Project
averaged GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
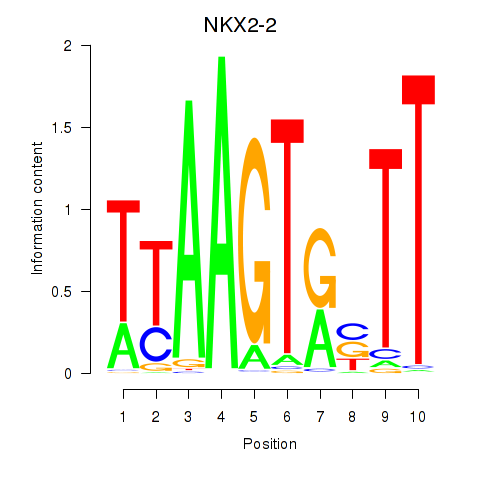
Results for NKX2-2
Z-value: 1.65
Transcription factors associated with NKX2-2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX2-2
|
ENSG00000125820.5 | NK2 homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NKX2-2 | hg19_v2_chr20_-_21494654_21494678 | -0.22 | 1.2e-03 | Click! |
Activity profile of NKX2-2 motif
Sorted Z-values of NKX2-2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_91926103 | 38.52 |
ENST00000314355.6
|
CKS2
|
CDC28 protein kinase regulatory subunit 2 |
| chr1_-_54405773 | 35.85 |
ENST00000371376.1
|
HSPB11
|
heat shock protein family B (small), member 11 |
| chr5_+_159848807 | 35.25 |
ENST00000352433.5
|
PTTG1
|
pituitary tumor-transforming 1 |
| chr9_+_131447342 | 34.53 |
ENST00000409104.3
|
SET
|
SET nuclear oncogene |
| chr2_+_170655322 | 33.68 |
ENST00000260956.4
ENST00000417292.1 |
SSB
|
Sjogren syndrome antigen B (autoantigen La) |
| chr5_+_159848854 | 33.62 |
ENST00000517480.1
ENST00000520452.1 ENST00000393964.1 |
PTTG1
|
pituitary tumor-transforming 1 |
| chr17_+_48823896 | 26.43 |
ENST00000511974.1
|
LUC7L3
|
LUC7-like 3 (S. cerevisiae) |
| chr8_-_121457332 | 26.41 |
ENST00000518918.1
|
MRPL13
|
mitochondrial ribosomal protein L13 |
| chr15_-_60690163 | 25.26 |
ENST00000558998.1
ENST00000560165.1 ENST00000557986.1 ENST00000559780.1 ENST00000559467.1 ENST00000559956.1 ENST00000332680.4 ENST00000396024.3 ENST00000421017.2 ENST00000560466.1 ENST00000558132.1 ENST00000559113.1 ENST00000557906.1 ENST00000558558.1 ENST00000560468.1 ENST00000559370.1 ENST00000558169.1 ENST00000559725.1 ENST00000558985.1 ENST00000451270.2 |
ANXA2
|
annexin A2 |
| chr8_-_101962777 | 24.93 |
ENST00000395951.3
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr12_+_69004619 | 24.57 |
ENST00000250559.9
ENST00000393436.5 ENST00000425247.2 ENST00000489473.2 ENST00000422358.2 ENST00000541167.1 ENST00000538283.1 ENST00000341355.5 ENST00000537460.1 ENST00000450214.2 ENST00000545270.1 ENST00000538980.1 ENST00000542018.1 ENST00000543393.1 |
RAP1B
|
RAP1B, member of RAS oncogene family |
| chr10_-_121296045 | 24.55 |
ENST00000392865.1
|
RGS10
|
regulator of G-protein signaling 10 |
| chr19_-_18433910 | 24.25 |
ENST00000594828.3
ENST00000593829.1 |
LSM4
|
LSM4 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr3_+_158991025 | 23.85 |
ENST00000337808.6
|
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr5_+_34757309 | 22.74 |
ENST00000397449.1
|
RAI14
|
retinoic acid induced 14 |
| chr22_-_42086477 | 22.72 |
ENST00000402458.1
|
NHP2L1
|
NHP2 non-histone chromosome protein 2-like 1 (S. cerevisiae) |
| chr19_+_47634039 | 21.80 |
ENST00000597808.1
ENST00000413379.3 ENST00000600706.1 ENST00000540850.1 ENST00000598840.1 ENST00000600753.1 ENST00000270225.7 ENST00000392776.3 |
SAE1
|
SUMO1 activating enzyme subunit 1 |
| chr17_+_48823975 | 21.40 |
ENST00000513969.1
ENST00000503728.1 |
LUC7L3
|
LUC7-like 3 (S. cerevisiae) |
| chr10_-_36813162 | 20.92 |
ENST00000440465.1
|
NAMPTL
|
nicotinamide phosphoribosyltransferase-like |
| chr14_-_100841670 | 20.60 |
ENST00000557297.1
ENST00000555813.1 ENST00000557135.1 ENST00000556698.1 ENST00000554509.1 ENST00000555410.1 |
WARS
|
tryptophanyl-tRNA synthetase |
| chr5_+_135394840 | 19.90 |
ENST00000503087.1
|
TGFBI
|
transforming growth factor, beta-induced, 68kDa |
| chr1_-_75198940 | 19.56 |
ENST00000417775.1
|
CRYZ
|
crystallin, zeta (quinone reductase) |
| chr2_-_151344172 | 19.43 |
ENST00000375734.2
ENST00000263895.4 ENST00000454202.1 |
RND3
|
Rho family GTPase 3 |
| chr15_+_65843130 | 19.23 |
ENST00000569894.1
|
PTPLAD1
|
protein tyrosine phosphatase-like A domain containing 1 |
| chr2_+_219110149 | 19.13 |
ENST00000456575.1
|
ARPC2
|
actin related protein 2/3 complex, subunit 2, 34kDa |
| chr8_-_121457608 | 18.80 |
ENST00000306185.3
|
MRPL13
|
mitochondrial ribosomal protein L13 |
| chr11_-_11374904 | 18.79 |
ENST00000528848.2
|
CSNK2A3
|
casein kinase 2, alpha 3 polypeptide |
| chr7_+_2394445 | 18.67 |
ENST00000360876.4
ENST00000413917.1 ENST00000397011.2 |
EIF3B
|
eukaryotic translation initiation factor 3, subunit B |
| chr3_+_23847432 | 18.42 |
ENST00000346855.3
|
UBE2E1
|
ubiquitin-conjugating enzyme E2E 1 |
| chrX_-_70474910 | 18.26 |
ENST00000373988.1
ENST00000373998.1 |
ZMYM3
|
zinc finger, MYM-type 3 |
| chr8_+_26150628 | 18.17 |
ENST00000523925.1
ENST00000315985.7 |
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha |
| chr19_-_14945933 | 17.98 |
ENST00000322301.3
|
OR7A5
|
olfactory receptor, family 7, subfamily A, member 5 |
| chr3_+_152017181 | 17.85 |
ENST00000498502.1
ENST00000324196.5 ENST00000545754.1 ENST00000357472.3 |
MBNL1
|
muscleblind-like splicing regulator 1 |
| chr8_+_71485681 | 17.79 |
ENST00000391684.1
|
AC120194.1
|
AC120194.1 |
| chr8_+_11666649 | 17.24 |
ENST00000528643.1
ENST00000525777.1 |
FDFT1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr19_+_39138320 | 17.21 |
ENST00000424234.2
ENST00000390009.3 ENST00000589528.1 |
ACTN4
|
actinin, alpha 4 |
| chr8_-_49834299 | 17.17 |
ENST00000396822.1
|
SNAI2
|
snail family zinc finger 2 |
| chr6_-_150067632 | 17.16 |
ENST00000460354.2
ENST00000367404.4 ENST00000543637.1 |
NUP43
|
nucleoporin 43kDa |
| chr11_-_107729887 | 17.12 |
ENST00000525815.1
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr18_+_29671812 | 16.50 |
ENST00000261593.3
ENST00000578914.1 |
RNF138
|
ring finger protein 138, E3 ubiquitin protein ligase |
| chr1_-_115259337 | 16.35 |
ENST00000369535.4
|
NRAS
|
neuroblastoma RAS viral (v-ras) oncogene homolog |
| chr5_-_43557791 | 15.94 |
ENST00000338972.4
ENST00000511321.1 ENST00000515338.1 |
PAIP1
|
poly(A) binding protein interacting protein 1 |
| chr15_+_79165372 | 15.84 |
ENST00000558502.1
|
MORF4L1
|
mortality factor 4 like 1 |
| chr14_-_100841930 | 15.80 |
ENST00000555031.1
ENST00000553395.1 ENST00000553545.1 ENST00000344102.5 ENST00000556338.1 ENST00000392882.2 ENST00000553934.1 |
WARS
|
tryptophanyl-tRNA synthetase |
| chr9_-_139361503 | 15.63 |
ENST00000453963.1
|
SEC16A
|
SEC16 homolog A (S. cerevisiae) |
| chr1_+_53480598 | 15.53 |
ENST00000430330.2
ENST00000408941.3 ENST00000478274.2 ENST00000484100.1 ENST00000435345.2 ENST00000488965.1 |
SCP2
|
sterol carrier protein 2 |
| chrX_-_23761317 | 15.51 |
ENST00000492081.1
ENST00000379303.5 ENST00000336430.7 |
ACOT9
|
acyl-CoA thioesterase 9 |
| chr3_+_23847394 | 15.35 |
ENST00000306627.3
|
UBE2E1
|
ubiquitin-conjugating enzyme E2E 1 |
| chr11_-_77850629 | 15.16 |
ENST00000376156.3
ENST00000525870.1 ENST00000530454.1 ENST00000525755.1 ENST00000527099.1 ENST00000525761.1 ENST00000299626.5 |
ALG8
|
ALG8, alpha-1,3-glucosyltransferase |
| chr6_-_150067696 | 15.01 |
ENST00000340413.2
ENST00000367403.3 |
NUP43
|
nucleoporin 43kDa |
| chr9_-_86593238 | 14.69 |
ENST00000351839.3
|
HNRNPK
|
heterogeneous nuclear ribonucleoprotein K |
| chr5_+_82767583 | 14.65 |
ENST00000512590.2
ENST00000513960.1 ENST00000513984.1 ENST00000502527.2 |
VCAN
|
versican |
| chr12_-_49582978 | 14.61 |
ENST00000301071.7
|
TUBA1A
|
tubulin, alpha 1a |
| chrX_+_123095546 | 14.44 |
ENST00000371157.3
ENST00000371145.3 ENST00000371144.3 |
STAG2
|
stromal antigen 2 |
| chr18_+_9103957 | 14.42 |
ENST00000400033.1
|
NDUFV2
|
NADH dehydrogenase (ubiquinone) flavoprotein 2, 24kDa |
| chr5_+_82767487 | 14.16 |
ENST00000343200.5
ENST00000342785.4 |
VCAN
|
versican |
| chrX_-_70474499 | 14.00 |
ENST00000353904.2
|
ZMYM3
|
zinc finger, MYM-type 3 |
| chr8_-_141810634 | 13.78 |
ENST00000521986.1
ENST00000523539.1 ENST00000538769.1 |
PTK2
|
protein tyrosine kinase 2 |
| chr8_-_49833978 | 13.44 |
ENST00000020945.1
|
SNAI2
|
snail family zinc finger 2 |
| chr22_-_36220420 | 13.27 |
ENST00000473487.2
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr12_-_120315074 | 13.23 |
ENST00000261833.7
ENST00000392521.2 |
CIT
|
citron (rho-interacting, serine/threonine kinase 21) |
| chr15_+_79165296 | 13.15 |
ENST00000558746.1
ENST00000558830.1 ENST00000559345.1 |
MORF4L1
|
mortality factor 4 like 1 |
| chr1_-_155880672 | 12.65 |
ENST00000609492.1
ENST00000368322.3 |
RIT1
|
Ras-like without CAAX 1 |
| chr9_+_71819927 | 12.47 |
ENST00000535702.1
|
TJP2
|
tight junction protein 2 |
| chr8_-_101719159 | 12.46 |
ENST00000520868.1
ENST00000522658.1 |
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr7_+_115862858 | 12.46 |
ENST00000393481.2
|
TES
|
testis derived transcript (3 LIM domains) |
| chr20_+_57226841 | 12.42 |
ENST00000358029.4
ENST00000361830.3 |
STX16
|
syntaxin 16 |
| chr9_+_71820057 | 11.92 |
ENST00000539225.1
|
TJP2
|
tight junction protein 2 |
| chr2_+_201994208 | 11.83 |
ENST00000440180.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr15_-_56209306 | 11.82 |
ENST00000506154.1
ENST00000338963.2 ENST00000508342.1 |
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4, E3 ubiquitin protein ligase |
| chr19_-_2427536 | 11.54 |
ENST00000591871.1
|
TIMM13
|
translocase of inner mitochondrial membrane 13 homolog (yeast) |
| chr18_+_55888767 | 11.49 |
ENST00000431212.2
ENST00000586268.1 ENST00000587190.1 |
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr3_+_183892635 | 11.47 |
ENST00000427072.1
ENST00000411763.2 ENST00000292807.5 ENST00000448139.1 ENST00000455925.1 |
AP2M1
|
adaptor-related protein complex 2, mu 1 subunit |
| chr3_+_179280668 | 11.46 |
ENST00000429709.2
ENST00000450518.2 ENST00000392662.1 ENST00000490364.1 |
ACTL6A
|
actin-like 6A |
| chr1_-_246670519 | 11.41 |
ENST00000388985.4
ENST00000490107.1 |
SMYD3
|
SET and MYND domain containing 3 |
| chr17_+_52978107 | 11.19 |
ENST00000445275.2
|
TOM1L1
|
target of myb1 (chicken)-like 1 |
| chr19_+_39138271 | 11.05 |
ENST00000252699.2
|
ACTN4
|
actinin, alpha 4 |
| chr1_-_36615051 | 10.54 |
ENST00000373163.1
|
TRAPPC3
|
trafficking protein particle complex 3 |
| chr11_+_9406169 | 10.42 |
ENST00000379719.3
ENST00000527431.1 |
IPO7
|
importin 7 |
| chr8_-_101718991 | 10.34 |
ENST00000517990.1
|
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr5_-_98262240 | 10.30 |
ENST00000284049.3
|
CHD1
|
chromodomain helicase DNA binding protein 1 |
| chr5_+_118812294 | 10.25 |
ENST00000509514.1
|
HSD17B4
|
hydroxysteroid (17-beta) dehydrogenase 4 |
| chr3_-_33686743 | 9.98 |
ENST00000333778.6
ENST00000539981.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr6_-_55739542 | 9.44 |
ENST00000446683.2
|
BMP5
|
bone morphogenetic protein 5 |
| chr1_-_36615065 | 8.98 |
ENST00000373166.3
ENST00000373159.1 ENST00000373162.1 |
TRAPPC3
|
trafficking protein particle complex 3 |
| chr10_+_13628921 | 8.70 |
ENST00000378572.3
|
PRPF18
|
pre-mRNA processing factor 18 |
| chr17_+_66521936 | 8.64 |
ENST00000592800.1
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr3_-_196911002 | 8.32 |
ENST00000452595.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr12_+_56546363 | 8.22 |
ENST00000551834.1
ENST00000552568.1 |
MYL6B
|
myosin, light chain 6B, alkali, smooth muscle and non-muscle |
| chr8_-_141774467 | 8.02 |
ENST00000520151.1
ENST00000519024.1 ENST00000519465.1 |
PTK2
|
protein tyrosine kinase 2 |
| chr14_+_96968707 | 8.00 |
ENST00000216277.8
ENST00000557320.1 ENST00000557471.1 |
PAPOLA
|
poly(A) polymerase alpha |
| chr11_+_64949899 | 7.99 |
ENST00000531068.1
ENST00000527699.1 ENST00000533909.1 ENST00000527323.1 |
CAPN1
|
calpain 1, (mu/I) large subunit |
| chr12_+_20963632 | 7.98 |
ENST00000540853.1
ENST00000261196.2 |
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr5_+_140602904 | 7.97 |
ENST00000515856.2
ENST00000239449.4 |
PCDHB14
|
protocadherin beta 14 |
| chr17_+_75315534 | 7.95 |
ENST00000590294.1
ENST00000329047.8 |
SEPT9
|
septin 9 |
| chr3_+_38206975 | 7.85 |
ENST00000446845.1
ENST00000311806.3 |
OXSR1
|
oxidative stress responsive 1 |
| chr6_+_158733692 | 7.81 |
ENST00000367094.2
ENST00000367097.3 |
TULP4
|
tubby like protein 4 |
| chr12_+_20963647 | 7.72 |
ENST00000381545.3
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr8_+_110552337 | 7.58 |
ENST00000337573.5
|
EBAG9
|
estrogen receptor binding site associated, antigen, 9 |
| chr2_+_162087577 | 7.57 |
ENST00000439442.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr5_+_167718604 | 7.30 |
ENST00000265293.4
|
WWC1
|
WW and C2 domain containing 1 |
| chr5_-_179047881 | 7.28 |
ENST00000521173.1
|
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 (H) |
| chr10_-_112678692 | 7.27 |
ENST00000605742.1
|
BBIP1
|
BBSome interacting protein 1 |
| chrX_+_11777671 | 7.26 |
ENST00000380693.3
ENST00000380692.2 |
MSL3
|
male-specific lethal 3 homolog (Drosophila) |
| chr13_+_50656307 | 7.22 |
ENST00000378180.4
|
DLEU1
|
deleted in lymphocytic leukemia 1 (non-protein coding) |
| chr2_+_189157536 | 7.16 |
ENST00000409580.1
ENST00000409637.3 |
GULP1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr8_+_11660227 | 7.16 |
ENST00000443614.2
ENST00000525900.1 |
FDFT1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr2_+_189157498 | 7.13 |
ENST00000359135.3
|
GULP1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr5_+_118812237 | 7.12 |
ENST00000513628.1
|
HSD17B4
|
hydroxysteroid (17-beta) dehydrogenase 4 |
| chr14_-_50154921 | 7.09 |
ENST00000553805.2
ENST00000554396.1 ENST00000216367.5 ENST00000539565.2 |
POLE2
|
polymerase (DNA directed), epsilon 2, accessory subunit |
| chr1_-_31538517 | 7.09 |
ENST00000440538.2
ENST00000423018.2 ENST00000424085.2 ENST00000426105.2 ENST00000257075.5 ENST00000373747.3 ENST00000525843.1 ENST00000373742.2 |
PUM1
|
pumilio RNA-binding family member 1 |
| chr1_-_227505289 | 7.07 |
ENST00000366765.3
|
CDC42BPA
|
CDC42 binding protein kinase alpha (DMPK-like) |
| chr15_+_79165151 | 6.97 |
ENST00000331268.5
|
MORF4L1
|
mortality factor 4 like 1 |
| chr10_+_13203543 | 6.91 |
ENST00000378714.3
ENST00000479669.1 ENST00000484800.2 |
MCM10
|
minichromosome maintenance complex component 10 |
| chr4_+_41937131 | 6.91 |
ENST00000504986.1
ENST00000508448.1 ENST00000513702.1 ENST00000325094.5 |
TMEM33
|
transmembrane protein 33 |
| chr16_+_30212378 | 6.70 |
ENST00000569485.1
|
SULT1A3
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3 |
| chr8_+_11660120 | 6.60 |
ENST00000220584.4
|
FDFT1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr2_-_55276320 | 6.58 |
ENST00000357376.3
|
RTN4
|
reticulon 4 |
| chr4_-_105416039 | 6.51 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chr2_+_120436733 | 6.15 |
ENST00000401466.1
ENST00000424086.1 ENST00000272521.6 |
TMEM177
|
transmembrane protein 177 |
| chr1_+_84630053 | 6.12 |
ENST00000394838.2
ENST00000370682.3 ENST00000432111.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr13_+_25875785 | 5.99 |
ENST00000381747.3
|
NUPL1
|
nucleoporin like 1 |
| chr6_+_32936353 | 5.97 |
ENST00000374825.4
|
BRD2
|
bromodomain containing 2 |
| chr16_+_30212050 | 5.81 |
ENST00000563322.1
|
SULT1A3
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3 |
| chr19_-_44174330 | 5.80 |
ENST00000340093.3
|
PLAUR
|
plasminogen activator, urokinase receptor |
| chr8_+_110552831 | 5.71 |
ENST00000530629.1
|
EBAG9
|
estrogen receptor binding site associated, antigen, 9 |
| chr8_-_122653630 | 5.50 |
ENST00000303924.4
|
HAS2
|
hyaluronan synthase 2 |
| chr20_-_13765526 | 5.37 |
ENST00000202816.1
|
ESF1
|
ESF1, nucleolar pre-rRNA processing protein, homolog (S. cerevisiae) |
| chrX_+_152953505 | 5.36 |
ENST00000253122.5
|
SLC6A8
|
solute carrier family 6 (neurotransmitter transporter), member 8 |
| chr14_-_20801427 | 5.31 |
ENST00000557665.1
ENST00000358932.4 ENST00000353689.4 |
CCNB1IP1
|
cyclin B1 interacting protein 1, E3 ubiquitin protein ligase |
| chr12_-_58240470 | 4.95 |
ENST00000548823.1
ENST00000398073.2 |
CTDSP2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr16_-_1821496 | 4.93 |
ENST00000564628.1
ENST00000563498.1 |
NME3
|
NME/NM23 nucleoside diphosphate kinase 3 |
| chr12_-_10955226 | 4.90 |
ENST00000240687.2
|
TAS2R7
|
taste receptor, type 2, member 7 |
| chr5_-_150521192 | 4.74 |
ENST00000523714.1
ENST00000521749.1 |
ANXA6
|
annexin A6 |
| chr10_-_104262460 | 4.65 |
ENST00000446605.2
ENST00000369905.4 ENST00000545684.1 |
ACTR1A
|
ARP1 actin-related protein 1 homolog A, centractin alpha (yeast) |
| chr7_+_79765071 | 4.61 |
ENST00000457358.2
|
GNAI1
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
| chr13_-_36944307 | 4.59 |
ENST00000355182.4
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome) |
| chr14_+_22962898 | 4.56 |
ENST00000390492.1
|
TRAJ45
|
T cell receptor alpha joining 45 |
| chr17_+_74372662 | 4.41 |
ENST00000591651.1
ENST00000545180.1 |
SPHK1
|
sphingosine kinase 1 |
| chrX_+_108779004 | 4.39 |
ENST00000218004.1
|
NXT2
|
nuclear transport factor 2-like export factor 2 |
| chr8_-_99954788 | 4.34 |
ENST00000523601.1
|
STK3
|
serine/threonine kinase 3 |
| chr12_-_10978957 | 4.34 |
ENST00000240619.2
|
TAS2R10
|
taste receptor, type 2, member 10 |
| chr5_+_125758865 | 4.26 |
ENST00000542322.1
ENST00000544396.1 |
GRAMD3
|
GRAM domain containing 3 |
| chrX_-_70473957 | 4.25 |
ENST00000373984.3
ENST00000314425.5 ENST00000373982.1 |
ZMYM3
|
zinc finger, MYM-type 3 |
| chr8_+_97597148 | 4.12 |
ENST00000521590.1
|
SDC2
|
syndecan 2 |
| chr12_-_54689532 | 4.02 |
ENST00000540264.2
ENST00000312156.4 |
NFE2
|
nuclear factor, erythroid 2 |
| chr3_-_113897899 | 3.80 |
ENST00000383673.2
ENST00000295881.7 |
DRD3
|
dopamine receptor D3 |
| chr4_+_56719782 | 3.72 |
ENST00000381295.2
ENST00000346134.7 ENST00000349598.6 |
EXOC1
|
exocyst complex component 1 |
| chr12_-_28124903 | 3.70 |
ENST00000395872.1
ENST00000354417.3 ENST00000201015.4 |
PTHLH
|
parathyroid hormone-like hormone |
| chr13_+_102104980 | 3.66 |
ENST00000545560.2
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr5_+_125758813 | 3.62 |
ENST00000285689.3
ENST00000515200.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr12_-_10875831 | 3.61 |
ENST00000279550.7
ENST00000228251.4 |
YBX3
|
Y box binding protein 3 |
| chr22_-_39268308 | 3.61 |
ENST00000407418.3
|
CBX6
|
chromobox homolog 6 |
| chr12_-_111926342 | 3.41 |
ENST00000389154.3
|
ATXN2
|
ataxin 2 |
| chr15_+_79165222 | 3.30 |
ENST00000559930.1
|
MORF4L1
|
mortality factor 4 like 1 |
| chr1_+_224301787 | 3.25 |
ENST00000366862.5
ENST00000424254.2 |
FBXO28
|
F-box protein 28 |
| chr17_-_42143963 | 3.24 |
ENST00000585388.1
ENST00000293406.3 |
LSM12
|
LSM12 homolog (S. cerevisiae) |
| chr19_-_39881669 | 3.13 |
ENST00000221266.7
|
PAF1
|
Paf1, RNA polymerase II associated factor, homolog (S. cerevisiae) |
| chr4_+_95376396 | 3.08 |
ENST00000508216.1
ENST00000514743.1 |
PDLIM5
|
PDZ and LIM domain 5 |
| chr9_+_125132803 | 3.05 |
ENST00000540753.1
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr7_+_150065278 | 3.02 |
ENST00000519397.1
ENST00000479668.1 ENST00000540729.1 |
REPIN1
|
replication initiator 1 |
| chr4_-_99578776 | 2.98 |
ENST00000515287.1
|
TSPAN5
|
tetraspanin 5 |
| chr14_+_39703084 | 2.94 |
ENST00000553728.1
|
RP11-407N17.3
|
cTAGE family member 5 isoform 4 |
| chr9_+_116263778 | 2.92 |
ENST00000394646.3
|
RGS3
|
regulator of G-protein signaling 3 |
| chr6_+_151561506 | 2.82 |
ENST00000253332.1
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr10_-_104262426 | 2.78 |
ENST00000487599.1
|
ACTR1A
|
ARP1 actin-related protein 1 homolog A, centractin alpha (yeast) |
| chr20_+_57226284 | 2.64 |
ENST00000458280.1
ENST00000355957.5 ENST00000361770.5 ENST00000312283.8 ENST00000412911.1 ENST00000359617.4 ENST00000371141.4 |
STX16
|
syntaxin 16 |
| chr9_+_116263639 | 2.62 |
ENST00000343817.5
|
RGS3
|
regulator of G-protein signaling 3 |
| chr2_+_152266392 | 2.60 |
ENST00000444746.2
ENST00000453091.2 ENST00000428287.2 ENST00000433166.2 ENST00000420714.3 ENST00000243326.5 ENST00000414861.2 |
RIF1
|
RAP1 interacting factor homolog (yeast) |
| chr12_-_68696652 | 2.45 |
ENST00000539972.1
|
MDM1
|
Mdm1 nuclear protein homolog (mouse) |
| chr11_-_72432950 | 2.43 |
ENST00000426523.1
ENST00000429686.1 |
ARAP1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr20_+_30697510 | 2.41 |
ENST00000217315.5
|
TM9SF4
|
transmembrane 9 superfamily protein member 4 |
| chr13_+_50570019 | 2.29 |
ENST00000442421.1
|
TRIM13
|
tripartite motif containing 13 |
| chr20_+_30697298 | 2.28 |
ENST00000398022.2
|
TM9SF4
|
transmembrane 9 superfamily protein member 4 |
| chr19_-_44174305 | 2.07 |
ENST00000601723.1
ENST00000339082.3 |
PLAUR
|
plasminogen activator, urokinase receptor |
| chr17_+_7591747 | 2.04 |
ENST00000534050.1
|
WRAP53
|
WD repeat containing, antisense to TP53 |
| chr12_-_71551652 | 1.97 |
ENST00000546561.1
|
TSPAN8
|
tetraspanin 8 |
| chr6_-_155776966 | 1.93 |
ENST00000159060.2
|
NOX3
|
NADPH oxidase 3 |
| chr6_-_11779014 | 1.88 |
ENST00000229583.5
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr6_+_151561085 | 1.87 |
ENST00000402676.2
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr4_-_111563076 | 1.62 |
ENST00000354925.2
ENST00000511990.1 |
PITX2
|
paired-like homeodomain 2 |
| chr5_+_148206156 | 1.62 |
ENST00000305988.4
|
ADRB2
|
adrenoceptor beta 2, surface |
| chr16_+_10837643 | 1.57 |
ENST00000574334.1
ENST00000283027.5 ENST00000433392.2 |
NUBP1
|
nucleotide binding protein 1 |
| chr11_-_26743546 | 1.53 |
ENST00000280467.6
ENST00000396005.3 |
SLC5A12
|
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 12 |
| chr17_-_38859996 | 1.51 |
ENST00000264651.2
|
KRT24
|
keratin 24 |
| chr3_-_170744498 | 1.46 |
ENST00000382808.4
ENST00000314251.3 |
SLC2A2
|
solute carrier family 2 (facilitated glucose transporter), member 2 |
| chr1_+_171217677 | 1.37 |
ENST00000402921.2
|
FMO1
|
flavin containing monooxygenase 1 |
| chr1_-_202936394 | 1.33 |
ENST00000367249.4
|
CYB5R1
|
cytochrome b5 reductase 1 |
| chr17_+_7591639 | 1.27 |
ENST00000396463.2
|
WRAP53
|
WD repeat containing, antisense to TP53 |
| chr4_-_110723335 | 1.25 |
ENST00000394634.2
|
CFI
|
complement factor I |
| chr2_-_40739501 | 1.22 |
ENST00000403092.1
ENST00000402441.1 ENST00000448531.1 |
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr2_-_214014959 | 1.16 |
ENST00000442445.1
ENST00000457361.1 ENST00000342002.2 |
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr11_+_107879459 | 1.09 |
ENST00000393094.2
|
CUL5
|
cullin 5 |
| chr8_+_76452097 | 1.05 |
ENST00000396423.2
|
HNF4G
|
hepatocyte nuclear factor 4, gamma |
| chr7_-_22234381 | 1.05 |
ENST00000458533.1
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr2_-_75938115 | 1.04 |
ENST00000321027.3
|
GCFC2
|
GC-rich sequence DNA-binding factor 2 |
| chr8_+_77593474 | 1.02 |
ENST00000455469.2
ENST00000050961.6 |
ZFHX4
|
zinc finger homeobox 4 |
| chrX_+_49091920 | 1.00 |
ENST00000376227.3
|
CCDC22
|
coiled-coil domain containing 22 |
| chr12_-_8803128 | 0.91 |
ENST00000543467.1
|
MFAP5
|
microfibrillar associated protein 5 |
| chrX_+_107288239 | 0.90 |
ENST00000217957.5
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX2-2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.2 | 30.6 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 9.4 | 28.3 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 9.3 | 37.1 | GO:0044111 | development involved in symbiotic interaction(GO:0044111) |
| 7.3 | 36.4 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 6.2 | 31.0 | GO:0008204 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 5.6 | 33.7 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 4.2 | 33.8 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 3.7 | 14.7 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 3.3 | 10.0 | GO:0071676 | negative regulation of mononuclear cell migration(GO:0071676) |
| 3.1 | 15.5 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 2.9 | 17.4 | GO:0036111 | very long-chain fatty-acyl-CoA metabolic process(GO:0036111) |
| 2.9 | 37.3 | GO:0070986 | left/right axis specification(GO:0070986) |
| 2.6 | 7.8 | GO:2000681 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 2.5 | 22.8 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 2.4 | 24.4 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 2.4 | 21.8 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 2.4 | 16.5 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 2.3 | 18.7 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 2.3 | 6.9 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 2.3 | 11.5 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 2.2 | 13.3 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 2.0 | 58.0 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 2.0 | 7.9 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 1.9 | 5.8 | GO:0071048 | nuclear mRNA surveillance of spliceosomal pre-mRNA splicing(GO:0071030) nuclear retention of unspliced pre-mRNA at the site of transcription(GO:0071048) |
| 1.8 | 12.5 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 1.7 | 24.9 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 1.5 | 74.2 | GO:0045143 | homologous chromosome segregation(GO:0045143) |
| 1.5 | 4.4 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 1.4 | 24.5 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 1.3 | 14.4 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 1.3 | 24.6 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 1.3 | 11.5 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 1.3 | 32.9 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 1.3 | 7.6 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 1.2 | 10.0 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 1.2 | 47.4 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 1.2 | 6.1 | GO:0097338 | response to clozapine(GO:0097338) |
| 1.2 | 7.3 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 1.2 | 27.7 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 1.2 | 8.3 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) |
| 1.2 | 28.8 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 1.1 | 4.6 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 1.1 | 11.3 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 1.1 | 3.3 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 1.1 | 5.5 | GO:0070295 | renal water absorption(GO:0070295) |
| 1.1 | 6.5 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 1.0 | 7.1 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 1.0 | 18.2 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.9 | 3.8 | GO:0050883 | negative regulation of sodium:proton antiporter activity(GO:0032416) gastric motility(GO:0035482) gastric emptying(GO:0035483) musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.9 | 15.2 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.9 | 19.6 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.9 | 15.1 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.9 | 19.1 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.9 | 4.3 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.8 | 3.3 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.8 | 38.5 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.7 | 11.2 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.7 | 32.2 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.7 | 6.6 | GO:0006999 | nuclear pore organization(GO:0006999) nuclear pore complex assembly(GO:0051292) endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.7 | 7.3 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.7 | 11.5 | GO:0032802 | low-density lipoprotein particle receptor catabolic process(GO:0032802) |
| 0.6 | 10.3 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.6 | 45.7 | GO:0070126 | mitochondrial translational elongation(GO:0070125) mitochondrial translational termination(GO:0070126) |
| 0.6 | 4.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.6 | 8.6 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.6 | 8.0 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.5 | 1.6 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.5 | 16.3 | GO:0072600 | establishment of protein localization to Golgi(GO:0072600) |
| 0.5 | 2.6 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.5 | 5.4 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.5 | 1.5 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.5 | 16.0 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.5 | 4.7 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.5 | 3.6 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.4 | 4.9 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.4 | 6.2 | GO:0072697 | protein localization to cell cortex(GO:0072697) |
| 0.4 | 8.0 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.4 | 17.9 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.3 | 3.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.3 | 3.7 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.3 | 4.7 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.3 | 13.2 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.3 | 7.9 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.3 | 12.9 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.3 | 14.0 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.3 | 1.9 | GO:0048840 | otolith development(GO:0048840) |
| 0.2 | 11.5 | GO:0048255 | mRNA stabilization(GO:0048255) |
| 0.2 | 7.8 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.2 | 19.0 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.2 | 4.7 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.2 | 2.5 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.2 | 3.0 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.2 | 1.5 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.2 | 25.1 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.2 | 5.5 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.2 | 1.2 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.1 | 15.0 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.1 | 2.3 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.1 | 17.4 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 4.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 8.0 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 0.7 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.1 | 7.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 3.1 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 2.5 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.1 | 7.4 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.1 | 5.0 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.1 | 1.0 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.1 | 6.3 | GO:0006810 | transport(GO:0006810) |
| 0.1 | 0.1 | GO:0035990 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.1 | 4.4 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.1 | 0.5 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 7.9 | GO:0030705 | cytoskeleton-dependent intracellular transport(GO:0030705) |
| 0.0 | 0.9 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 5.2 | GO:0002062 | chondrocyte differentiation(GO:0002062) |
| 0.0 | 4.4 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.3 | GO:0042797 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.0 | 0.1 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.0 | 13.3 | GO:0001558 | regulation of cell growth(GO:0001558) |
| 0.0 | 0.9 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 2.5 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 2.3 | GO:0006606 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) |
| 0.0 | 3.7 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 3.0 | GO:0006260 | DNA replication(GO:0006260) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.4 | 19.1 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 5.5 | 21.8 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 5.1 | 25.3 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 2.9 | 14.4 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 2.2 | 24.2 | GO:0005688 | U6 snRNP(GO:0005688) |
| 2.2 | 47.8 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 2.1 | 18.7 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 2.0 | 32.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 1.9 | 5.8 | GO:0071020 | post-spliceosomal complex(GO:0071020) |
| 1.8 | 58.0 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 1.6 | 35.8 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 1.6 | 19.5 | GO:0030008 | TRAPP complex(GO:0030008) |
| 1.4 | 11.5 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 1.4 | 7.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 1.2 | 7.4 | GO:0002177 | manchette(GO:0002177) |
| 1.1 | 45.7 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 1.0 | 28.3 | GO:0031143 | pseudopodium(GO:0031143) |
| 1.0 | 3.0 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 1.0 | 10.0 | GO:0045180 | basal cortex(GO:0045180) |
| 0.9 | 3.7 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.8 | 7.3 | GO:0034464 | BBSome(GO:0034464) |
| 0.8 | 14.9 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.8 | 6.9 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.7 | 14.8 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.7 | 63.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.7 | 7.9 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.7 | 8.3 | GO:0097025 | lateral loop(GO:0043219) MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.7 | 38.5 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.7 | 19.1 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.6 | 11.3 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.6 | 11.5 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.5 | 18.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.5 | 8.2 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.5 | 28.3 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.4 | 24.6 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.4 | 6.6 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.4 | 1.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.4 | 8.0 | GO:0031105 | septin complex(GO:0031105) |
| 0.4 | 11.8 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.4 | 5.3 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.3 | 3.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.3 | 11.5 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.3 | 1.9 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.3 | 39.8 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.2 | 14.6 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.2 | 33.7 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.2 | 28.0 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.2 | 25.6 | GO:0042641 | actomyosin(GO:0042641) |
| 0.2 | 36.6 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.2 | 2.3 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.2 | 4.6 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.2 | 19.9 | GO:0005604 | basement membrane(GO:0005604) |
| 0.2 | 21.5 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.2 | 16.3 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.2 | 24.5 | GO:0043679 | axon terminus(GO:0043679) |
| 0.1 | 21.0 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 1.9 | GO:0012505 | endomembrane system(GO:0012505) |
| 0.1 | 12.8 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.1 | 3.3 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 0.5 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 10.4 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 24.4 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.1 | 14.4 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.1 | 3.6 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.1 | 9.9 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.1 | 1.3 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 3.8 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.9 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 3.1 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.3 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 4.3 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 1.2 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.8 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 1.5 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 11.0 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 8.6 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 1.3 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 1.3 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.3 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 12.0 | GO:0005654 | nucleoplasm(GO:0005654) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.7 | 31.0 | GO:0004310 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 7.3 | 36.4 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 5.9 | 17.8 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 5.6 | 33.8 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 5.5 | 38.5 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 5.2 | 15.5 | GO:0070538 | oleic acid binding(GO:0070538) |
| 5.1 | 15.2 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 4.2 | 25.3 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 3.9 | 19.6 | GO:0070404 | NADH binding(GO:0070404) |
| 3.6 | 21.8 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 2.9 | 23.3 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 2.9 | 17.4 | GO:0033989 | 3alpha,7alpha,12alpha-trihydroxy-5beta-cholest-24-enoyl-CoA hydratase activity(GO:0033989) 17-beta-hydroxysteroid dehydrogenase (NAD+) activity(GO:0044594) |
| 2.6 | 7.9 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 2.5 | 63.8 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 2.4 | 24.2 | GO:0042731 | PH domain binding(GO:0042731) |
| 2.1 | 12.5 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 1.9 | 5.8 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 1.8 | 5.5 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 1.3 | 15.9 | GO:0008494 | translation activator activity(GO:0008494) |
| 1.3 | 21.8 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 1.3 | 11.4 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 1.3 | 7.6 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 1.0 | 8.0 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 1.0 | 5.0 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 1.0 | 15.5 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 1.0 | 18.2 | GO:0048156 | tau protein binding(GO:0048156) |
| 1.0 | 28.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.9 | 68.9 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.8 | 9.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.8 | 24.5 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.8 | 3.0 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.8 | 8.3 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.7 | 4.4 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.7 | 34.5 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.7 | 15.7 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.7 | 14.4 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.6 | 11.5 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.6 | 19.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.6 | 28.3 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.6 | 13.3 | GO:0016505 | peptidase activator activity involved in apoptotic process(GO:0016505) |
| 0.5 | 3.8 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.5 | 1.6 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.5 | 1.5 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.5 | 16.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.5 | 6.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.4 | 18.7 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.4 | 8.6 | GO:0008603 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.4 | 6.9 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.4 | 1.2 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.4 | 11.3 | GO:0005123 | death receptor binding(GO:0005123) cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.4 | 4.6 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.4 | 19.5 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.4 | 10.0 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.4 | 94.0 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.4 | 17.9 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.3 | 24.6 | GO:0019003 | GDP binding(GO:0019003) |
| 0.3 | 15.1 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.3 | 5.4 | GO:0072349 | modified amino acid transmembrane transporter activity(GO:0072349) |
| 0.3 | 4.7 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.3 | 3.3 | GO:0031702 | angiotensin receptor binding(GO:0031701) type 1 angiotensin receptor binding(GO:0031702) |
| 0.3 | 11.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.3 | 10.3 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.3 | 6.9 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.3 | 7.1 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.3 | 36.1 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.2 | 21.6 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.2 | 10.4 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.2 | 4.9 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.2 | 61.5 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.2 | 0.7 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.2 | 3.7 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.2 | 11.2 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.2 | 2.3 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.2 | 3.7 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.2 | 8.0 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.2 | 27.5 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.2 | 6.0 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.2 | 1.5 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.2 | 4.2 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.2 | 4.3 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.2 | 1.9 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 14.7 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.1 | 3.1 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 3.3 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.1 | 7.3 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 7.3 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.1 | 1.3 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 1.3 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 0.3 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 0.5 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.1 | 1.9 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.1 | 14.9 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.1 | 1.0 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 26.7 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 0.8 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 3.6 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.1 | 5.1 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.1 | 2.5 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 1.6 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 6.5 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 4.0 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 1.3 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 46.4 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.7 | 16.3 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.7 | 23.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.7 | 39.7 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.5 | 23.8 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.5 | 11.5 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.4 | 9.0 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.3 | 35.5 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.3 | 13.2 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.3 | 29.7 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.3 | 11.8 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.3 | 11.3 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.3 | 26.8 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.3 | 5.5 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.3 | 5.0 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.3 | 7.7 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.3 | 19.1 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.3 | 14.6 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.3 | 8.3 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.3 | 11.5 | PID MYC PATHWAY | C-MYC pathway |
| 0.3 | 16.3 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.2 | 12.0 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.2 | 21.1 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.2 | 9.2 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.2 | 8.0 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.2 | 3.7 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.2 | 3.7 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.2 | 28.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 8.1 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 1.2 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.8 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.9 | PID NOTCH PATHWAY | Notch signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 32.9 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 1.7 | 42.3 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 1.6 | 24.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 1.3 | 33.8 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 1.3 | 24.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 1.3 | 29.0 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 1.1 | 38.7 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 1.1 | 33.7 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 1.0 | 24.9 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 1.0 | 36.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 1.0 | 31.0 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 1.0 | 27.0 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.9 | 28.3 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.9 | 34.0 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.9 | 68.9 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF MITOTIC PROTEINS | Genes involved in APC/C:Cdc20 mediated degradation of mitotic proteins |
| 0.8 | 12.3 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.6 | 11.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.6 | 11.5 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.6 | 15.2 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.6 | 8.0 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.5 | 14.6 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.5 | 15.1 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.5 | 4.6 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.5 | 22.7 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.4 | 14.8 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.4 | 12.1 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.4 | 7.1 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.4 | 11.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.4 | 17.6 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.3 | 8.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.3 | 5.5 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.3 | 18.2 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.3 | 7.9 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.3 | 22.0 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.3 | 11.8 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.2 | 18.7 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.2 | 7.6 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.2 | 3.3 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.2 | 4.1 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.2 | 6.9 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.2 | 11.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.2 | 14.4 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.2 | 14.4 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.2 | 3.7 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.2 | 27.0 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 4.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 7.1 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 5.0 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 1.5 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 5.0 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 6.9 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 1.2 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 1.3 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.3 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 2.7 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.5 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |