Project
averaged GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
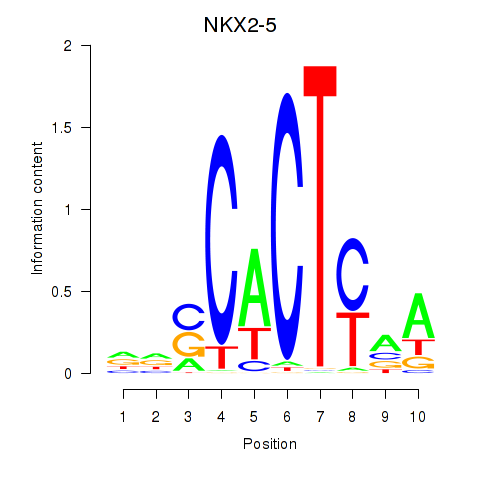
Results for NKX2-5
Z-value: 0.85
Transcription factors associated with NKX2-5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX2-5
|
ENSG00000183072.9 | NK2 homeobox 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NKX2-5 | hg19_v2_chr5_-_172662303_172662360 | -0.11 | 1.1e-01 | Click! |
Activity profile of NKX2-5 motif
Sorted Z-values of NKX2-5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_-_13835147 | 15.12 |
ENST00000493677.1
ENST00000355135.2 |
GPM6B
|
glycoprotein M6B |
| chr3_+_125694347 | 11.37 |
ENST00000505382.1
ENST00000511082.1 |
ROPN1B
|
rhophilin associated tail protein 1B |
| chr15_+_42696992 | 9.68 |
ENST00000561817.1
|
CAPN3
|
calpain 3, (p94) |
| chr15_+_42697018 | 9.56 |
ENST00000397204.4
|
CAPN3
|
calpain 3, (p94) |
| chr5_-_149792295 | 9.33 |
ENST00000518797.1
ENST00000524315.1 ENST00000009530.7 ENST00000377795.3 |
CD74
|
CD74 molecule, major histocompatibility complex, class II invariant chain |
| chr9_+_17579084 | 8.81 |
ENST00000380607.4
|
SH3GL2
|
SH3-domain GRB2-like 2 |
| chr8_-_120651020 | 8.79 |
ENST00000522826.1
ENST00000520066.1 ENST00000259486.6 ENST00000075322.6 |
ENPP2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr3_+_125687987 | 8.10 |
ENST00000514116.1
ENST00000251776.4 ENST00000504401.1 ENST00000513830.1 ENST00000508088.1 |
ROPN1B
|
rhophilin associated tail protein 1B |
| chr3_-_123710893 | 8.08 |
ENST00000467907.1
ENST00000459660.1 ENST00000495093.1 ENST00000460743.1 ENST00000405845.3 ENST00000484329.1 ENST00000479867.1 ENST00000496145.1 |
ROPN1
|
rhophilin associated tail protein 1 |
| chr15_+_42696954 | 7.56 |
ENST00000337571.4
ENST00000569136.1 |
CAPN3
|
calpain 3, (p94) |
| chr17_-_29624343 | 6.69 |
ENST00000247271.4
|
OMG
|
oligodendrocyte myelin glycoprotein |
| chr22_-_38577782 | 6.42 |
ENST00000430886.1
ENST00000332509.3 ENST00000447598.2 ENST00000435484.1 ENST00000402064.1 ENST00000436218.1 |
PLA2G6
|
phospholipase A2, group VI (cytosolic, calcium-independent) |
| chr6_-_112115074 | 6.13 |
ENST00000368667.2
|
FYN
|
FYN oncogene related to SRC, FGR, YES |
| chr22_-_38577731 | 6.07 |
ENST00000335539.3
|
PLA2G6
|
phospholipase A2, group VI (cytosolic, calcium-independent) |
| chr7_+_99775520 | 5.51 |
ENST00000317296.5
ENST00000422690.1 ENST00000439782.1 |
STAG3
|
stromal antigen 3 |
| chr7_+_99775366 | 5.47 |
ENST00000394018.2
ENST00000416412.1 |
STAG3
|
stromal antigen 3 |
| chr22_-_50523760 | 5.44 |
ENST00000395876.2
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr22_-_50524298 | 5.31 |
ENST00000311597.5
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr7_-_37488834 | 4.98 |
ENST00000310758.4
|
ELMO1
|
engulfment and cell motility 1 |
| chr2_-_175499294 | 4.81 |
ENST00000392547.2
|
WIPF1
|
WAS/WASL interacting protein family, member 1 |
| chr19_+_16999654 | 4.77 |
ENST00000248076.3
|
F2RL3
|
coagulation factor II (thrombin) receptor-like 3 |
| chr21_+_44313375 | 4.76 |
ENST00000354250.2
ENST00000340344.4 |
NDUFV3
|
NADH dehydrogenase (ubiquinone) flavoprotein 3, 10kDa |
| chr15_-_81282133 | 4.53 |
ENST00000261758.4
|
MESDC2
|
mesoderm development candidate 2 |
| chr14_-_21516590 | 4.37 |
ENST00000555026.1
|
NDRG2
|
NDRG family member 2 |
| chr17_-_29641084 | 4.24 |
ENST00000544462.1
|
EVI2B
|
ecotropic viral integration site 2B |
| chr5_-_74062930 | 4.24 |
ENST00000509430.1
ENST00000345239.2 ENST00000427854.2 ENST00000506778.1 |
GFM2
|
G elongation factor, mitochondrial 2 |
| chr15_-_42840961 | 4.19 |
ENST00000563454.1
ENST00000397130.3 ENST00000570160.1 ENST00000323443.2 |
LRRC57
|
leucine rich repeat containing 57 |
| chr10_+_111967345 | 3.98 |
ENST00000332674.5
ENST00000453116.1 |
MXI1
|
MAX interactor 1, dimerization protein |
| chr17_-_19771216 | 3.84 |
ENST00000395544.4
|
ULK2
|
unc-51 like autophagy activating kinase 2 |
| chr3_-_187388173 | 3.77 |
ENST00000287641.3
|
SST
|
somatostatin |
| chr8_+_133879193 | 3.74 |
ENST00000377869.1
ENST00000220616.4 |
TG
|
thyroglobulin |
| chrX_-_49965663 | 3.73 |
ENST00000376056.2
ENST00000376058.2 ENST00000358526.2 |
AKAP4
|
A kinase (PRKA) anchor protein 4 |
| chr17_-_29641104 | 3.61 |
ENST00000577894.1
ENST00000330927.4 |
EVI2B
|
ecotropic viral integration site 2B |
| chr14_-_106963409 | 3.51 |
ENST00000390621.2
|
IGHV1-45
|
immunoglobulin heavy variable 1-45 |
| chr3_-_15374659 | 3.40 |
ENST00000426925.1
|
SH3BP5
|
SH3-domain binding protein 5 (BTK-associated) |
| chr11_+_108093839 | 3.39 |
ENST00000452508.2
|
ATM
|
ataxia telangiectasia mutated |
| chr5_+_140729649 | 3.27 |
ENST00000523390.1
|
PCDHGB1
|
protocadherin gamma subfamily B, 1 |
| chr4_+_113970772 | 3.25 |
ENST00000504454.1
ENST00000394537.3 ENST00000357077.4 ENST00000264366.6 |
ANK2
|
ankyrin 2, neuronal |
| chr16_+_57769635 | 3.24 |
ENST00000379661.3
ENST00000562592.1 ENST00000566726.1 |
KATNB1
|
katanin p80 (WD repeat containing) subunit B 1 |
| chr15_+_81589254 | 3.23 |
ENST00000394652.2
|
IL16
|
interleukin 16 |
| chr13_-_41240717 | 2.98 |
ENST00000379561.5
|
FOXO1
|
forkhead box O1 |
| chr17_+_27055798 | 2.98 |
ENST00000268766.6
|
NEK8
|
NIMA-related kinase 8 |
| chr20_+_57226284 | 2.98 |
ENST00000458280.1
ENST00000355957.5 ENST00000361770.5 ENST00000312283.8 ENST00000412911.1 ENST00000359617.4 ENST00000371141.4 |
STX16
|
syntaxin 16 |
| chr16_-_30134266 | 2.96 |
ENST00000484663.1
ENST00000478356.1 |
MAPK3
|
mitogen-activated protein kinase 3 |
| chr12_+_123717458 | 2.90 |
ENST00000253233.1
|
C12orf65
|
chromosome 12 open reading frame 65 |
| chr14_-_106830057 | 2.79 |
ENST00000390616.2
|
IGHV4-34
|
immunoglobulin heavy variable 4-34 |
| chr2_-_201936302 | 2.75 |
ENST00000453765.1
ENST00000452799.1 ENST00000446678.1 ENST00000418596.3 |
FAM126B
|
family with sequence similarity 126, member B |
| chr17_+_58755184 | 2.66 |
ENST00000589222.1
ENST00000407086.3 ENST00000390652.5 |
BCAS3
|
breast carcinoma amplified sequence 3 |
| chr15_+_42841008 | 2.55 |
ENST00000260372.3
ENST00000568876.1 ENST00000568846.2 ENST00000562398.1 |
HAUS2
|
HAUS augmin-like complex, subunit 2 |
| chr10_-_81320151 | 2.51 |
ENST00000372325.2
ENST00000372327.5 ENST00000417041.1 |
SFTPA2
|
surfactant protein A2 |
| chr17_+_40811283 | 2.50 |
ENST00000251412.7
|
TUBG2
|
tubulin, gamma 2 |
| chr17_+_46048376 | 2.49 |
ENST00000338399.4
|
CDK5RAP3
|
CDK5 regulatory subunit associated protein 3 |
| chr11_+_17756279 | 2.41 |
ENST00000265969.6
|
KCNC1
|
potassium voltage-gated channel, Shaw-related subfamily, member 1 |
| chr17_+_48624450 | 2.33 |
ENST00000006658.6
ENST00000356488.4 ENST00000393244.3 |
SPATA20
|
spermatogenesis associated 20 |
| chr19_+_5681011 | 2.30 |
ENST00000581893.1
ENST00000411793.2 ENST00000301382.4 ENST00000581773.1 ENST00000423665.2 ENST00000583928.1 ENST00000342970.2 ENST00000422535.2 ENST00000581521.1 ENST00000339423.2 |
HSD11B1L
|
hydroxysteroid (11-beta) dehydrogenase 1-like |
| chr10_-_49813090 | 2.30 |
ENST00000249601.4
|
ARHGAP22
|
Rho GTPase activating protein 22 |
| chr14_-_106406090 | 2.26 |
ENST00000390593.2
|
IGHV6-1
|
immunoglobulin heavy variable 6-1 |
| chr4_+_156680518 | 2.26 |
ENST00000513437.1
|
GUCY1B3
|
guanylate cyclase 1, soluble, beta 3 |
| chr19_-_5680499 | 2.26 |
ENST00000587589.1
|
C19orf70
|
chromosome 19 open reading frame 70 |
| chr8_-_10588010 | 2.25 |
ENST00000304501.1
|
SOX7
|
SRY (sex determining region Y)-box 7 |
| chr4_-_2264015 | 2.25 |
ENST00000337190.2
|
MXD4
|
MAX dimerization protein 4 |
| chr6_+_33378738 | 2.24 |
ENST00000374512.3
ENST00000374516.3 |
PHF1
|
PHD finger protein 1 |
| chr1_+_154301264 | 2.17 |
ENST00000341822.2
|
ATP8B2
|
ATPase, aminophospholipid transporter, class I, type 8B, member 2 |
| chr12_+_4918342 | 2.11 |
ENST00000280684.3
ENST00000433855.1 |
KCNA6
|
potassium voltage-gated channel, shaker-related subfamily, member 6 |
| chr10_-_101841588 | 1.98 |
ENST00000370418.3
|
CPN1
|
carboxypeptidase N, polypeptide 1 |
| chr16_+_77233294 | 1.96 |
ENST00000378644.4
|
SYCE1L
|
synaptonemal complex central element protein 1-like |
| chr12_+_93772402 | 1.92 |
ENST00000546925.1
|
NUDT4
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4 |
| chr1_+_211433275 | 1.87 |
ENST00000367005.4
|
RCOR3
|
REST corepressor 3 |
| chr5_-_171615315 | 1.87 |
ENST00000176763.5
|
STK10
|
serine/threonine kinase 10 |
| chr16_-_30134524 | 1.86 |
ENST00000395202.1
ENST00000395199.3 ENST00000263025.4 ENST00000322266.5 ENST00000403394.1 |
MAPK3
|
mitogen-activated protein kinase 3 |
| chr20_-_44485835 | 1.85 |
ENST00000457981.1
ENST00000426915.1 ENST00000217455.4 |
ACOT8
|
acyl-CoA thioesterase 8 |
| chr19_+_41313017 | 1.72 |
ENST00000595621.1
ENST00000595051.1 |
EGLN2
|
egl-9 family hypoxia-inducible factor 2 |
| chr11_-_118436606 | 1.66 |
ENST00000530872.1
|
IFT46
|
intraflagellar transport 46 homolog (Chlamydomonas) |
| chr18_+_42260861 | 1.61 |
ENST00000282030.5
|
SETBP1
|
SET binding protein 1 |
| chr11_-_111175739 | 1.56 |
ENST00000532918.1
|
COLCA1
|
colorectal cancer associated 1 |
| chr15_-_72563585 | 1.51 |
ENST00000287196.9
ENST00000260376.7 |
PARP6
|
poly (ADP-ribose) polymerase family, member 6 |
| chr4_+_2470664 | 1.51 |
ENST00000314289.8
ENST00000541204.1 ENST00000502316.1 ENST00000507247.1 ENST00000509258.1 ENST00000511859.1 |
RNF4
|
ring finger protein 4 |
| chrX_-_30326445 | 1.48 |
ENST00000378963.1
|
NR0B1
|
nuclear receptor subfamily 0, group B, member 1 |
| chr11_-_118436707 | 1.46 |
ENST00000264020.2
ENST00000264021.3 |
IFT46
|
intraflagellar transport 46 homolog (Chlamydomonas) |
| chr5_+_140810132 | 1.40 |
ENST00000252085.3
|
PCDHGA12
|
protocadherin gamma subfamily A, 12 |
| chr12_+_111051832 | 1.40 |
ENST00000550703.2
ENST00000551590.1 |
TCTN1
|
tectonic family member 1 |
| chr10_-_99531709 | 1.40 |
ENST00000266066.3
|
SFRP5
|
secreted frizzled-related protein 5 |
| chrX_+_11777671 | 1.39 |
ENST00000380693.3
ENST00000380692.2 |
MSL3
|
male-specific lethal 3 homolog (Drosophila) |
| chr13_-_95364389 | 1.39 |
ENST00000376945.2
|
SOX21
|
SRY (sex determining region Y)-box 21 |
| chr10_+_76586348 | 1.39 |
ENST00000372724.1
ENST00000287239.4 ENST00000372714.1 |
KAT6B
|
K(lysine) acetyltransferase 6B |
| chr4_-_168155730 | 1.33 |
ENST00000502330.1
ENST00000357154.3 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr12_-_371994 | 1.30 |
ENST00000343164.4
ENST00000436453.1 ENST00000445055.2 ENST00000546319.1 |
SLC6A13
|
solute carrier family 6 (neurotransmitter transporter), member 13 |
| chr5_+_140345820 | 1.30 |
ENST00000289269.5
|
PCDHAC2
|
protocadherin alpha subfamily C, 2 |
| chr9_+_116263778 | 1.27 |
ENST00000394646.3
|
RGS3
|
regulator of G-protein signaling 3 |
| chr9_-_117692697 | 1.23 |
ENST00000223795.2
|
TNFSF8
|
tumor necrosis factor (ligand) superfamily, member 8 |
| chr17_+_53342311 | 1.20 |
ENST00000226067.5
|
HLF
|
hepatic leukemia factor |
| chr13_-_53422640 | 1.17 |
ENST00000338862.4
ENST00000377942.3 |
PCDH8
|
protocadherin 8 |
| chr12_+_111051902 | 1.16 |
ENST00000397655.3
ENST00000471804.2 ENST00000377654.3 ENST00000397659.4 |
TCTN1
|
tectonic family member 1 |
| chr20_+_57226841 | 1.11 |
ENST00000358029.4
ENST00000361830.3 |
STX16
|
syntaxin 16 |
| chr8_-_95220775 | 1.10 |
ENST00000441892.2
ENST00000521491.1 ENST00000027335.3 |
CDH17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr9_+_116263639 | 1.04 |
ENST00000343817.5
|
RGS3
|
regulator of G-protein signaling 3 |
| chr14_+_22963806 | 1.04 |
ENST00000390493.1
|
TRAJ44
|
T cell receptor alpha joining 44 |
| chr3_+_42201653 | 1.03 |
ENST00000341421.3
ENST00000396175.1 |
TRAK1
|
trafficking protein, kinesin binding 1 |
| chr1_-_11907829 | 1.02 |
ENST00000376480.3
|
NPPA
|
natriuretic peptide A |
| chr3_+_108321623 | 1.02 |
ENST00000497905.1
ENST00000463306.1 |
DZIP3
|
DAZ interacting zinc finger protein 3 |
| chr20_-_41818373 | 1.02 |
ENST00000373187.1
ENST00000356100.2 ENST00000373184.1 ENST00000373190.1 |
PTPRT
|
protein tyrosine phosphatase, receptor type, T |
| chr11_+_118754475 | 1.02 |
ENST00000292174.4
|
CXCR5
|
chemokine (C-X-C motif) receptor 5 |
| chr3_-_47950745 | 1.02 |
ENST00000429422.1
|
MAP4
|
microtubule-associated protein 4 |
| chr1_-_22109682 | 1.00 |
ENST00000400301.1
ENST00000532737.1 |
USP48
|
ubiquitin specific peptidase 48 |
| chr12_-_51422017 | 0.97 |
ENST00000394904.3
|
SLC11A2
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr2_+_219646462 | 0.95 |
ENST00000258415.4
|
CYP27A1
|
cytochrome P450, family 27, subfamily A, polypeptide 1 |
| chr1_+_22962948 | 0.92 |
ENST00000374642.3
|
C1QA
|
complement component 1, q subcomponent, A chain |
| chr4_-_168155700 | 0.91 |
ENST00000357545.4
ENST00000512648.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr14_+_81421355 | 0.87 |
ENST00000541158.2
|
TSHR
|
thyroid stimulating hormone receptor |
| chr17_-_46667594 | 0.83 |
ENST00000476342.1
ENST00000460160.1 ENST00000472863.1 |
HOXB3
|
homeobox B3 |
| chr17_+_29421987 | 0.82 |
ENST00000431387.4
|
NF1
|
neurofibromin 1 |
| chr9_-_138393580 | 0.78 |
ENST00000371791.1
|
C9orf116
|
chromosome 9 open reading frame 116 |
| chr7_+_129906660 | 0.77 |
ENST00000222481.4
|
CPA2
|
carboxypeptidase A2 (pancreatic) |
| chr7_+_129007964 | 0.75 |
ENST00000460109.1
ENST00000474594.1 ENST00000446212.1 |
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chr3_-_194072019 | 0.74 |
ENST00000429275.1
ENST00000323830.3 |
CPN2
|
carboxypeptidase N, polypeptide 2 |
| chrX_-_14047996 | 0.74 |
ENST00000380523.4
ENST00000398355.3 |
GEMIN8
|
gem (nuclear organelle) associated protein 8 |
| chrX_-_154299501 | 0.74 |
ENST00000369476.3
ENST00000369484.3 |
MTCP1
CMC4
|
mature T-cell proliferation 1 C-x(9)-C motif containing 4 |
| chr7_-_81399411 | 0.73 |
ENST00000423064.2
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr17_-_46667628 | 0.73 |
ENST00000498678.1
|
HOXB3
|
homeobox B3 |
| chr16_+_33006369 | 0.69 |
ENST00000425181.3
|
IGHV3OR16-10
|
immunoglobulin heavy variable 3/OR16-10 (non-functional) |
| chr1_+_32671236 | 0.68 |
ENST00000537469.1
ENST00000291358.6 |
IQCC
|
IQ motif containing C |
| chr1_-_202858227 | 0.67 |
ENST00000367262.3
|
RABIF
|
RAB interacting factor |
| chr22_+_40322623 | 0.67 |
ENST00000399090.2
|
GRAP2
|
GRB2-related adaptor protein 2 |
| chr2_-_85895295 | 0.66 |
ENST00000428225.1
ENST00000519937.2 |
SFTPB
|
surfactant protein B |
| chr16_+_68344877 | 0.64 |
ENST00000566657.1
ENST00000565745.1 ENST00000569571.1 ENST00000569047.3 ENST00000449359.3 |
PRMT7
|
protein arginine methyltransferase 7 |
| chr22_+_40322595 | 0.62 |
ENST00000420971.1
ENST00000544756.1 |
GRAP2
|
GRB2-related adaptor protein 2 |
| chr9_+_134065506 | 0.57 |
ENST00000483497.2
|
NUP214
|
nucleoporin 214kDa |
| chr16_+_765092 | 0.54 |
ENST00000568223.2
|
METRN
|
meteorin, glial cell differentiation regulator |
| chr12_-_49259643 | 0.50 |
ENST00000309739.5
|
RND1
|
Rho family GTPase 1 |
| chr4_-_168155577 | 0.50 |
ENST00000541354.1
ENST00000509854.1 ENST00000512681.1 ENST00000421836.2 ENST00000510741.1 ENST00000510403.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr1_+_114472222 | 0.47 |
ENST00000369558.1
ENST00000369561.4 |
HIPK1
|
homeodomain interacting protein kinase 1 |
| chr7_-_81399329 | 0.47 |
ENST00000453411.1
ENST00000444829.2 |
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr6_-_41715128 | 0.46 |
ENST00000356667.4
ENST00000373025.3 ENST00000425343.2 |
PGC
|
progastricsin (pepsinogen C) |
| chr7_+_130020180 | 0.44 |
ENST00000481342.1
ENST00000011292.3 ENST00000604896.1 |
CPA1
|
carboxypeptidase A1 (pancreatic) |
| chr2_+_152214098 | 0.43 |
ENST00000243347.3
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6 |
| chr17_-_7493390 | 0.41 |
ENST00000538513.2
ENST00000570788.1 ENST00000250055.2 |
SOX15
|
SRY (sex determining region Y)-box 15 |
| chr4_-_74853897 | 0.41 |
ENST00000296028.3
|
PPBP
|
pro-platelet basic protein (chemokine (C-X-C motif) ligand 7) |
| chr1_-_149982624 | 0.40 |
ENST00000417191.1
ENST00000369135.4 |
OTUD7B
|
OTU domain containing 7B |
| chr16_-_67997947 | 0.39 |
ENST00000537830.2
|
SLC12A4
|
solute carrier family 12 (potassium/chloride transporter), member 4 |
| chr5_+_78407602 | 0.38 |
ENST00000274353.5
ENST00000524080.1 |
BHMT
|
betaine--homocysteine S-methyltransferase |
| chr1_-_23520755 | 0.36 |
ENST00000314113.3
|
HTR1D
|
5-hydroxytryptamine (serotonin) receptor 1D, G protein-coupled |
| chr6_-_27279949 | 0.36 |
ENST00000444565.1
ENST00000377451.2 |
POM121L2
|
POM121 transmembrane nucleoporin-like 2 |
| chr14_+_81421861 | 0.33 |
ENST00000298171.2
|
TSHR
|
thyroid stimulating hormone receptor |
| chr5_+_75699040 | 0.32 |
ENST00000274364.6
|
IQGAP2
|
IQ motif containing GTPase activating protein 2 |
| chr17_+_66521936 | 0.32 |
ENST00000592800.1
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr16_+_68344981 | 0.30 |
ENST00000441236.1
ENST00000348497.4 ENST00000339507.5 |
PRMT7
|
protein arginine methyltransferase 7 |
| chr17_+_45973516 | 0.29 |
ENST00000376741.4
|
SP2
|
Sp2 transcription factor |
| chr16_+_8814563 | 0.29 |
ENST00000425191.2
ENST00000569156.1 |
ABAT
|
4-aminobutyrate aminotransferase |
| chr7_-_87342564 | 0.27 |
ENST00000265724.3
ENST00000416177.1 |
ABCB1
|
ATP-binding cassette, sub-family B (MDR/TAP), member 1 |
| chr14_+_21467414 | 0.26 |
ENST00000554422.1
ENST00000298681.4 |
SLC39A2
|
solute carrier family 39 (zinc transporter), member 2 |
| chr7_-_128415844 | 0.25 |
ENST00000249389.2
|
OPN1SW
|
opsin 1 (cone pigments), short-wave-sensitive |
| chr12_+_14518598 | 0.25 |
ENST00000261168.4
ENST00000538511.1 ENST00000545723.1 ENST00000543189.1 ENST00000536444.1 |
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr10_+_73156664 | 0.22 |
ENST00000398809.4
ENST00000398842.3 ENST00000461841.3 ENST00000299366.7 |
CDH23
|
cadherin-related 23 |
| chrX_+_86772707 | 0.13 |
ENST00000373119.4
|
KLHL4
|
kelch-like family member 4 |
| chr14_+_81421710 | 0.11 |
ENST00000342443.6
|
TSHR
|
thyroid stimulating hormone receptor |
| chrX_+_115567767 | 0.06 |
ENST00000371900.4
|
SLC6A14
|
solute carrier family 6 (amino acid transporter), member 14 |
| chr14_+_81421921 | 0.04 |
ENST00000554263.1
ENST00000554435.1 |
TSHR
|
thyroid stimulating hormone receptor |
| chr1_+_154300217 | 0.02 |
ENST00000368489.3
|
ATP8B2
|
ATPase, aminophospholipid transporter, class I, type 8B, member 2 |
| chr3_+_45984947 | 0.02 |
ENST00000304552.4
|
CXCR6
|
chemokine (C-X-C motif) receptor 6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX2-5
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.5 | 26.8 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 3.1 | 12.5 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 1.9 | 9.3 | GO:0002906 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 1.5 | 15.1 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 1.1 | 3.4 | GO:1904884 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 1.1 | 3.2 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 1.1 | 19.5 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 1.0 | 8.8 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 1.0 | 4.8 | GO:0000189 | MAPK import into nucleus(GO:0000189) |
| 0.9 | 3.8 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.8 | 3.2 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.8 | 11.0 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.8 | 12.9 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.8 | 2.3 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.6 | 2.6 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.6 | 6.1 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.6 | 3.0 | GO:0035947 | regulation of gluconeogenesis by regulation of transcription from RNA polymerase II promoter(GO:0035947) |
| 0.6 | 4.8 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.6 | 8.8 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.5 | 1.1 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.5 | 3.8 | GO:0075071 | autophagy of host cells involved in interaction with symbiont(GO:0075044) autophagy involved in symbiotic interaction(GO:0075071) |
| 0.5 | 4.2 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.5 | 3.4 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.4 | 3.7 | GO:0015705 | iodide transport(GO:0015705) |
| 0.4 | 0.4 | GO:0014717 | regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014717) positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) satellite cell activation involved in skeletal muscle regeneration(GO:0014901) |
| 0.4 | 2.0 | GO:0030070 | insulin processing(GO:0030070) |
| 0.4 | 1.5 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.4 | 2.2 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.3 | 1.4 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.3 | 1.3 | GO:1904588 | cellular response to glycoprotein(GO:1904588) cellular response to thyrotropin-releasing hormone(GO:1905229) |
| 0.3 | 0.9 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.3 | 1.6 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.3 | 2.5 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.3 | 4.4 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.3 | 0.8 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.3 | 3.0 | GO:0035330 | regulation of hippo signaling(GO:0035330) |
| 0.2 | 4.1 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.2 | 1.7 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.2 | 1.0 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.2 | 1.0 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 2.7 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 3.2 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.1 | 1.2 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.1 | 1.8 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.1 | 1.5 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.1 | 5.0 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.1 | 0.6 | GO:1901911 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 3.3 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.1 | 4.0 | GO:0042994 | cytoplasmic sequestering of transcription factor(GO:0042994) |
| 0.1 | 3.4 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.1 | 0.7 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 1.0 | GO:0019896 | axonal transport of mitochondrion(GO:0019896) |
| 0.1 | 1.6 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.1 | 2.0 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 6.5 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 2.2 | GO:0045332 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.1 | 6.5 | GO:0050771 | negative regulation of axonogenesis(GO:0050771) |
| 0.1 | 3.1 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 | 0.5 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.1 | 1.2 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 2.5 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.1 | 4.5 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.1 | 0.4 | GO:0031335 | regulation of sulfur amino acid metabolic process(GO:0031335) regulation of homocysteine metabolic process(GO:0050666) |
| 0.1 | 2.3 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 1.9 | GO:2000401 | regulation of lymphocyte migration(GO:2000401) |
| 0.1 | 0.5 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.1 | 1.0 | GO:0048535 | lymph node development(GO:0048535) |
| 0.1 | 8.4 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.4 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 1.4 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 6.8 | GO:0002433 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 0.0 | 0.4 | GO:0014827 | intestine smooth muscle contraction(GO:0014827) |
| 0.0 | 0.8 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.0 | 1.0 | GO:0070646 | protein deubiquitination(GO:0016579) protein modification by small protein removal(GO:0070646) |
| 0.0 | 3.2 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.0 | 2.9 | GO:0070126 | mitochondrial translational termination(GO:0070126) |
| 0.0 | 0.5 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 2.2 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.0 | 0.4 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 2.0 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.7 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 0.9 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.0 | 0.3 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 1.0 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 1.3 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 1.4 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.4 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.7 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.4 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 0.9 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 1.0 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.2 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.3 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.6 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 11.0 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 1.3 | 9.3 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.4 | 4.7 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.4 | 30.0 | GO:0030315 | T-tubule(GO:0030315) |
| 0.3 | 3.7 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.3 | 2.6 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.2 | 1.0 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.2 | 1.4 | GO:0072487 | MSL complex(GO:0072487) |
| 0.2 | 1.0 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.2 | 0.6 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.2 | 0.9 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 0.2 | 2.6 | GO:0036038 | MKS complex(GO:0036038) |
| 0.2 | 3.8 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.2 | 4.8 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.2 | 3.2 | GO:0042599 | lamellar body(GO:0042599) |
| 0.2 | 22.6 | GO:0031514 | motile cilium(GO:0031514) |
| 0.1 | 1.4 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 1.5 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 8.8 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.1 | 6.5 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 2.5 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.1 | 10.7 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 2.7 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 0.7 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.1 | 2.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 3.3 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 2.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 2.4 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.1 | 11.3 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 2.9 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 0.3 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.1 | 2.0 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.1 | 3.4 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.1 | 4.1 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.8 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.4 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 1.0 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 2.0 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 5.7 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 1.8 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 5.8 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 1.0 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 41.6 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 2.1 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 1.9 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 1.9 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 3.4 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 1.2 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.4 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.0 | 0.7 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.2 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 1.0 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 4.8 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 1.3 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 8.8 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 1.9 | 26.8 | GO:0031432 | titin binding(GO:0031432) |
| 1.9 | 9.3 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 1.4 | 12.5 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 1.0 | 6.1 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 1.0 | 2.9 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.9 | 2.7 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.8 | 4.8 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.6 | 3.4 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.5 | 3.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.5 | 19.5 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.5 | 1.5 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.3 | 0.9 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.3 | 1.1 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.3 | 4.8 | GO:0005522 | profilin binding(GO:0005522) |
| 0.2 | 4.8 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.2 | 1.3 | GO:0015185 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.2 | 3.2 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.2 | 1.0 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.2 | 1.0 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.2 | 1.3 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.2 | 2.5 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.2 | 1.7 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.2 | 1.8 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.2 | 4.3 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.2 | 4.2 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.2 | 4.5 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.1 | 3.9 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.4 | GO:0047150 | betaine-homocysteine S-methyltransferase activity(GO:0047150) |
| 0.1 | 0.7 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.6 | GO:0052842 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.1 | 6.5 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 3.0 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 2.5 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 1.0 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.1 | 2.3 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 4.1 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 1.0 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 11.3 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.1 | 2.0 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.3 | GO:0032145 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.1 | 4.0 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.1 | 8.5 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 0.8 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 1.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 8.4 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 2.6 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 1.0 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 3.6 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 1.5 | GO:0000217 | DNA secondary structure binding(GO:0000217) |
| 0.0 | 1.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.4 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 2.0 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.4 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.4 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 1.2 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.4 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 1.0 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 8.3 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.4 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 1.2 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.2 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.7 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 1.4 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.5 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 1.6 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 1.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 1.0 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.8 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.3 | 4.8 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.2 | 11.1 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 8.8 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 3.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 3.0 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 1.5 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 5.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 1.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 4.3 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 2.7 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 2.3 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 3.4 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.1 | 2.5 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 2.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 3.1 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.6 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.8 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 2.3 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.0 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 4.2 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 12.0 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.4 | 8.8 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.4 | 6.1 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.3 | 4.8 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.3 | 3.4 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.2 | 2.5 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 4.1 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 5.0 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.1 | 2.8 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.1 | 4.8 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.1 | 9.5 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 4.3 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 1.3 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.1 | 0.9 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 8.9 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 2.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 6.7 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.1 | 1.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 1.7 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 1.3 | REACTOME CD28 CO STIMULATION | Genes involved in CD28 co-stimulation |
| 0.0 | 2.6 | REACTOME RECRUITMENT OF MITOTIC CENTROSOME PROTEINS AND COMPLEXES | Genes involved in Recruitment of mitotic centrosome proteins and complexes |
| 0.0 | 0.5 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 1.6 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.0 | 0.4 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 1.0 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 5.0 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 2.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.3 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 2.3 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.3 | REACTOME GLUCAGON SIGNALING IN METABOLIC REGULATION | Genes involved in Glucagon signaling in metabolic regulation |
| 0.0 | 1.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.6 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |