 | 3.2 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.5 | 2.1 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.5 | 1.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.5 | 14.8 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.5 | 5.8 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.5 | 2.6 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.5 | 2.6 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.5 | 2.6 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.5 | 3.1 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.5 | 33.6 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.5 | 9.6 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.5 | 10.5 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.5 | 8.0 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.5 | 5.5 | GO:0030334 | regulation of cell migration(GO:0030334) |
| 0.5 | 1.0 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.5 | 1.5 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.5 | 1.5 | GO:0048852 | hypophysis morphogenesis(GO:0048850) diencephalon morphogenesis(GO:0048852) |
| 0.5 | 1.5 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.5 | 1.9 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.5 | 1.4 | GO:0036378 | calcitriol biosynthetic process from calciol(GO:0036378) |
| 0.5 | 1.9 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.5 | 6.2 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.5 | 2.4 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.5 | 1.4 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 0.5 | 1.4 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.5 | 4.2 | GO:0090037 | positive regulation of protein kinase C signaling(GO:0090037) |
| 0.5 | 1.8 | GO:0003335 | corneocyte development(GO:0003335) |
| 0.5 | 1.4 | GO:0045359 | interferon-beta biosynthetic process(GO:0045350) regulation of interferon-beta biosynthetic process(GO:0045357) positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.5 | 1.4 | GO:0061386 | closure of optic fissure(GO:0061386) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.5 | 2.7 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.4 | 2.7 | GO:0097091 | synaptic vesicle clustering(GO:0097091) |
| 0.4 | 1.3 | GO:0097049 | motor neuron apoptotic process(GO:0097049) |
| 0.4 | 3.9 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.4 | 10.0 | GO:0046856 | phospholipid dephosphorylation(GO:0046839) phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.4 | 2.6 | GO:0006540 | glutamate decarboxylation to succinate(GO:0006540) |
| 0.4 | 2.2 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.4 | 5.6 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.4 | 0.9 | GO:0071320 | cellular response to cAMP(GO:0071320) |
| 0.4 | 2.6 | GO:0046015 | regulation of transcription by glucose(GO:0046015) |
| 0.4 | 10.6 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.4 | 2.5 | GO:1901563 | response to camptothecin(GO:1901563) |
| 0.4 | 2.1 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.4 | 1.3 | GO:0052314 | serotonin biosynthetic process(GO:0042427) phytoalexin metabolic process(GO:0052314) |
| 0.4 | 4.2 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.4 | 7.8 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.4 | 3.7 | GO:0086073 | cardiac muscle cell-cardiac muscle cell adhesion(GO:0086042) bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.4 | 2.0 | GO:0050686 | negative regulation of mRNA processing(GO:0050686) |
| 0.4 | 2.8 | GO:0018377 | protein myristoylation(GO:0018377) |
| 0.4 | 1.6 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.4 | 2.4 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.4 | 7.6 | GO:0002407 | dendritic cell chemotaxis(GO:0002407) |
| 0.4 | 2.4 | GO:0035384 | thioester biosynthetic process(GO:0035384) acyl-CoA biosynthetic process(GO:0071616) |
| 0.4 | 2.8 | GO:0042126 | nitrate metabolic process(GO:0042126) |
| 0.4 | 6.7 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.4 | 1.6 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.4 | 1.2 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.4 | 2.0 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.4 | 9.8 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.4 | 2.3 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.4 | 8.9 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.4 | 1.2 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.4 | 2.7 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.4 | 1.2 | GO:0042414 | epinephrine metabolic process(GO:0042414) epinephrine biosynthetic process(GO:0042418) |
| 0.4 | 9.2 | GO:0006972 | hyperosmotic response(GO:0006972) |
| 0.4 | 2.3 | GO:0032466 | negative regulation of cytokinesis(GO:0032466) |
| 0.4 | 4.6 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.4 | 2.3 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.4 | 5.3 | GO:0043306 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.4 | 1.5 | GO:0001188 | RNA polymerase I transcriptional preinitiation complex assembly(GO:0001188) RNA polymerase I transcriptional preinitiation complex assembly at the promoter for the nuclear large rRNA transcript(GO:0001189) |
| 0.4 | 3.4 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.4 | 1.1 | GO:0061101 | neuroendocrine cell differentiation(GO:0061101) |
| 0.4 | 2.3 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.4 | 0.8 | GO:2001183 | negative regulation of interleukin-10 secretion(GO:2001180) negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.4 | 1.5 | GO:0032096 | negative regulation of response to food(GO:0032096) negative regulation of appetite(GO:0032099) somatostatin secretion(GO:0070253) |
| 0.4 | 8.2 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.4 | 13.4 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.4 | 1.9 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.4 | 1.5 | GO:0048842 | positive regulation of axon extension involved in axon guidance(GO:0048842) |
| 0.4 | 8.5 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.4 | 3.3 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.4 | 5.5 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.4 | 3.3 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.4 | 1.1 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.4 | 2.2 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.4 | 1.4 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.4 | 2.1 | GO:0051321 | meiotic cell cycle(GO:0051321) |
| 0.4 | 5.3 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.4 | 2.1 | GO:0008595 | tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.4 | 2.1 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.4 | 1.8 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.4 | 12.7 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.4 | 1.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.4 | 2.8 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.3 | 0.7 | GO:0042026 | protein refolding(GO:0042026) |
| 0.3 | 2.1 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.3 | 0.7 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) negative regulation of membrane depolarization(GO:1904180) |
| 0.3 | 3.8 | GO:0017085 | response to insecticide(GO:0017085) |
| 0.3 | 1.7 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.3 | 2.7 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.3 | 1.4 | GO:0043632 | ubiquitin-dependent protein catabolic process(GO:0006511) modification-dependent macromolecule catabolic process(GO:0043632) |
| 0.3 | 2.7 | GO:0045732 | positive regulation of protein catabolic process(GO:0045732) |
| 0.3 | 3.7 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.3 | 5.4 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.3 | 0.7 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.3 | 6.7 | GO:0003341 | cilium movement(GO:0003341) |
| 0.3 | 2.0 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.3 | 2.3 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.3 | 4.6 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.3 | 3.3 | GO:0060326 | cell chemotaxis(GO:0060326) |
| 0.3 | 1.3 | GO:0060214 | stem cell fate commitment(GO:0048865) endocardium formation(GO:0060214) |
| 0.3 | 5.2 | GO:0010002 | cardioblast differentiation(GO:0010002) |
| 0.3 | 9.4 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.3 | 1.3 | GO:0090210 | regulation of establishment of blood-brain barrier(GO:0090210) negative regulation of establishment of blood-brain barrier(GO:0090212) |
| 0.3 | 3.2 | GO:0019627 | urea cycle(GO:0000050) urea metabolic process(GO:0019627) nitrogen cycle metabolic process(GO:0071941) |
| 0.3 | 2.6 | GO:0060689 | cell differentiation involved in salivary gland development(GO:0060689) |
| 0.3 | 2.5 | GO:0015747 | urate transport(GO:0015747) |
| 0.3 | 2.2 | GO:0019720 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.3 | 3.1 | GO:1902221 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.3 | 5.0 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.3 | 5.3 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.3 | 6.5 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.3 | 1.5 | GO:0032380 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.3 | 0.3 | GO:0060157 | urinary bladder development(GO:0060157) |
| 0.3 | 3.9 | GO:0007512 | adult heart development(GO:0007512) |
| 0.3 | 12.4 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.3 | 0.9 | GO:1903753 | negative regulation of p38MAPK cascade(GO:1903753) |
| 0.3 | 25.0 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.3 | 1.5 | GO:0033274 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.3 | 0.3 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.3 | 2.6 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.3 | 1.2 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.3 | 4.6 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.3 | 1.4 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.3 | 1.9 | GO:0060841 | venous blood vessel development(GO:0060841) |
| 0.3 | 0.8 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.3 | 0.5 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.3 | 3.0 | GO:1904903 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.3 | 4.9 | GO:0035635 | entry of bacterium into host cell(GO:0035635) |
| 0.3 | 1.3 | GO:0001975 | response to amphetamine(GO:0001975) |
| 0.3 | 2.1 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.3 | 0.8 | GO:0052047 | positive regulation of fibrinolysis(GO:0051919) interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.3 | 1.0 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.3 | 0.8 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.3 | 3.1 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.3 | 5.5 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.3 | 1.8 | GO:0090110 | cargo loading into vesicle(GO:0035459) cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.3 | 1.5 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 0.3 | 1.3 | GO:1901660 | calcium ion export(GO:1901660) calcium ion export from cell(GO:1990034) |
| 0.3 | 2.0 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.3 | 6.3 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.3 | 1.8 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.2 | 3.5 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.2 | 2.2 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.2 | 1.7 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.2 | 1.0 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.2 | 4.4 | GO:0019835 | cytolysis(GO:0019835) |
| 0.2 | 2.9 | GO:0032495 | response to muramyl dipeptide(GO:0032495) |
| 0.2 | 2.7 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.2 | 1.2 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.2 | 5.6 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.2 | 1.0 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.2 | 1.4 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.2 | 0.5 | GO:0023035 | CD40 signaling pathway(GO:0023035) |
| 0.2 | 1.2 | GO:0038170 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.2 | 4.7 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.2 | 1.9 | GO:0035413 | positive regulation of catenin import into nucleus(GO:0035413) |
| 0.2 | 2.8 | GO:0001771 | immunological synapse formation(GO:0001771) |
| 0.2 | 1.6 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.2 | 0.7 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.2 | 3.2 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.2 | 0.7 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.2 | 1.6 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.2 | 0.4 | GO:0097104 | postsynaptic membrane assembly(GO:0097104) |
| 0.2 | 2.4 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.2 | 1.5 | GO:0019724 | B cell mediated immunity(GO:0019724) |
| 0.2 | 3.5 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.2 | 2.9 | GO:1904294 | positive regulation of ERAD pathway(GO:1904294) |
| 0.2 | 0.4 | GO:0051106 | regulation of DNA ligation(GO:0051105) positive regulation of DNA ligation(GO:0051106) |
| 0.2 | 1.1 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.2 | 3.4 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.2 | 0.6 | GO:0033078 | extrathymic T cell differentiation(GO:0033078) extrathymic T cell selection(GO:0045062) |
| 0.2 | 3.0 | GO:2000696 | regulation of epithelial cell differentiation involved in kidney development(GO:2000696) |
| 0.2 | 1.7 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.2 | 0.6 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.2 | 1.3 | GO:0042116 | macrophage activation(GO:0042116) |
| 0.2 | 1.5 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.2 | 3.5 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.2 | 18.9 | GO:0007286 | spermatid development(GO:0007286) |
| 0.2 | 1.2 | GO:0033089 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.2 | 4.1 | GO:0097503 | sialylation(GO:0097503) |
| 0.2 | 0.6 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.2 | 0.8 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.2 | 0.6 | GO:0035814 | negative regulation of renal sodium excretion(GO:0035814) |
| 0.2 | 0.8 | GO:0098838 | methotrexate transport(GO:0051958) reduced folate transmembrane transport(GO:0098838) |
| 0.2 | 1.8 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.2 | 2.2 | GO:0060252 | positive regulation of glial cell proliferation(GO:0060252) |
| 0.2 | 2.6 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.2 | 1.0 | GO:0002860 | natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of response to tumor cell(GO:0002834) positive regulation of response to tumor cell(GO:0002836) regulation of immune response to tumor cell(GO:0002837) positive regulation of immune response to tumor cell(GO:0002839) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.2 | 1.4 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.2 | 1.4 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.2 | 1.2 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.2 | 1.7 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.2 | 7.0 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.2 | 1.5 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.2 | 3.5 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.2 | 0.8 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) regulation of keratinocyte differentiation(GO:0045616) |
| 0.2 | 0.8 | GO:0003418 | growth plate cartilage chondrocyte differentiation(GO:0003418) |
| 0.2 | 1.3 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.2 | 1.9 | GO:0001696 | gastric acid secretion(GO:0001696) |
| 0.2 | 1.7 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.2 | 0.9 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.2 | 0.7 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.2 | 3.3 | GO:0007140 | male meiosis(GO:0007140) |
| 0.2 | 5.0 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.2 | 0.9 | GO:0008306 | associative learning(GO:0008306) |
| 0.2 | 5.3 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.2 | 0.9 | GO:0010755 | regulation of plasminogen activation(GO:0010755) negative regulation of plasminogen activation(GO:0010757) |
| 0.2 | 0.7 | GO:0051492 | regulation of stress fiber assembly(GO:0051492) |
| 0.2 | 1.8 | GO:0015074 | DNA integration(GO:0015074) |
| 0.2 | 3.2 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.2 | 3.0 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.2 | 3.6 | GO:0071902 | positive regulation of protein serine/threonine kinase activity(GO:0071902) |
| 0.2 | 1.8 | GO:0045023 | G0 to G1 transition(GO:0045023) |
| 0.2 | 1.1 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.2 | 1.6 | GO:0006555 | methionine metabolic process(GO:0006555) |
| 0.2 | 1.1 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.2 | 2.1 | GO:0042355 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.2 | 0.9 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.2 | 5.4 | GO:0008214 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.2 | 0.5 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.2 | 5.2 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.2 | 12.5 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.2 | 2.0 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.2 | 3.0 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.2 | 0.3 | GO:0033341 | regulation of collagen binding(GO:0033341) |
| 0.2 | 0.7 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.2 | 3.7 | GO:0050709 | negative regulation of protein secretion(GO:0050709) |
| 0.2 | 2.3 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.2 | 1.8 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.2 | 1.0 | GO:0006898 | receptor-mediated endocytosis(GO:0006898) |
| 0.2 | 0.8 | GO:0097090 | presynaptic membrane organization(GO:0097090) presynaptic membrane assembly(GO:0097105) |
| 0.2 | 3.0 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.2 | 0.6 | GO:0032571 | response to vitamin K(GO:0032571) |
| 0.2 | 7.9 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.2 | 2.4 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.2 | 1.9 | GO:0018065 | protein-cofactor linkage(GO:0018065) |
| 0.2 | 0.2 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.2 | 4.8 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.2 | 1.9 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.2 | 0.3 | GO:0034447 | very-low-density lipoprotein particle clearance(GO:0034447) |
| 0.2 | 8.0 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.1 | 0.9 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.1 | 0.7 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.1 | 2.8 | GO:0007269 | neurotransmitter secretion(GO:0007269) signal release from synapse(GO:0099643) |
| 0.1 | 1.0 | GO:0060745 | mammary gland branching involved in pregnancy(GO:0060745) |
| 0.1 | 4.0 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.1 | 3.0 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) |
| 0.1 | 0.9 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 4.7 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 2.5 | GO:0048147 | negative regulation of fibroblast proliferation(GO:0048147) |
| 0.1 | 1.0 | GO:1902750 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) negative regulation of cell cycle G2/M phase transition(GO:1902750) |
| 0.1 | 1.1 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.1 | 1.8 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.1 | 0.4 | GO:0061365 | positive regulation of triglyceride lipase activity(GO:0061365) |
| 0.1 | 0.7 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.1 | 0.3 | GO:0042369 | vitamin D catabolic process(GO:0042369) |
| 0.1 | 1.1 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.1 | 2.2 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 2.2 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.1 | 0.3 | GO:0001694 | histamine metabolic process(GO:0001692) histamine biosynthetic process(GO:0001694) |
| 0.1 | 1.2 | GO:0048511 | rhythmic process(GO:0048511) |
| 0.1 | 0.4 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 0.1 | 5.5 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.1 | 1.7 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.1 | 2.9 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.1 | 0.3 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.1 | 0.5 | GO:1905225 | thyroid-stimulating hormone signaling pathway(GO:0038194) cellular response to glycoprotein(GO:1904588) response to thyrotropin-releasing hormone(GO:1905225) cellular response to thyrotropin-releasing hormone(GO:1905229) |
| 0.1 | 5.7 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 2.4 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.1 | 0.7 | GO:0032695 | negative regulation of interleukin-12 production(GO:0032695) |
| 0.1 | 1.1 | GO:0036155 | acylglycerol acyl-chain remodeling(GO:0036155) |
| 0.1 | 6.7 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.1 | 1.9 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) |
| 0.1 | 0.3 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.1 | 3.3 | GO:0009083 | branched-chain amino acid metabolic process(GO:0009081) branched-chain amino acid catabolic process(GO:0009083) |
| 0.1 | 1.1 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.1 | 0.8 | GO:0001554 | luteolysis(GO:0001554) |
| 0.1 | 6.0 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.1 | 1.3 | GO:0046473 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.1 | 1.6 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.1 | 7.1 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 0.4 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.1 | 0.6 | GO:0098915 | membrane repolarization during ventricular cardiac muscle cell action potential(GO:0098915) |
| 0.1 | 0.4 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.1 | 2.0 | GO:0010719 | negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.1 | 0.4 | GO:0090625 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing by siRNA(GO:0090625) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.1 | 14.8 | GO:0007601 | visual perception(GO:0007601) |
| 0.1 | 2.5 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.1 | 1.3 | GO:0035878 | nail development(GO:0035878) |
| 0.1 | 1.5 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 1.4 | GO:0033198 | response to ATP(GO:0033198) |
| 0.1 | 0.2 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.1 | 2.0 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.1 | 2.3 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
| 0.1 | 1.1 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.1 | 0.2 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.1 | 2.6 | GO:0055010 | ventricular cardiac muscle tissue morphogenesis(GO:0055010) |
| 0.1 | 1.1 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.1 | 1.5 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.1 | 3.2 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 3.3 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.1 | 5.2 | GO:0006835 | dicarboxylic acid transport(GO:0006835) |
| 0.1 | 2.9 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 1.7 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.1 | 0.2 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.1 | 1.4 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.1 | 1.6 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.1 | 0.7 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.1 | 0.3 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.1 | 0.4 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) hexose mediated signaling(GO:0009757) sugar mediated signaling pathway(GO:0010182) glucose mediated signaling pathway(GO:0010255) regulation of glucose mediated signaling pathway(GO:1902659) |
| 0.1 | 0.6 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.1 | 1.4 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.1 | 2.1 | GO:0048146 | positive regulation of fibroblast proliferation(GO:0048146) |
| 0.1 | 0.6 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 5.2 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.1 | 2.8 | GO:0097192 | signal transduction in absence of ligand(GO:0038034) extrinsic apoptotic signaling pathway in absence of ligand(GO:0097192) |
| 0.1 | 1.2 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.1 | 0.8 | GO:1901018 | positive regulation of potassium ion transmembrane transporter activity(GO:1901018) |
| 0.1 | 1.5 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.1 | 1.3 | GO:0045840 | positive regulation of mitotic nuclear division(GO:0045840) |
| 0.1 | 0.2 | GO:2001056 | activation of cysteine-type endopeptidase activity(GO:0097202) positive regulation of cysteine-type endopeptidase activity(GO:2001056) |
| 0.1 | 0.7 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.1 | 0.2 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.1 | 1.4 | GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) |
| 0.1 | 0.5 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.1 | 1.4 | GO:0002279 | mast cell activation involved in immune response(GO:0002279) mast cell degranulation(GO:0043303) |
| 0.1 | 1.1 | GO:0042102 | positive regulation of T cell proliferation(GO:0042102) |
| 0.1 | 0.9 | GO:0006751 | glutathione catabolic process(GO:0006751) leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.1 | 0.6 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.1 | 0.2 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 1.7 | GO:0090102 | cochlea development(GO:0090102) |
| 0.1 | 1.8 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 0.5 | GO:0042220 | response to cocaine(GO:0042220) |
| 0.1 | 0.2 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.1 | 1.1 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.1 | 0.3 | GO:0007270 | neuron-neuron synaptic transmission(GO:0007270) |
| 0.1 | 0.6 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.1 | 0.3 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.1 | 1.1 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.1 | 0.4 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.1 | 1.5 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.8 | GO:0006544 | glycine metabolic process(GO:0006544) |
| 0.0 | 2.3 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.0 | 1.0 | GO:0002042 | cell migration involved in sprouting angiogenesis(GO:0002042) |
| 0.0 | 0.5 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 1.3 | GO:0070303 | negative regulation of stress-activated MAPK cascade(GO:0032873) negative regulation of stress-activated protein kinase signaling cascade(GO:0070303) |
| 0.0 | 0.2 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.8 | GO:0005980 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.4 | GO:0098789 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 2.1 | GO:1990266 | neutrophil migration(GO:1990266) |
| 0.0 | 0.3 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 1.8 | GO:0001892 | embryonic placenta development(GO:0001892) |
| 0.0 | 0.1 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.0 | 1.0 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 1.1 | GO:0050672 | negative regulation of mononuclear cell proliferation(GO:0032945) negative regulation of lymphocyte proliferation(GO:0050672) |
| 0.0 | 3.0 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.1 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 | 0.2 | GO:0061081 | positive regulation of macrophage cytokine production(GO:0060907) positive regulation of myeloid leukocyte cytokine production involved in immune response(GO:0061081) |
| 0.0 | 1.1 | GO:0009311 | oligosaccharide metabolic process(GO:0009311) |
| 0.0 | 0.5 | GO:0070873 | regulation of glycogen metabolic process(GO:0070873) |
| 0.0 | 0.2 | GO:0090278 | negative regulation of peptide secretion(GO:0002792) negative regulation of peptide hormone secretion(GO:0090278) |
| 0.0 | 2.7 | GO:0015698 | inorganic anion transport(GO:0015698) |
| 0.0 | 0.1 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 0.8 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.3 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.1 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.0 | 3.8 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.0 | 0.3 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.0 | 0.7 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.1 | GO:2000505 | regulation of energy homeostasis(GO:2000505) |
| 0.0 | 0.6 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.6 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.3 | GO:0032480 | negative regulation of type I interferon production(GO:0032480) |
| 0.0 | 0.0 | GO:0071073 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) positive regulation of phospholipid biosynthetic process(GO:0071073) |
| 0.0 | 0.2 | GO:0006661 | phosphatidylinositol biosynthetic process(GO:0006661) |
| 0.0 | 1.2 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.3 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.2 | GO:0070528 | protein kinase C signaling(GO:0070528) |
| 0.0 | 0.4 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.3 | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway(GO:0010803) |
| 0.0 | 0.2 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.7 | 42.8 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 9.1 | 27.3 | GO:0071748 | IgA immunoglobulin complex(GO:0071745) IgA immunoglobulin complex, circulating(GO:0071746) monomeric IgA immunoglobulin complex(GO:0071748) polymeric IgA immunoglobulin complex(GO:0071749) secretory IgA immunoglobulin complex(GO:0071751) |
| 4.8 | 9.5 | GO:0031461 | cullin-RING ubiquitin ligase complex(GO:0031461) |
| 4.4 | 13.3 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 4.2 | 21.1 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 3.4 | 13.5 | GO:0043293 | apoptosome(GO:0043293) |
| 3.1 | 12.4 | GO:0030914 | STAGA complex(GO:0030914) |
| 2.6 | 10.5 | GO:1990745 | EARP complex(GO:1990745) |
| 2.5 | 20.4 | GO:0035976 | AP1 complex(GO:0035976) |
| 2.1 | 98.5 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 1.9 | 7.8 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 1.9 | 9.7 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 1.9 | 20.7 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 1.9 | 7.5 | GO:1990246 | uniplex complex(GO:1990246) |
| 1.9 | 7.5 | GO:0031417 | NatC complex(GO:0031417) |
| 1.8 | 22.2 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 1.8 | 5.4 | GO:0020003 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 1.8 | 7.2 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 1.6 | 8.2 | GO:0036128 | CatSper complex(GO:0036128) |
| 1.5 | 7.5 | GO:0036398 | TCR signalosome(GO:0036398) |
| 1.4 | 5.6 | GO:0097361 | CIA complex(GO:0097361) |
| 1.4 | 5.6 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 1.4 | 1.4 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 1.4 | 1.4 | GO:0030870 | Mre11 complex(GO:0030870) |
| 1.4 | 9.5 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 1.3 | 9.3 | GO:0072487 | MSL complex(GO:0072487) |
| 1.2 | 3.6 | GO:0097181 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 1.2 | 3.5 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 1.1 | 5.7 | GO:0000801 | central element(GO:0000801) |
| 1.1 | 19.8 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 1.1 | 8.6 | GO:0032010 | phagolysosome(GO:0032010) |
| 1.0 | 8.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 1.0 | 3.0 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 1.0 | 7.0 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 1.0 | 2.0 | GO:0005652 | nuclear lamina(GO:0005652) |
| 1.0 | 16.3 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 1.0 | 3.8 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.9 | 8.5 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.9 | 4.7 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.9 | 4.5 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.9 | 4.5 | GO:0089701 | U2AF(GO:0089701) |
| 0.9 | 10.3 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 0.8 | 8.5 | GO:0070938 | contractile ring(GO:0070938) |
| 0.8 | 5.0 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.8 | 5.8 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.8 | 3.2 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.8 | 5.5 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 0.8 | 10.6 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.8 | 4.5 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.7 | 10.3 | GO:0031045 | dense core granule(GO:0031045) |
| 0.7 | 2.2 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.7 | 2.9 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.7 | 14.9 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.7 | 4.2 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.7 | 9.9 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.7 | 4.2 | GO:0043196 | varicosity(GO:0043196) |
| 0.7 | 3.4 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.7 | 12.1 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.7 | 7.3 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.7 | 13.1 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.7 | 2.6 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.7 | 8.5 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.6 | 2.6 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.6 | 14.2 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.6 | 10.9 | GO:0005922 | connexon complex(GO:0005922) |
| 0.6 | 3.6 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.6 | 4.7 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.6 | 4.0 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.6 | 5.1 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.6 | 3.4 | GO:0033063 | DNA recombinase mediator complex(GO:0033061) Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.6 | 1.7 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.6 | 3.9 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.6 | 3.3 | GO:0002177 | manchette(GO:0002177) |
| 0.6 | 2.2 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.5 | 13.7 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.5 | 2.7 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.5 | 12.9 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.5 | 2.6 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 0.5 | 9.8 | GO:0044447 | axoneme part(GO:0044447) |
| 0.5 | 10.6 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.5 | 2.0 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.5 | 1.9 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.5 | 8.1 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.5 | 4.2 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.5 | 2.3 | GO:0001652 | granular component(GO:0001652) |
| 0.5 | 2.8 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.5 | 3.7 | GO:0090576 | RNA polymerase III transcription factor complex(GO:0090576) |
| 0.5 | 3.2 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.5 | 6.4 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.5 | 0.5 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.5 | 18.9 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.4 | 4.5 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.4 | 1.8 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.4 | 1.3 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.4 | 2.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.4 | 6.3 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.4 | 3.4 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.4 | 4.5 | GO:0032797 | SMN complex(GO:0032797) |
| 0.4 | 2.5 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.4 | 28.5 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.4 | 2.8 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.4 | 11.6 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.4 | 0.8 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.4 | 5.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.4 | 2.4 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.4 | 4.2 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.4 | 3.1 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.4 | 6.5 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.4 | 28.9 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.4 | 4.8 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.4 | 1.5 | GO:0038038 | G-protein coupled receptor homodimeric complex(GO:0038038) |
| 0.4 | 4.8 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.4 | 20.5 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.4 | 3.6 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.4 | 2.5 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.4 | 1.8 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.3 | 3.1 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.3 | 3.0 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.3 | 3.3 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.3 | 2.0 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.3 | 2.6 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.3 | 6.1 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.3 | 1.9 | GO:0034464 | BBSome(GO:0034464) |
| 0.3 | 1.8 | GO:0000779 | condensed chromosome, centromeric region(GO:0000779) |
| 0.3 | 49.5 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.3 | 12.4 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.3 | 1.5 | GO:0097123 | cyclin A1-CDK2 complex(GO:0097123) |
| 0.3 | 1.2 | GO:0070081 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.3 | 3.4 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.3 | 2.6 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.3 | 0.3 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.3 | 2.4 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.2 | 1.2 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.2 | 1.0 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.2 | 12.9 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.2 | 27.3 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.2 | 2.4 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.2 | 5.2 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.2 | 5.6 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.2 | 2.3 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.2 | 2.9 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.2 | 1.9 | GO:0016342 | catenin complex(GO:0016342) |
| 0.2 | 0.8 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.2 | 2.4 | GO:0000782 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.2 | 2.2 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.2 | 0.8 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.2 | 1.3 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.2 | 1.0 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.2 | 25.6 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.2 | 2.0 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.2 | 13.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.2 | 6.0 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.2 | 4.3 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.2 | 4.4 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.2 | 5.2 | GO:0005776 | autophagosome(GO:0005776) |
| 0.2 | 4.0 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.2 | 5.1 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.2 | 1.3 | GO:0045179 | apical cortex(GO:0045179) |
| 0.2 | 0.9 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.2 | 0.6 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.2 | 17.9 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 7.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 0.9 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 14.5 | GO:0031514 | motile cilium(GO:0031514) |
| 0.1 | 4.0 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.1 | 1.5 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 2.9 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 1.2 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.1 | 4.1 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 2.5 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 1.6 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.1 | 4.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 10.8 | GO:0001726 | ruffle(GO:0001726) |
| 0.1 | 13.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 1.7 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 3.9 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 1.4 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 1.7 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.1 | 2.8 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.1 | 0.7 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 2.5 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 1.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 11.4 | GO:0043679 | axon terminus(GO:0043679) |
| 0.1 | 1.3 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 3.7 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 1.6 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 0.9 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 6.5 | GO:0042641 | actomyosin(GO:0042641) |
| 0.1 | 5.3 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 1.5 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 3.4 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 1.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.1 | 3.9 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.1 | 0.9 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 1.4 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 3.0 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 4.3 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 0.5 | GO:0042825 | TAP complex(GO:0042825) |
| 0.1 | 0.8 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 20.7 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.1 | 2.1 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 1.0 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 10.4 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.1 | 0.6 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.1 | 0.4 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 7.7 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 0.4 | GO:0070522 | ERCC4-ERCC1 complex(GO:0070522) |
| 0.1 | 0.3 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.1 | 1.2 | GO:0009986 | cell surface(GO:0009986) |
| 0.1 | 1.1 | GO:0019908 | nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.1 | 11.1 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 0.7 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.4 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.5 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.9 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 1.4 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.7 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 3.2 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.6 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 0.6 | GO:0097223 | sperm part(GO:0097223) |
| 0.0 | 0.9 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 1.9 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 15.2 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 1.7 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.7 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.2 | GO:0070069 | cytochrome complex(GO:0070069) |
| 0.0 | 0.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.1 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.5 | 28.6 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 6.6 | 26.3 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 6.1 | 18.4 | GO:0015439 | heme-transporting ATPase activity(GO:0015439) |
| 4.6 | 13.8 | GO:0001861 | complement component C4b receptor activity(GO:0001861) complement component C3b receptor activity(GO:0004877) |
| 4.5 | 13.6 | GO:0005457 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 4.2 | 12.5 | GO:0032093 | SAM domain binding(GO:0032093) |
| 3.8 | 15.2 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 3.8 | 11.3 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 3.6 | 21.5 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 3.4 | 24.0 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 3.3 | 10.0 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 3.3 | 42.8 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 3.2 | 18.9 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 3.0 | 11.8 | GO:0051120 | hepoxilin A3 synthase activity(GO:0051120) |
| 2.9 | 14.7 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 2.9 | 8.8 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 2.8 | 8.5 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 2.6 | 7.8 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 2.5 | 7.6 | GO:0004488 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 2.4 | 7.3 | GO:0003863 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 2.4 | 7.3 | GO:0034584 | piRNA binding(GO:0034584) |
| 2.4 | 131.1 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 2.3 | 11.3 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 2.2 | 9.0 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 2.1 | 38.5 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 2.1 | 6.4 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 2.1 | 4.2 | GO:0050473 | arachidonate 15-lipoxygenase activity(GO:0050473) |
| 2.1 | 12.5 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 2.1 | 10.4 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 2.0 | 7.9 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 2.0 | 7.8 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 1.9 | 9.5 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 1.9 | 15.0 | GO:0032396 | inhibitory MHC class I receptor activity(GO:0032396) |
| 1.8 | 14.8 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 1.8 | 22.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 1.8 | 5.4 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 1.8 | 5.4 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 1.8 | 5.3 | GO:0047322 | [hydroxymethylglutaryl-CoA reductase (NADPH)] kinase activity(GO:0047322) [acetyl-CoA carboxylase] kinase activity(GO:0050405) |
| 1.7 | 5.2 | GO:0015361 | low-affinity sodium:dicarboxylate symporter activity(GO:0015361) |
| 1.7 | 5.0 | GO:0061663 | NEDD8 ligase activity(GO:0061663) |
| 1.6 | 12.8 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 1.6 | 4.7 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 1.6 | 4.7 | GO:0052894 | norspermine:oxygen oxidoreductase activity(GO:0052894) N1-acetylspermine:oxygen oxidoreductase (N1-acetylspermidine-forming) activity(GO:0052895) |
| 1.5 | 19.8 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 1.5 | 11.8 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 1.5 | 13.3 | GO:0003682 | chromatin binding(GO:0003682) |
| 1.4 | 4.3 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 1.4 | 2.9 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 1.4 | 12.9 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 1.4 | 4.2 | GO:0051139 | metal ion:proton antiporter activity(GO:0051139) |
| 1.4 | 4.2 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) |
| 1.4 | 4.2 | GO:0019961 | interferon binding(GO:0019961) |
| 1.4 | 8.4 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 1.4 | 9.8 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 1.4 | 5.5 | GO:0016880 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 1.4 | 5.4 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 1.3 | 4.0 | GO:0004913 | interleukin-4 receptor activity(GO:0004913) interleukin-7 receptor activity(GO:0004917) |
| 1.3 | 6.4 | GO:0086077 | gap junction channel activity involved in SA node cell-atrial cardiac muscle cell electrical coupling(GO:0086020) gap junction channel activity involved in AV node cell-bundle of His cell electrical coupling(GO:0086077) |
| 1.3 | 5.1 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 1.3 | 7.6 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 1.3 | 7.6 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 1.2 | 3.7 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 1.2 | 3.7 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 1.2 | 4.8 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 1.2 | 17.9 | GO:0008391 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 1.2 | 5.8 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 1.1 | 12.4 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 1.1 | 3.3 | GO:0047150 | betaine-homocysteine S-methyltransferase activity(GO:0047150) |
| 1.1 | 5.6 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 1.1 | 4.4 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 1.1 | 10.9 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 1.1 | 3.3 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 1.1 | 10.8 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 1.1 | 3.2 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 1.1 | 4.3 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 1.1 | 3.2 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 1.1 | 14.8 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 1.0 | 13.6 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 1.0 | 3.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 1.0 | 6.3 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 1.0 | 3.1 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 1.0 | 3.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 1.0 | 3.0 | GO:0004961 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 1.0 | 13.9 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 1.0 | 2.0 | GO:0009374 | biotin binding(GO:0009374) |
| 1.0 | 3.9 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 1.0 | 4.9 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 1.0 | 10.6 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 1.0 | 2.9 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 1.0 | 3.8 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 1.0 | 2.9 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.9 | 4.7 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.9 | 5.6 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.9 | 10.3 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.9 | 2.8 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.9 | 2.7 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.9 | 1.8 | GO:0016597 | amino acid binding(GO:0016597) modified amino acid binding(GO:0072341) |
| 0.9 | 3.6 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.9 | 4.5 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.9 | 2.7 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.9 | 3.5 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.8 | 4.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.8 | 5.9 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.8 | 8.4 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.8 | 3.4 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.8 | 5.0 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.8 | 5.8 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.8 | 8.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.8 | 3.2 | GO:1901474 | azole transmembrane transporter activity(GO:1901474) |
| 0.8 | 4.8 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.8 | 3.2 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.8 | 8.7 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.8 | 4.8 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.8 | 4.0 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.8 | 8.5 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.8 | 2.3 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.8 | 5.4 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.7 | 3.0 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.7 | 3.7 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.7 | 4.5 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.7 | 3.7 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.7 | 1.5 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.7 | 12.6 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.7 | 5.9 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.7 | 2.2 | GO:0005330 | dopamine:sodium symporter activity(GO:0005330) |
| 0.7 | 4.4 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.7 | 36.5 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.7 | 3.6 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.7 | 2.9 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.7 | 2.9 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.7 | 21.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.7 | 5.7 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.7 | 2.8 | GO:0008940 | nitrate reductase activity(GO:0008940) |
| 0.7 | 13.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.7 | 4.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.7 | 4.8 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.7 | 7.5 | GO:0008061 | chitinase activity(GO:0004568) chitin binding(GO:0008061) |
| 0.7 | 4.8 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.7 | 6.7 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.7 | 2.7 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.7 | 12.5 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.7 | 5.9 | GO:0043426 | MRF binding(GO:0043426) |
| 0.7 | 2.6 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.6 | 2.6 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.6 | 1.9 | GO:0004915 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.6 | 8.9 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.6 | 2.6 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.6 | 2.5 | GO:0035473 | lipase binding(GO:0035473) |
| 0.6 | 4.4 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.6 | 1.9 | GO:0016782 | transferase activity, transferring sulfur-containing groups(GO:0016782) |
| 0.6 | 1.9 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.6 | 3.7 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.6 | 4.3 | GO:0004459 | lactate dehydrogenase activity(GO:0004457) L-lactate dehydrogenase activity(GO:0004459) |
| 0.6 | 4.9 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.6 | 1.8 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 0.6 | 7.2 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.6 | 3.6 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.6 | 7.7 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.6 | 7.7 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.6 | 4.7 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.6 | 1.8 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.6 | 7.5 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.6 | 2.9 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.6 | 7.4 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.6 | 6.2 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.6 | 9.0 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.6 | 3.9 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.6 | 1.7 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.6 | 2.2 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.5 | 3.8 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.5 | 3.7 | GO:0016713 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.5 | 6.9 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.5 | 1.6 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.5 | 16.6 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.5 | 3.1 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.5 | 1.5 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.5 | 2.6 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.5 | 9.2 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.5 | 5.6 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.5 | 7.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.5 | 10.6 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.5 | 4.0 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.5 | 2.5 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.5 | 10.9 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.5 | 2.5 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.5 | 2.4 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.5 | 17.4 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.5 | 1.4 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.5 | 9.5 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.5 | 1.4 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.5 | 19.9 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.5 | 1.4 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.5 | 11.7 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.5 | 2.3 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.5 | 2.8 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.5 | 2.3 | GO:0070728 | leucine binding(GO:0070728) |
| 0.5 | 1.4 | GO:0000035 | acyl binding(GO:0000035) |
| 0.5 | 2.3 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.5 | 5.0 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.5 | 13.2 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.4 | 4.5 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.4 | 9.2 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.4 | 34.1 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.4 | 1.7 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.4 | 17.0 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.4 | 1.7 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.4 | 1.7 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.4 | 1.7 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.4 | 4.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.4 | 1.7 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.4 | 1.2 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) NAD(P)+ nucleosidase activity(GO:0050135) |
| 0.4 | 6.1 | GO:0015250 | water channel activity(GO:0015250) |
| 0.4 | 3.7 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.4 | 8.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.4 | 2.0 | GO:0070905 | serine binding(GO:0070905) |
| 0.4 | 1.6 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.4 | 3.6 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.4 | 4.7 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.4 | 1.6 | GO:0044877 | macromolecular complex binding(GO:0044877) |
| 0.4 | 1.2 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.4 | 1.6 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.4 | 7.0 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.4 | 1.2 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.4 | 11.6 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.4 | 2.3 | GO:0052658 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.4 | 1.2 | GO:0004603 | phenylethanolamine N-methyltransferase activity(GO:0004603) |
| 0.4 | 2.3 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.4 | 2.3 | GO:0004331 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.4 | 3.4 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.4 | 2.3 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.4 | 3.7 | GO:0030882 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.4 | 8.2 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.4 | 1.1 | GO:0019107 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) myristoyltransferase activity(GO:0019107) |
| 0.4 | 2.6 | GO:0099589 | G-protein coupled serotonin receptor activity(GO:0004993) serotonin receptor activity(GO:0099589) |
| 0.4 | 3.3 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.4 | 6.5 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.4 | 3.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.4 | 2.5 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.4 | 12.4 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.4 | 1.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.4 | 3.9 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.3 | 8.0 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.3 | 0.7 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.3 | 3.1 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.3 | 42.2 | GO:0003823 | antigen binding(GO:0003823) |
| 0.3 | 3.7 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.3 | 1.0 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.3 | 1.7 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.3 | 1.7 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.3 | 2.7 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.3 | 1.7 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.3 | 1.3 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.3 | 1.6 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.3 | 5.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.3 | 1.3 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.3 | 1.3 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.3 | 3.5 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.3 | 3.1 | GO:0022821 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) potassium ion antiporter activity(GO:0022821) |
| 0.3 | 17.2 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.3 | 0.9 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.3 | 0.3 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.3 | 2.1 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.3 | 6.4 | GO:0099604 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.3 | 1.8 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.3 | 1.5 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.3 | 6.7 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.3 | 5.5 | GO:0008026 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) ATP-dependent helicase activity(GO:0008026) RNA-dependent ATPase activity(GO:0008186) purine NTP-dependent helicase activity(GO:0070035) |
| 0.3 | 3.4 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.3 | 4.8 | GO:0070513 | death domain binding(GO:0070513) |
| 0.3 | 1.1 | GO:0034618 | nitric-oxide synthase activity(GO:0004517) arginine binding(GO:0034618) |
| 0.3 | 3.3 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.3 | 1.9 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.3 | 1.4 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.3 | 3.0 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.3 | 3.0 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.3 | 1.1 | GO:0052795 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.3 | 2.7 | GO:0003909 | DNA ligase activity(GO:0003909) |
| 0.3 | 2.7 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.3 | 5.3 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.3 | 7.9 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.3 | 1.3 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.3 | 9.8 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.3 | 2.3 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.2 | 3.2 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.2 | 2.0 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.2 | 2.5 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.2 | 1.2 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.2 | 2.7 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.2 | 5.9 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.2 | 2.9 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.2 | 4.3 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.2 | 3.1 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.2 | 26.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.2 | 1.2 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.2 | 3.3 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.2 | 2.3 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.2 | 2.3 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.2 | 3.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.2 | 8.9 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.2 | 0.7 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.2 | 8.1 | GO:0032451 | demethylase activity(GO:0032451) |
| 0.2 | 1.4 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.2 | 0.7 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.2 | 1.1 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.2 | 1.8 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.2 | 0.9 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.2 | 2.0 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.2 | 3.3 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.2 | 2.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.2 | 5.0 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.2 | 5.4 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.2 | 1.5 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.2 | 1.7 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.2 | 1.5 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.2 | 2.5 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.2 | 0.8 | GO:0008518 | reduced folate carrier activity(GO:0008518) methotrexate transporter activity(GO:0015350) |
| 0.2 | 1.4 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.2 | 0.8 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.2 | 4.3 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.2 | 2.5 | GO:0016866 | intramolecular transferase activity(GO:0016866) |
| 0.2 | 0.8 | GO:0042020 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.2 | 1.4 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.2 | 1.0 | GO:0047374 | methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.2 | 30.3 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.2 | 2.5 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.2 | 2.6 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.2 | 1.1 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.2 | 4.8 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.2 | 3.3 | GO:0016502 | purinergic nucleotide receptor activity(GO:0001614) nucleotide receptor activity(GO:0016502) |
| 0.2 | 5.9 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 1.5 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.2 | 7.2 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.2 | 2.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.2 | 0.9 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.2 | 0.9 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.2 | 5.8 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.2 | 2.9 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.2 | 1.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.2 | 210.3 | GO:0001071 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.2 | 2.5 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.2 | 2.8 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.2 | 0.5 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.2 | 6.9 | GO:0016278 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.2 | 0.7 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.2 | 1.1 | GO:0030250 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.2 | 1.5 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.2 | 3.8 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.2 | 0.5 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.2 | 0.6 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.2 | 1.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.2 | 2.4 | GO:0019864 | IgG binding(GO:0019864) |
| 0.1 | 3.1 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.1 | 7.0 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.1 | 2.7 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 5.0 | GO:0008235 | metalloexopeptidase activity(GO:0008235) |
| 0.1 | 1.0 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.1 | 1.8 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 2.5 | GO:0016758 | transferase activity, transferring hexosyl groups(GO:0016758) |
| 0.1 | 1.9 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 2.7 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.1 | 2.6 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 2.6 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 0.8 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.1 | 2.9 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.1 | 4.5 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 0.7 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 0.3 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.1 | 6.5 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 3.3 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 2.6 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 5.8 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.1 | 0.6 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 5.4 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.1 | 1.5 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 0.5 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.1 | 3.6 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 5.4 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 1.4 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 1.7 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.1 | 0.4 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 5.9 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.1 | 2.8 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 1.2 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 4.3 | GO:0034212 | peptide N-acetyltransferase activity(GO:0034212) |
| 0.1 | 1.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 2.0 | GO:0010857 | calcium-dependent protein kinase activity(GO:0010857) |
| 0.1 | 1.6 | GO:0016594 | glycine binding(GO:0016594) |
| 0.1 | 2.1 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 1.1 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 0.8 | GO:0005113 | patched binding(GO:0005113) |
| 0.1 | 6.7 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) |
| 0.1 | 8.7 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.1 | 0.7 | GO:0004844 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.1 | 3.4 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 0.4 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.1 | 0.6 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.1 | 3.2 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 0.9 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.1 | 0.3 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.1 | 1.5 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 2.5 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.1 | 2.0 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.1 | 0.2 | GO:0005549 | odorant binding(GO:0005549) |
| 0.1 | 2.0 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 2.7 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 1.2 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 1.4 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.8 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 1.4 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 2.4 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 3.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 4.7 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 0.9 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.5 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 0.9 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.1 | 0.2 | GO:0070403 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) NAD+ binding(GO:0070403) |
| 0.1 | 1.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 0.2 | GO:0043028 | cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.1 | 0.7 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 0.8 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 9.6 | GO:0005088 | Ras guanyl-nucleotide exchange factor activity(GO:0005088) |
| 0.1 | 0.6 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 0.7 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.1 | 1.4 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 0.4 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 0.9 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.1 | 0.8 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.1 | 1.0 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 1.3 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 4.5 | GO:0005261 | cation channel activity(GO:0005261) |
| 0.1 | 0.1 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.1 | 1.5 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 0.2 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.1 | 0.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.8 | GO:0043394 | proteoglycan binding(GO:0043394) |
| 0.0 | 3.4 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.4 | GO:0005539 | glycosaminoglycan binding(GO:0005539) |
| 0.0 | 1.3 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 1.1 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.2 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.6 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 0.9 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 1.0 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.2 | GO:0019862 | IgA binding(GO:0019862) |
| 0.0 | 2.4 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.5 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 1.0 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 1.4 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 6.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.1 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.0 | 0.8 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.2 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.9 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.1 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.6 | GO:1901476 | carbohydrate transmembrane transporter activity(GO:0015144) carbohydrate transporter activity(GO:1901476) |
| 0.0 | 2.4 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.2 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.3 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.0 | 0.4 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 1.1 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.0 | 0.2 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.0 | 0.1 | GO:0005126 | cytokine receptor binding(GO:0005126) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 6.3 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 1.1 | 5.3 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 1.0 | 54.8 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.9 | 2.6 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.8 | 23.3 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.7 | 3.5 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.7 | 28.3 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.7 | 9.1 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.6 | 7.7 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.6 | 60.8 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.6 | 4.2 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.6 | 32.2 | PID ATM PATHWAY | ATM pathway |
| 0.5 | 17.2 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.5 | 14.0 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.4 | 37.3 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.4 | 3.9 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.4 | 17.0 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.4 | 11.8 | PID EPO PATHWAY | EPO signaling pathway |
| 0.4 | 9.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.4 | 2.4 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.4 | 19.7 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.4 | 12.9 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.3 | 3.0 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.3 | 1.9 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.3 | 4.9 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.3 | 6.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.3 | 1.8 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.3 | 1.3 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.2 | 3.6 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.2 | 9.1 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.2 | 3.0 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.2 | 36.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.2 | 2.8 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.2 | 2.0 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.2 | 2.4 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.2 | 11.5 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.2 | 6.9 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.2 | 4.8 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.2 | 5.6 | PID ATR PATHWAY | ATR signaling pathway |
| 0.2 | 1.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.2 | 4.5 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.2 | 1.3 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.2 | 1.7 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.2 | 3.1 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.2 | 5.2 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.2 | 0.8 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 8.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 9.2 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 2.1 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.1 | 0.5 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 1.7 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 3.1 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 3.8 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 1.8 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.1 | 1.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 1.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 1.8 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.1 | 3.3 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 5.8 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 2.6 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 1.9 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 1.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.1 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 1.8 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.4 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 2.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.3 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 2.0 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.0 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 1.9 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.9 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 12.1 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.5 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 1.0 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.8 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.9 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.4 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.4 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.2 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 2.9 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 2.2 | 4.3 | REACTOME FORMATION OF THE HIV1 EARLY ELONGATION COMPLEX | Genes involved in Formation of the HIV-1 Early Elongation Complex |
| 1.9 | 1.9 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 1.5 | 32.7 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 1.5 | 7.3 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 1.3 | 2.6 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 1.2 | 24.6 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 1.2 | 1.2 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 1.1 | 48.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 1.1 | 15.0 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 1.0 | 24.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 1.0 | 20.0 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.9 | 78.5 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.9 | 10.0 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.8 | 12.6 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.8 | 11.6 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.8 | 8.9 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.8 | 10.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.7 | 7.2 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.7 | 12.6 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.7 | 10.5 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.7 | 8.8 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.7 | 8.5 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.6 | 10.9 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.6 | 14.4 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.6 | 1.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.5 | 6.6 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.5 | 4.8 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.5 | 22.6 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.5 | 9.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.5 | 6.6 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.5 | 8.0 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.5 | 7.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.5 | 1.9 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.5 | 13.4 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.5 | 5.4 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.4 | 9.9 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.4 | 20.0 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.4 | 10.1 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.4 | 16.5 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.4 | 161.9 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.4 | 5.7 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.4 | 8.1 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.4 | 13.5 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.4 | 5.9 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.4 | 7.8 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.4 | 7.1 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.4 | 11.1 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.4 | 3.9 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.3 | 10.0 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.3 | 5.6 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.3 | 1.0 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.3 | 7.5 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.3 | 6.6 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.3 | 2.8 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.3 | 3.1 | REACTOME TRANSCRIPTION COUPLED NER TC NER | Genes involved in Transcription-coupled NER (TC-NER) |
| 0.3 | 6.8 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.3 | 8.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.3 | 3.4 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.3 | 4.2 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.3 | 6.1 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.3 | 7.7 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.3 | 7.3 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.3 | 2.6 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.3 | 4.9 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.2 | 5.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.2 | 13.5 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.2 | 2.7 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.2 | 13.1 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.2 | 5.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.2 | 2.5 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.2 | 3.7 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.2 | 8.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.2 | 10.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.2 | 1.9 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.2 | 1.7 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.2 | 6.9 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.2 | 10.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.2 | 1.6 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.2 | 3.7 | REACTOME PRE NOTCH EXPRESSION AND PROCESSING | Genes involved in Pre-NOTCH Expression and Processing |
| 0.2 | 3.5 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.2 | 2.2 | REACTOME EARLY PHASE OF HIV LIFE CYCLE | Genes involved in Early Phase of HIV Life Cycle |
| 0.2 | 7.1 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.2 | 5.1 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.2 | 8.2 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.2 | 3.7 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.2 | 3.8 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.2 | 0.2 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.2 | 7.4 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.2 | 1.9 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.2 | 3.4 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.2 | 13.7 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 2.5 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.1 | 2.7 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 9.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.1 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.1 | 4.8 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 3.7 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 2.8 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.1 | 13.4 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 2.1 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.1 | 3.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 3.3 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.1 | 1.2 | REACTOME OPSINS | Genes involved in Opsins |
| 0.1 | 1.2 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 13.4 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 0.5 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.1 | 1.8 | REACTOME PI3K EVENTS IN ERBB4 SIGNALING | Genes involved in PI3K events in ERBB4 signaling |
| 0.1 | 3.7 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 1.9 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 0.9 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.1 | 0.5 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 21.5 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.1 | 1.1 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 0.6 | REACTOME CELL DEATH SIGNALLING VIA NRAGE NRIF AND NADE | Genes involved in Cell death signalling via NRAGE, NRIF and NADE |
| 0.1 | 1.5 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 0.6 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 1.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.6 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 1.2 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.1 | 0.4 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 0.6 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 0.6 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 1.5 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.5 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 1.3 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.5 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.0 | 0.8 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 1.1 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 1.3 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 1.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.7 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.4 | REACTOME LIPID DIGESTION MOBILIZATION AND TRANSPORT | Genes involved in Lipid digestion, mobilization, and transport |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.1 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |