Project
averaged GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
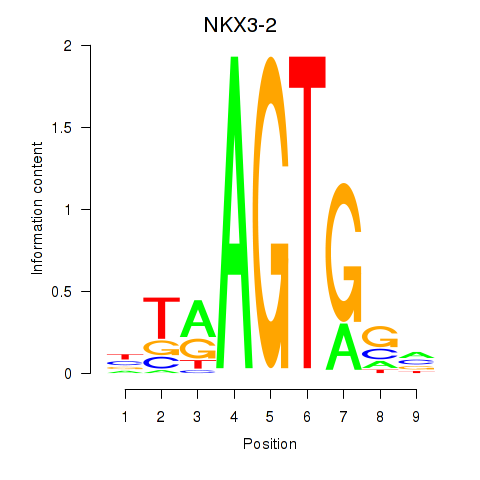
Results for NKX3-2
Z-value: 1.14
Transcription factors associated with NKX3-2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX3-2
|
ENSG00000109705.7 | NK3 homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NKX3-2 | hg19_v2_chr4_-_13546632_13546674 | 0.29 | 1.8e-05 | Click! |
Activity profile of NKX3-2 motif
Sorted Z-values of NKX3-2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_+_38420783 | 17.68 |
ENST00000422612.2
ENST00000286824.6 ENST00000545599.1 |
TSPAN7
|
tetraspanin 7 |
| chr1_-_156399184 | 17.02 |
ENST00000368243.1
ENST00000357975.4 ENST00000310027.5 ENST00000400991.2 |
C1orf61
|
chromosome 1 open reading frame 61 |
| chr12_-_54978086 | 16.29 |
ENST00000553113.1
|
PPP1R1A
|
protein phosphatase 1, regulatory (inhibitor) subunit 1A |
| chrX_+_38420623 | 15.37 |
ENST00000378482.2
|
TSPAN7
|
tetraspanin 7 |
| chr8_-_27115931 | 14.22 |
ENST00000523048.1
|
STMN4
|
stathmin-like 4 |
| chr2_+_17721920 | 13.52 |
ENST00000295156.4
|
VSNL1
|
visinin-like 1 |
| chr17_+_38083977 | 12.52 |
ENST00000578802.1
ENST00000578478.1 ENST00000582263.1 |
RP11-387H17.4
|
RP11-387H17.4 |
| chr8_-_27115903 | 12.30 |
ENST00000350889.4
ENST00000519997.1 ENST00000519614.1 ENST00000522908.1 ENST00000265770.7 |
STMN4
|
stathmin-like 4 |
| chr1_+_160097462 | 11.10 |
ENST00000447527.1
|
ATP1A2
|
ATPase, Na+/K+ transporting, alpha 2 polypeptide |
| chr19_+_16999654 | 10.96 |
ENST00000248076.3
|
F2RL3
|
coagulation factor II (thrombin) receptor-like 3 |
| chr5_-_149792295 | 10.21 |
ENST00000518797.1
ENST00000524315.1 ENST00000009530.7 ENST00000377795.3 |
CD74
|
CD74 molecule, major histocompatibility complex, class II invariant chain |
| chr19_-_3028354 | 10.18 |
ENST00000586422.1
|
TLE2
|
transducin-like enhancer of split 2 (E(sp1) homolog, Drosophila) |
| chr2_-_208489707 | 9.46 |
ENST00000448007.2
ENST00000432416.1 ENST00000411432.1 |
METTL21A
|
methyltransferase like 21A |
| chr4_+_156588249 | 8.84 |
ENST00000393832.3
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3 |
| chr19_-_6720686 | 8.58 |
ENST00000245907.6
|
C3
|
complement component 3 |
| chr5_+_156696362 | 8.55 |
ENST00000377576.3
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2 |
| chr14_-_60337684 | 7.95 |
ENST00000267484.5
|
RTN1
|
reticulon 1 |
| chr8_-_90769422 | 7.77 |
ENST00000524190.1
ENST00000523859.1 |
RP11-37B2.1
|
RP11-37B2.1 |
| chr22_+_38453207 | 7.72 |
ENST00000404072.3
ENST00000424694.1 |
PICK1
|
protein interacting with PRKCA 1 |
| chr2_-_175869936 | 7.58 |
ENST00000409900.3
|
CHN1
|
chimerin 1 |
| chr6_-_32557610 | 7.48 |
ENST00000360004.5
|
HLA-DRB1
|
major histocompatibility complex, class II, DR beta 1 |
| chr19_-_17185848 | 7.42 |
ENST00000593360.1
|
HAUS8
|
HAUS augmin-like complex, subunit 8 |
| chrX_+_48660287 | 7.39 |
ENST00000444343.2
ENST00000376610.2 ENST00000334136.5 ENST00000376619.2 |
HDAC6
|
histone deacetylase 6 |
| chr7_-_99573677 | 7.30 |
ENST00000292401.4
|
AZGP1
|
alpha-2-glycoprotein 1, zinc-binding |
| chr2_-_175870085 | 7.24 |
ENST00000409156.3
|
CHN1
|
chimerin 1 |
| chr11_-_111175739 | 7.23 |
ENST00000532918.1
|
COLCA1
|
colorectal cancer associated 1 |
| chr4_+_156587979 | 7.07 |
ENST00000511507.1
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3 |
| chr2_+_25016282 | 6.97 |
ENST00000260662.1
|
CENPO
|
centromere protein O |
| chr2_+_220379052 | 6.92 |
ENST00000347842.3
ENST00000358078.4 |
ASIC4
|
acid-sensing (proton-gated) ion channel family member 4 |
| chr15_+_99645277 | 6.70 |
ENST00000336292.6
ENST00000328642.7 |
SYNM
|
synemin, intermediate filament protein |
| chrX_+_44732757 | 6.67 |
ENST00000377967.4
ENST00000536777.1 ENST00000382899.4 ENST00000543216.1 |
KDM6A
|
lysine (K)-specific demethylase 6A |
| chr10_-_98031265 | 6.62 |
ENST00000224337.5
ENST00000371176.2 |
BLNK
|
B-cell linker |
| chrX_-_49965663 | 6.60 |
ENST00000376056.2
ENST00000376058.2 ENST00000358526.2 |
AKAP4
|
A kinase (PRKA) anchor protein 4 |
| chr10_-_98031310 | 6.53 |
ENST00000427367.2
ENST00000413476.2 |
BLNK
|
B-cell linker |
| chr3_-_149688502 | 6.45 |
ENST00000481767.1
ENST00000475518.1 |
PFN2
|
profilin 2 |
| chr1_-_173793458 | 6.40 |
ENST00000356198.2
|
CENPL
|
centromere protein L |
| chrX_-_71526813 | 6.37 |
ENST00000246139.5
|
CITED1
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 1 |
| chr14_-_106963409 | 6.32 |
ENST00000390621.2
|
IGHV1-45
|
immunoglobulin heavy variable 1-45 |
| chr11_-_22851367 | 6.11 |
ENST00000354193.4
|
SVIP
|
small VCP/p97-interacting protein |
| chr14_-_74551172 | 5.96 |
ENST00000553458.1
|
ALDH6A1
|
aldehyde dehydrogenase 6 family, member A1 |
| chr5_+_78407602 | 5.91 |
ENST00000274353.5
ENST00000524080.1 |
BHMT
|
betaine--homocysteine S-methyltransferase |
| chr16_-_21289627 | 5.91 |
ENST00000396023.2
ENST00000415987.2 |
CRYM
|
crystallin, mu |
| chr4_+_156680518 | 5.87 |
ENST00000513437.1
|
GUCY1B3
|
guanylate cyclase 1, soluble, beta 3 |
| chr3_-_49907323 | 5.86 |
ENST00000296471.7
ENST00000488336.1 ENST00000467248.1 ENST00000466940.1 ENST00000463537.1 ENST00000480398.2 |
CAMKV
|
CaM kinase-like vesicle-associated |
| chr5_+_156712372 | 5.69 |
ENST00000541131.1
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2 |
| chr17_-_19771216 | 5.68 |
ENST00000395544.4
|
ULK2
|
unc-51 like autophagy activating kinase 2 |
| chr10_+_101088836 | 5.57 |
ENST00000356713.4
|
CNNM1
|
cyclin M1 |
| chr14_-_90798418 | 5.56 |
ENST00000354366.3
|
NRDE2
|
NRDE-2, necessary for RNA interference, domain containing |
| chr22_+_38453378 | 5.48 |
ENST00000437453.1
ENST00000356976.3 |
PICK1
|
protein interacting with PRKCA 1 |
| chr10_-_75571566 | 5.45 |
ENST00000299641.4
|
NDST2
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 2 |
| chr4_-_141348789 | 5.41 |
ENST00000414773.1
|
CLGN
|
calmegin |
| chr10_+_7745303 | 5.33 |
ENST00000429820.1
ENST00000379587.4 |
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr10_+_101089107 | 5.31 |
ENST00000446890.1
ENST00000370528.3 |
CNNM1
|
cyclin M1 |
| chr19_+_36266417 | 5.24 |
ENST00000378944.5
ENST00000007510.4 |
ARHGAP33
|
Rho GTPase activating protein 33 |
| chr2_+_25015968 | 5.23 |
ENST00000380834.2
ENST00000473706.1 |
CENPO
|
centromere protein O |
| chr4_+_156588115 | 5.21 |
ENST00000455639.2
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3 |
| chr4_+_113970772 | 5.19 |
ENST00000504454.1
ENST00000394537.3 ENST00000357077.4 ENST00000264366.6 |
ANK2
|
ankyrin 2, neuronal |
| chr15_+_81475047 | 5.14 |
ENST00000559388.1
|
IL16
|
interleukin 16 |
| chr10_-_99531709 | 4.89 |
ENST00000266066.3
|
SFRP5
|
secreted frizzled-related protein 5 |
| chr7_+_129906660 | 4.87 |
ENST00000222481.4
|
CPA2
|
carboxypeptidase A2 (pancreatic) |
| chr10_-_75571341 | 4.84 |
ENST00000309979.6
|
NDST2
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 2 |
| chr10_+_7745232 | 4.84 |
ENST00000358415.4
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr17_-_617949 | 4.75 |
ENST00000401468.3
ENST00000574029.1 ENST00000291074.5 ENST00000571805.1 ENST00000437048.2 ENST00000446250.2 |
VPS53
|
vacuolar protein sorting 53 homolog (S. cerevisiae) |
| chr20_+_42839722 | 4.73 |
ENST00000442383.1
ENST00000435163.1 |
OSER1-AS1
|
OSER1 antisense RNA 1 (head to head) |
| chr17_-_74137374 | 4.70 |
ENST00000322957.6
|
FOXJ1
|
forkhead box J1 |
| chrX_+_76709648 | 4.67 |
ENST00000439435.1
|
FGF16
|
fibroblast growth factor 16 |
| chr17_+_45810594 | 4.66 |
ENST00000177694.1
|
TBX21
|
T-box 21 |
| chr11_-_119993979 | 4.61 |
ENST00000524816.3
ENST00000525327.1 |
TRIM29
|
tripartite motif containing 29 |
| chr5_-_160279207 | 4.46 |
ENST00000327245.5
|
ATP10B
|
ATPase, class V, type 10B |
| chr19_-_5680202 | 4.29 |
ENST00000590389.1
|
C19orf70
|
chromosome 19 open reading frame 70 |
| chr4_+_156588350 | 4.24 |
ENST00000296518.7
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3 |
| chr8_+_86376081 | 4.23 |
ENST00000285379.5
|
CA2
|
carbonic anhydrase II |
| chr2_+_79347577 | 4.21 |
ENST00000233735.1
|
REG1A
|
regenerating islet-derived 1 alpha |
| chr17_+_58755184 | 4.19 |
ENST00000589222.1
ENST00000407086.3 ENST00000390652.5 |
BCAS3
|
breast carcinoma amplified sequence 3 |
| chr7_+_29237354 | 4.15 |
ENST00000546235.1
|
CHN2
|
chimerin 2 |
| chr1_-_153013588 | 4.13 |
ENST00000360379.3
|
SPRR2D
|
small proline-rich protein 2D |
| chr5_+_40909354 | 4.11 |
ENST00000313164.9
|
C7
|
complement component 7 |
| chr3_+_38537960 | 4.08 |
ENST00000453767.1
|
EXOG
|
endo/exonuclease (5'-3'), endonuclease G-like |
| chr19_-_6767516 | 4.07 |
ENST00000245908.6
|
SH2D3A
|
SH2 domain containing 3A |
| chr11_+_2421718 | 4.06 |
ENST00000380996.5
ENST00000333256.6 ENST00000380992.1 ENST00000437110.1 ENST00000435795.1 |
TSSC4
|
tumor suppressing subtransferable candidate 4 |
| chr11_-_102401469 | 4.05 |
ENST00000260227.4
|
MMP7
|
matrix metallopeptidase 7 (matrilysin, uterine) |
| chr1_+_205473720 | 4.01 |
ENST00000429964.2
ENST00000506784.1 ENST00000360066.2 |
CDK18
|
cyclin-dependent kinase 18 |
| chr1_+_152974218 | 3.99 |
ENST00000331860.3
ENST00000443178.1 ENST00000295367.4 |
SPRR3
|
small proline-rich protein 3 |
| chrX_+_100333709 | 3.94 |
ENST00000372930.4
|
TMEM35
|
transmembrane protein 35 |
| chr5_-_115872142 | 3.89 |
ENST00000510263.1
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr12_-_21927736 | 3.89 |
ENST00000240662.2
|
KCNJ8
|
potassium inwardly-rectifying channel, subfamily J, member 8 |
| chr3_-_128712955 | 3.88 |
ENST00000265068.5
|
KIAA1257
|
KIAA1257 |
| chr17_-_46507537 | 3.86 |
ENST00000336915.6
|
SKAP1
|
src kinase associated phosphoprotein 1 |
| chr3_+_38537763 | 3.84 |
ENST00000287675.5
ENST00000358249.2 ENST00000422077.2 |
EXOG
|
endo/exonuclease (5'-3'), endonuclease G-like |
| chr12_+_57984965 | 3.83 |
ENST00000540759.2
ENST00000551772.1 ENST00000550465.1 ENST00000354947.5 |
PIP4K2C
|
phosphatidylinositol-5-phosphate 4-kinase, type II, gamma |
| chr7_+_16828866 | 3.79 |
ENST00000597084.1
|
AC073333.1
|
Uncharacterized protein |
| chr15_+_75639951 | 3.73 |
ENST00000564784.1
ENST00000569035.1 |
NEIL1
|
nei endonuclease VIII-like 1 (E. coli) |
| chr10_-_95360983 | 3.73 |
ENST00000371464.3
|
RBP4
|
retinol binding protein 4, plasma |
| chr3_+_111697843 | 3.69 |
ENST00000534857.1
ENST00000273359.3 ENST00000494817.1 |
ABHD10
|
abhydrolase domain containing 10 |
| chr1_+_151739131 | 3.68 |
ENST00000400999.1
|
OAZ3
|
ornithine decarboxylase antizyme 3 |
| chr4_-_11430221 | 3.61 |
ENST00000514690.1
|
HS3ST1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 1 |
| chr3_-_50541028 | 3.61 |
ENST00000266039.3
ENST00000435965.1 ENST00000395083.1 |
CACNA2D2
|
calcium channel, voltage-dependent, alpha 2/delta subunit 2 |
| chr3_-_149688896 | 3.59 |
ENST00000239940.7
|
PFN2
|
profilin 2 |
| chr7_-_99573640 | 3.55 |
ENST00000411734.1
|
AZGP1
|
alpha-2-glycoprotein 1, zinc-binding |
| chr14_-_23770683 | 3.54 |
ENST00000561437.1
ENST00000559942.1 ENST00000560913.1 ENST00000559314.1 ENST00000558058.1 |
PPP1R3E
|
protein phosphatase 1, regulatory subunit 3E |
| chr6_-_136788001 | 3.52 |
ENST00000544465.1
|
MAP7
|
microtubule-associated protein 7 |
| chr1_-_182573514 | 3.51 |
ENST00000367558.5
|
RGS16
|
regulator of G-protein signaling 16 |
| chr3_+_125694347 | 3.46 |
ENST00000505382.1
ENST00000511082.1 |
ROPN1B
|
rhophilin associated tail protein 1B |
| chr10_-_115933942 | 3.41 |
ENST00000369285.3
ENST00000369287.3 ENST00000369286.1 |
C10orf118
|
chromosome 10 open reading frame 118 |
| chr17_-_9929581 | 3.38 |
ENST00000437099.2
ENST00000396115.2 |
GAS7
|
growth arrest-specific 7 |
| chrX_+_51928002 | 3.38 |
ENST00000375626.3
|
MAGED4
|
melanoma antigen family D, 4 |
| chrX_+_153665248 | 3.36 |
ENST00000447750.2
|
GDI1
|
GDP dissociation inhibitor 1 |
| chr12_+_6961279 | 3.35 |
ENST00000229268.8
ENST00000389231.5 ENST00000542087.1 |
USP5
|
ubiquitin specific peptidase 5 (isopeptidase T) |
| chr19_+_48867652 | 3.31 |
ENST00000344846.2
|
SYNGR4
|
synaptogyrin 4 |
| chr11_-_1036706 | 3.29 |
ENST00000421673.2
|
MUC6
|
mucin 6, oligomeric mucus/gel-forming |
| chr5_+_79331164 | 3.27 |
ENST00000350881.2
|
THBS4
|
thrombospondin 4 |
| chr7_-_135412925 | 3.23 |
ENST00000354042.4
|
SLC13A4
|
solute carrier family 13 (sodium/sulfate symporter), member 4 |
| chr16_+_57406368 | 3.23 |
ENST00000006053.6
ENST00000563383.1 |
CX3CL1
|
chemokine (C-X3-C motif) ligand 1 |
| chr1_-_216978709 | 3.19 |
ENST00000360012.3
|
ESRRG
|
estrogen-related receptor gamma |
| chr6_-_11382478 | 3.19 |
ENST00000397378.3
ENST00000513989.1 ENST00000508546.1 ENST00000504387.1 |
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9 |
| chr3_+_52454971 | 3.18 |
ENST00000465863.1
|
PHF7
|
PHD finger protein 7 |
| chr6_+_42928485 | 3.17 |
ENST00000372808.3
|
GNMT
|
glycine N-methyltransferase |
| chr11_-_64511789 | 3.08 |
ENST00000419843.1
ENST00000394430.1 |
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr19_+_36545833 | 3.05 |
ENST00000401500.2
ENST00000270301.7 ENST00000427823.2 |
WDR62
|
WD repeat domain 62 |
| chr9_+_34458771 | 3.04 |
ENST00000437363.1
ENST00000242317.4 |
DNAI1
|
dynein, axonemal, intermediate chain 1 |
| chr6_-_150039249 | 3.02 |
ENST00000543571.1
|
LATS1
|
large tumor suppressor kinase 1 |
| chr1_+_173793777 | 3.01 |
ENST00000239457.5
|
DARS2
|
aspartyl-tRNA synthetase 2, mitochondrial |
| chr14_-_24711470 | 3.01 |
ENST00000559969.1
|
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr9_-_131644202 | 2.98 |
ENST00000320665.6
ENST00000436267.2 |
CCBL1
|
cysteine conjugate-beta lyase, cytoplasmic |
| chr1_+_22307592 | 2.93 |
ENST00000400277.2
|
CELA3B
|
chymotrypsin-like elastase family, member 3B |
| chr18_-_6414884 | 2.92 |
ENST00000317931.7
ENST00000284898.6 ENST00000400104.3 |
L3MBTL4
|
l(3)mbt-like 4 (Drosophila) |
| chr19_-_36297348 | 2.91 |
ENST00000589835.1
|
PRODH2
|
proline dehydrogenase (oxidase) 2 |
| chr19_-_44809121 | 2.91 |
ENST00000591609.1
ENST00000589799.1 ENST00000291182.4 ENST00000589248.1 |
ZNF235
|
zinc finger protein 235 |
| chr6_+_26156551 | 2.88 |
ENST00000304218.3
|
HIST1H1E
|
histone cluster 1, H1e |
| chr12_-_120765565 | 2.86 |
ENST00000423423.3
ENST00000308366.4 |
PLA2G1B
|
phospholipase A2, group IB (pancreas) |
| chr3_-_185216766 | 2.84 |
ENST00000296254.3
|
TMEM41A
|
transmembrane protein 41A |
| chr11_-_207221 | 2.82 |
ENST00000486280.1
ENST00000332865.6 ENST00000529614.2 ENST00000325147.9 ENST00000410108.1 ENST00000382762.3 |
BET1L
|
Bet1 golgi vesicular membrane trafficking protein-like |
| chr11_-_63381823 | 2.82 |
ENST00000323646.5
|
PLA2G16
|
phospholipase A2, group XVI |
| chr1_+_66258846 | 2.80 |
ENST00000341517.4
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr12_+_56473910 | 2.77 |
ENST00000411731.2
|
ERBB3
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr1_+_60280458 | 2.76 |
ENST00000455990.1
ENST00000371208.3 |
HOOK1
|
hook microtubule-tethering protein 1 |
| chr1_+_158223923 | 2.73 |
ENST00000289429.5
|
CD1A
|
CD1a molecule |
| chr11_+_128563652 | 2.73 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr11_-_119993734 | 2.72 |
ENST00000533302.1
|
TRIM29
|
tripartite motif containing 29 |
| chr16_+_83986827 | 2.71 |
ENST00000393306.1
ENST00000565123.1 |
OSGIN1
|
oxidative stress induced growth inhibitor 1 |
| chr22_-_21356375 | 2.69 |
ENST00000215742.4
ENST00000399133.2 |
THAP7
|
THAP domain containing 7 |
| chr4_+_62067860 | 2.68 |
ENST00000514591.1
|
LPHN3
|
latrophilin 3 |
| chr9_-_23821842 | 2.68 |
ENST00000544538.1
|
ELAVL2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr11_-_64512469 | 2.68 |
ENST00000377485.1
|
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr19_+_57999079 | 2.67 |
ENST00000426954.2
ENST00000354197.4 ENST00000523882.1 ENST00000520540.1 ENST00000519310.1 ENST00000442920.2 ENST00000523312.1 ENST00000424930.2 |
ZNF419
|
zinc finger protein 419 |
| chr12_+_50794730 | 2.64 |
ENST00000523389.1
ENST00000518561.1 ENST00000347328.5 ENST00000550260.1 |
LARP4
|
La ribonucleoprotein domain family, member 4 |
| chr10_-_17171817 | 2.63 |
ENST00000377833.4
|
CUBN
|
cubilin (intrinsic factor-cobalamin receptor) |
| chr11_-_118047376 | 2.63 |
ENST00000278947.5
|
SCN2B
|
sodium channel, voltage-gated, type II, beta subunit |
| chr15_+_42694573 | 2.62 |
ENST00000397200.4
ENST00000569827.1 |
CAPN3
|
calpain 3, (p94) |
| chr12_-_10151773 | 2.62 |
ENST00000298527.6
ENST00000348658.4 |
CLEC1B
|
C-type lectin domain family 1, member B |
| chr19_+_57999101 | 2.60 |
ENST00000347466.6
ENST00000523138.1 ENST00000415379.2 ENST00000521754.1 ENST00000221735.7 ENST00000518999.1 ENST00000521137.1 |
ZNF419
|
zinc finger protein 419 |
| chr4_+_75023816 | 2.55 |
ENST00000395759.2
ENST00000331145.6 ENST00000359107.5 ENST00000325278.6 |
MTHFD2L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2-like |
| chr12_-_122296755 | 2.53 |
ENST00000289004.4
|
HPD
|
4-hydroxyphenylpyruvate dioxygenase |
| chr12_-_54019755 | 2.53 |
ENST00000588078.1
ENST00000551480.2 ENST00000548118.2 ENST00000456903.4 ENST00000591834.1 |
ATF7
RP11-793H13.10
|
activating transcription factor 7 Uncharacterized protein |
| chr11_-_128737163 | 2.51 |
ENST00000324003.3
ENST00000392665.2 |
KCNJ1
|
potassium inwardly-rectifying channel, subfamily J, member 1 |
| chrX_+_18725758 | 2.46 |
ENST00000472826.1
ENST00000544635.1 ENST00000496075.2 |
PPEF1
|
protein phosphatase, EF-hand calcium binding domain 1 |
| chr2_-_24270217 | 2.46 |
ENST00000295148.4
ENST00000406895.3 |
C2orf44
|
chromosome 2 open reading frame 44 |
| chr14_-_98444386 | 2.46 |
ENST00000556462.1
ENST00000556138.1 |
C14orf64
|
chromosome 14 open reading frame 64 |
| chr4_+_144303093 | 2.46 |
ENST00000505913.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr10_+_5005445 | 2.42 |
ENST00000380872.4
|
AKR1C1
|
aldo-keto reductase family 1, member C1 |
| chr16_-_2318055 | 2.42 |
ENST00000561518.1
ENST00000561718.1 ENST00000567147.1 ENST00000562690.1 ENST00000569598.2 |
RNPS1
|
RNA binding protein S1, serine-rich domain |
| chr1_+_210001309 | 2.41 |
ENST00000491415.2
|
DIEXF
|
digestive organ expansion factor homolog (zebrafish) |
| chr14_+_21467414 | 2.38 |
ENST00000554422.1
ENST00000298681.4 |
SLC39A2
|
solute carrier family 39 (zinc transporter), member 2 |
| chr10_+_18629628 | 2.38 |
ENST00000377329.4
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr20_+_53092123 | 2.36 |
ENST00000262593.5
|
DOK5
|
docking protein 5 |
| chr16_+_55690002 | 2.36 |
ENST00000414754.3
|
SLC6A2
|
solute carrier family 6 (neurotransmitter transporter), member 2 |
| chr14_-_106830057 | 2.34 |
ENST00000390616.2
|
IGHV4-34
|
immunoglobulin heavy variable 4-34 |
| chr11_-_128737259 | 2.30 |
ENST00000440599.2
ENST00000392666.1 ENST00000324036.3 |
KCNJ1
|
potassium inwardly-rectifying channel, subfamily J, member 1 |
| chr19_+_16435625 | 2.28 |
ENST00000248071.5
ENST00000592003.1 |
KLF2
|
Kruppel-like factor 2 |
| chr16_-_27561209 | 2.27 |
ENST00000356183.4
ENST00000561623.1 |
GTF3C1
|
general transcription factor IIIC, polypeptide 1, alpha 220kDa |
| chr20_+_53092232 | 2.26 |
ENST00000395939.1
|
DOK5
|
docking protein 5 |
| chr5_-_59481406 | 2.23 |
ENST00000546160.1
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr5_+_56111361 | 2.21 |
ENST00000399503.3
|
MAP3K1
|
mitogen-activated protein kinase kinase kinase 1, E3 ubiquitin protein ligase |
| chr12_+_4382917 | 2.20 |
ENST00000261254.3
|
CCND2
|
cyclin D2 |
| chr4_-_141348999 | 2.19 |
ENST00000325617.5
|
CLGN
|
calmegin |
| chr9_+_131062367 | 2.18 |
ENST00000601297.1
|
AL359091.2
|
CDNA: FLJ21673 fis, clone COL09042; HCG2036511; Uncharacterized protein |
| chr1_+_36690011 | 2.17 |
ENST00000354618.5
ENST00000469141.2 ENST00000478853.1 |
THRAP3
|
thyroid hormone receptor associated protein 3 |
| chr7_+_130020180 | 2.13 |
ENST00000481342.1
ENST00000011292.3 ENST00000604896.1 |
CPA1
|
carboxypeptidase A1 (pancreatic) |
| chr15_-_88799661 | 2.13 |
ENST00000360948.2
ENST00000357724.2 ENST00000355254.2 ENST00000317501.3 |
NTRK3
|
neurotrophic tyrosine kinase, receptor, type 3 |
| chr5_+_140810132 | 2.12 |
ENST00000252085.3
|
PCDHGA12
|
protocadherin gamma subfamily A, 12 |
| chr18_+_72167096 | 2.12 |
ENST00000324301.8
|
CNDP2
|
CNDP dipeptidase 2 (metallopeptidase M20 family) |
| chr4_+_144258021 | 2.12 |
ENST00000262994.4
|
GAB1
|
GRB2-associated binding protein 1 |
| chr1_+_119957554 | 2.12 |
ENST00000543831.1
ENST00000433745.1 ENST00000369416.3 |
HSD3B2
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2 |
| chr1_-_202858227 | 2.11 |
ENST00000367262.3
|
RABIF
|
RAB interacting factor |
| chr6_-_32731243 | 2.09 |
ENST00000427449.1
ENST00000411527.1 |
HLA-DQB2
|
major histocompatibility complex, class II, DQ beta 2 |
| chr11_+_67250490 | 2.08 |
ENST00000528641.2
ENST00000279146.3 |
AIP
|
aryl hydrocarbon receptor interacting protein |
| chr2_-_182545603 | 2.06 |
ENST00000295108.3
|
NEUROD1
|
neuronal differentiation 1 |
| chr7_+_110731062 | 2.05 |
ENST00000308478.5
ENST00000451085.1 ENST00000422987.3 ENST00000421101.1 |
LRRN3
|
leucine rich repeat neuronal 3 |
| chr14_-_21567009 | 2.04 |
ENST00000556174.1
ENST00000554478.1 ENST00000553980.1 ENST00000421093.2 |
ZNF219
|
zinc finger protein 219 |
| chr19_-_51920952 | 2.03 |
ENST00000356298.5
ENST00000339313.5 ENST00000529627.1 ENST00000439889.2 ENST00000353836.5 ENST00000432469.2 |
SIGLEC10
|
sialic acid binding Ig-like lectin 10 |
| chr10_+_35415719 | 1.99 |
ENST00000474362.1
ENST00000374721.3 |
CREM
|
cAMP responsive element modulator |
| chr10_-_47173994 | 1.99 |
ENST00000414655.2
ENST00000545298.1 ENST00000359178.4 ENST00000358140.4 ENST00000503031.1 |
ANXA8L1
LINC00842
|
annexin A8-like 1 long intergenic non-protein coding RNA 842 |
| chr2_+_102927962 | 1.97 |
ENST00000233954.1
ENST00000393393.3 ENST00000410040.1 |
IL1RL1
IL18R1
|
interleukin 1 receptor-like 1 interleukin 18 receptor 1 |
| chr11_-_59633951 | 1.96 |
ENST00000257264.3
|
TCN1
|
transcobalamin I (vitamin B12 binding protein, R binder family) |
| chr6_-_32784687 | 1.96 |
ENST00000447394.1
ENST00000438763.2 |
HLA-DOB
|
major histocompatibility complex, class II, DO beta |
| chr20_+_2821340 | 1.96 |
ENST00000380445.3
ENST00000380469.3 |
VPS16
|
vacuolar protein sorting 16 homolog (S. cerevisiae) |
| chr4_-_109087872 | 1.95 |
ENST00000510624.1
|
LEF1
|
lymphoid enhancer-binding factor 1 |
| chr10_-_7708918 | 1.94 |
ENST00000256861.6
ENST00000397146.2 ENST00000446830.2 ENST00000397145.2 |
ITIH5
|
inter-alpha-trypsin inhibitor heavy chain family, member 5 |
| chr7_-_79082867 | 1.94 |
ENST00000419488.1
ENST00000354212.4 |
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr17_+_68071389 | 1.94 |
ENST00000283936.1
ENST00000392671.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX3-2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 25.4 | GO:0052565 | response to defense-related nitric oxide production by other organism involved in symbiotic interaction(GO:0052551) response to defense-related host nitric oxide production(GO:0052565) |
| 2.9 | 8.6 | GO:0001798 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 2.5 | 7.4 | GO:0090034 | regulation of chaperone-mediated protein complex assembly(GO:0090034) positive regulation of chaperone-mediated protein complex assembly(GO:0090035) |
| 2.2 | 6.7 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 2.2 | 11.1 | GO:0051941 | regulation of amino acid uptake involved in synaptic transmission(GO:0051941) regulation of glutamate uptake involved in transmission of nerve impulse(GO:0051946) regulation of L-glutamate import(GO:1900920) |
| 2.0 | 10.2 | GO:0002906 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 2.0 | 6.1 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 2.0 | 5.9 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 1.9 | 13.2 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 1.8 | 14.2 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 1.7 | 5.2 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 1.7 | 6.7 | GO:0031443 | fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 1.6 | 4.9 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 1.6 | 4.7 | GO:0002661 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 1.5 | 7.5 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 1.5 | 6.0 | GO:0019859 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 1.4 | 4.2 | GO:0042938 | dipeptide transport(GO:0042938) |
| 1.4 | 11.0 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 1.3 | 2.6 | GO:0030220 | platelet formation(GO:0030220) |
| 1.2 | 4.9 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 1.1 | 3.2 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 1.1 | 6.4 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 1.0 | 5.9 | GO:0031335 | regulation of sulfur amino acid metabolic process(GO:0031335) regulation of homocysteine metabolic process(GO:0050666) |
| 1.0 | 1.9 | GO:0003402 | planar cell polarity pathway involved in axis elongation(GO:0003402) |
| 1.0 | 4.8 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.9 | 9.5 | GO:0018022 | peptidyl-lysine methylation(GO:0018022) |
| 0.9 | 4.5 | GO:0034204 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.9 | 10.3 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.8 | 0.8 | GO:0051935 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.8 | 10.0 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.8 | 5.7 | GO:0075044 | autophagy of host cells involved in interaction with symbiont(GO:0075044) autophagy involved in symbiotic interaction(GO:0075071) |
| 0.8 | 2.3 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.7 | 3.7 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.7 | 3.7 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.7 | 3.6 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.7 | 2.9 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.7 | 2.1 | GO:0052026 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.7 | 4.7 | GO:1904849 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.7 | 2.0 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.6 | 1.2 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.6 | 3.0 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.6 | 3.0 | GO:0001826 | inner cell mass cell differentiation(GO:0001826) |
| 0.6 | 4.6 | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.5 | 5.9 | GO:0006554 | lysine catabolic process(GO:0006554) |
| 0.5 | 5.2 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.5 | 2.6 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.5 | 1.5 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.5 | 2.5 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 0.5 | 3.0 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.5 | 3.0 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.5 | 1.5 | GO:0045229 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.5 | 1.5 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.5 | 2.9 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.5 | 2.9 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.5 | 1.4 | GO:1901594 | detection of temperature stimulus involved in thermoception(GO:0050960) response to capsazepine(GO:1901594) |
| 0.5 | 3.7 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.5 | 4.6 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.5 | 15.9 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.5 | 14.1 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.4 | 0.9 | GO:0048867 | stem cell fate determination(GO:0048867) |
| 0.4 | 2.6 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.4 | 3.4 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.4 | 5.0 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.4 | 19.6 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.4 | 2.1 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.4 | 2.8 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.4 | 2.4 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.4 | 0.8 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 0.4 | 1.9 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.4 | 6.1 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.4 | 2.2 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.4 | 2.5 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.4 | 9.3 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.4 | 3.9 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.3 | 1.0 | GO:0061009 | midbrain-hindbrain boundary morphogenesis(GO:0021555) trochlear nerve development(GO:0021558) regulation of timing of neuron differentiation(GO:0060164) common bile duct development(GO:0061009) |
| 0.3 | 13.5 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.3 | 2.4 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.3 | 2.1 | GO:0051344 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.3 | 8.4 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.3 | 2.0 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.3 | 2.3 | GO:0042791 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.3 | 1.9 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.3 | 3.5 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.3 | 5.6 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.3 | 4.2 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.3 | 3.5 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.3 | 3.2 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.3 | 12.4 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.3 | 3.3 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.3 | 1.4 | GO:0044026 | DNA hypermethylation(GO:0044026) |
| 0.2 | 5.1 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.2 | 2.0 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.2 | 3.3 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.2 | 1.2 | GO:0086053 | SA node cell to atrial cardiac muscle cell communication by electrical coupling(GO:0086021) AV node cell to bundle of His cell communication by electrical coupling(GO:0086053) |
| 0.2 | 2.0 | GO:0051963 | regulation of synapse assembly(GO:0051963) positive regulation of synapse assembly(GO:0051965) |
| 0.2 | 5.7 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.2 | 1.6 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) |
| 0.2 | 0.9 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 0.2 | 0.6 | GO:0051710 | regulation of cytolysis in other organism(GO:0051710) |
| 0.2 | 10.2 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.2 | 0.6 | GO:1900371 | regulation of cyclic nucleotide metabolic process(GO:0030799) positive regulation of cyclic nucleotide metabolic process(GO:0030801) regulation of cyclic nucleotide biosynthetic process(GO:0030802) positive regulation of cyclic nucleotide biosynthetic process(GO:0030804) regulation of nucleotide biosynthetic process(GO:0030808) positive regulation of nucleotide biosynthetic process(GO:0030810) regulation of cGMP metabolic process(GO:0030823) positive regulation of cGMP metabolic process(GO:0030825) regulation of cGMP biosynthetic process(GO:0030826) positive regulation of cGMP biosynthetic process(GO:0030828) regulation of purine nucleotide biosynthetic process(GO:1900371) positive regulation of purine nucleotide biosynthetic process(GO:1900373) |
| 0.2 | 23.5 | GO:0005977 | glycogen metabolic process(GO:0005977) |
| 0.2 | 12.1 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.2 | 7.6 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.2 | 1.4 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.2 | 1.6 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.2 | 4.3 | GO:0016246 | RNA interference(GO:0016246) |
| 0.2 | 1.9 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.2 | 1.1 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.2 | 1.5 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.2 | 10.5 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.2 | 4.1 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.2 | 1.1 | GO:0043045 | DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.2 | 3.9 | GO:0015874 | norepinephrine transport(GO:0015874) |
| 0.2 | 1.5 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.2 | 1.4 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.2 | 2.8 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.2 | 1.1 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.2 | 0.8 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.2 | 2.7 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.2 | 1.5 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 0.4 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) negative regulation of retinal ganglion cell axon guidance(GO:0090260) |
| 0.1 | 0.9 | GO:0003413 | chondrocyte differentiation involved in endochondral bone morphogenesis(GO:0003413) |
| 0.1 | 2.1 | GO:0006705 | androgen biosynthetic process(GO:0006702) mineralocorticoid biosynthetic process(GO:0006705) mineralocorticoid metabolic process(GO:0008212) |
| 0.1 | 0.9 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 0.9 | GO:0010001 | glial cell differentiation(GO:0010001) |
| 0.1 | 1.2 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.1 | 0.5 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 8.4 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 0.9 | GO:1902659 | regulation of glucose mediated signaling pathway(GO:1902659) |
| 0.1 | 0.8 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.1 | 7.2 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.1 | 1.4 | GO:0097190 | apoptotic signaling pathway(GO:0097190) |
| 0.1 | 0.8 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.1 | 2.4 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 4.7 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 2.1 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.1 | 1.7 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.1 | 2.2 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.1 | 2.2 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 3.3 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.1 | 0.5 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.1 | 2.2 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.1 | 13.5 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.1 | 1.6 | GO:0043486 | histone exchange(GO:0043486) |
| 0.1 | 5.8 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.1 | 0.5 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.1 | 3.0 | GO:0098534 | centriole replication(GO:0007099) centriole assembly(GO:0098534) |
| 0.1 | 2.8 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.1 | 1.6 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.1 | 1.3 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.1 | 0.8 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.1 | 2.4 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.1 | 0.7 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.1 | 0.6 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.1 | 1.3 | GO:0010876 | lipid localization(GO:0010876) |
| 0.1 | 1.3 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 0.1 | 5.7 | GO:0044764 | multi-organism cellular process(GO:0044764) |
| 0.0 | 4.4 | GO:0031295 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 2.4 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.0 | GO:2000969 | positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.0 | 0.8 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.2 | GO:0034344 | detection of virus(GO:0009597) type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.0 | 1.2 | GO:1902593 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.0 | 3.5 | GO:0006970 | response to osmotic stress(GO:0006970) |
| 0.0 | 0.5 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 2.8 | GO:0072676 | lymphocyte migration(GO:0072676) |
| 0.0 | 1.8 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.1 | GO:0086100 | endothelin receptor signaling pathway(GO:0086100) |
| 0.0 | 1.2 | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0035774) |
| 0.0 | 3.9 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 1.7 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.0 | 1.1 | GO:0033198 | response to ATP(GO:0033198) |
| 0.0 | 0.7 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 2.8 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.4 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.8 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 3.5 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 2.3 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 1.3 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 1.7 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.0 | 0.2 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 1.6 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 2.9 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 4.9 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 5.4 | GO:0090305 | nucleic acid phosphodiester bond hydrolysis(GO:0090305) |
| 0.0 | 3.1 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.6 | GO:0043403 | skeletal muscle tissue regeneration(GO:0043403) |
| 0.0 | 1.1 | GO:0060135 | maternal process involved in female pregnancy(GO:0060135) |
| 0.0 | 3.9 | GO:0072659 | protein localization to plasma membrane(GO:0072659) |
| 0.0 | 0.8 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 1.7 | GO:0006959 | humoral immune response(GO:0006959) |
| 0.0 | 3.2 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.9 | GO:0060349 | bone morphogenesis(GO:0060349) |
| 0.0 | 1.5 | GO:0000910 | cytokinesis(GO:0000910) |
| 0.0 | 2.7 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.0 | 1.3 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 2.4 | GO:0007254 | JNK cascade(GO:0007254) |
| 0.0 | 0.4 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.2 | GO:0010830 | regulation of myotube differentiation(GO:0010830) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 6.7 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 1.5 | 10.2 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 1.3 | 3.9 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 1.3 | 31.9 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 1.2 | 4.7 | GO:1990745 | EARP complex(GO:1990745) |
| 1.2 | 1.2 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.9 | 8.2 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.8 | 7.4 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.8 | 1.5 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.7 | 2.8 | GO:0070695 | FHF complex(GO:0070695) |
| 0.6 | 12.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.6 | 6.7 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.6 | 3.0 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.6 | 6.6 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.6 | 11.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.5 | 2.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.5 | 1.8 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.5 | 4.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.4 | 2.6 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.4 | 1.2 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.4 | 3.5 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.4 | 3.0 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.4 | 20.6 | GO:0016235 | aggresome(GO:0016235) |
| 0.4 | 2.2 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.3 | 2.3 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.3 | 1.9 | GO:1990130 | EGO complex(GO:0034448) Iml1 complex(GO:1990130) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.3 | 1.2 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.3 | 0.9 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.3 | 0.9 | GO:0002079 | inner acrosomal membrane(GO:0002079) outer acrosomal membrane(GO:0002081) |
| 0.3 | 2.4 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.3 | 1.5 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.2 | 2.5 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 0.2 | 5.4 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.2 | 1.9 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.2 | 2.2 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.2 | 2.6 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.2 | 1.6 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.2 | 2.0 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.2 | 2.9 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.2 | 11.0 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.2 | 8.4 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.2 | 4.2 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.2 | 2.6 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.2 | 2.8 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 6.9 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 22.0 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 10.0 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 6.4 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 3.9 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.1 | 23.3 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 1.4 | GO:0000243 | commitment complex(GO:0000243) |
| 0.1 | 5.9 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 2.2 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 0.6 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.6 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 0.1 | 2.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 0.8 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 1.2 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 2.4 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.1 | 2.8 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 8.6 | GO:0019866 | mitochondrial inner membrane(GO:0005743) organelle inner membrane(GO:0019866) |
| 0.1 | 0.2 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 1.1 | GO:0001741 | XY body(GO:0001741) |
| 0.1 | 1.5 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 4.9 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 2.8 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 3.0 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 1.4 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 2.5 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.1 | 10.1 | GO:0000922 | spindle pole(GO:0000922) |
| 0.1 | 4.2 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 1.3 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 2.8 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 1.9 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 4.7 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 4.2 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 3.4 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 4.1 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 3.5 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 3.5 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 2.9 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 1.7 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 2.8 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 2.0 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.7 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.8 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 43.4 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 1.1 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 1.7 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 39.5 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.3 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 9.6 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 2.7 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 4.6 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 1.3 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 10.3 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 2.2 | 6.7 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 2.0 | 10.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 2.0 | 16.3 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 2.0 | 12.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 2.0 | 5.9 | GO:0047150 | betaine-homocysteine S-methyltransferase activity(GO:0047150) |
| 1.8 | 7.4 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 1.6 | 13.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 1.6 | 9.5 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 1.5 | 5.9 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 1.4 | 4.2 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 1.3 | 3.8 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 1.3 | 31.9 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 1.0 | 4.9 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.8 | 2.5 | GO:0004488 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.8 | 11.1 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.8 | 2.4 | GO:0005330 | dopamine:sodium symporter activity(GO:0005330) |
| 0.8 | 2.3 | GO:0000995 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) transcription factor activity, core RNA polymerase III binding(GO:0000995) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) |
| 0.7 | 3.7 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.7 | 2.9 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.7 | 2.1 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.7 | 2.1 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) steroid delta-isomerase activity(GO:0004769) |
| 0.7 | 13.2 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.7 | 2.0 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.7 | 0.7 | GO:0001007 | transcription factor activity, RNA polymerase III transcription factor binding(GO:0001007) |
| 0.6 | 1.9 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.6 | 1.9 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.6 | 2.4 | GO:0047718 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) androsterone dehydrogenase activity(GO:0047023) indanol dehydrogenase activity(GO:0047718) |
| 0.6 | 3.6 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.6 | 2.8 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.6 | 1.7 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.5 | 4.2 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.5 | 2.6 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.5 | 1.5 | GO:0005333 | norepinephrine transmembrane transporter activity(GO:0005333) |
| 0.5 | 1.5 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.5 | 2.9 | GO:0032564 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.5 | 1.4 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.5 | 6.4 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.4 | 6.7 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.4 | 7.9 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.4 | 4.6 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.4 | 7.9 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.4 | 11.5 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.4 | 3.2 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.4 | 2.4 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.4 | 6.6 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.4 | 3.5 | GO:0030882 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.3 | 1.4 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.3 | 5.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.3 | 1.2 | GO:0043559 | insulin binding(GO:0043559) |
| 0.3 | 1.5 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.3 | 2.4 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.3 | 13.8 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.3 | 2.0 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.3 | 2.9 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.3 | 0.8 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.3 | 1.4 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.3 | 2.7 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.2 | 3.7 | GO:0016918 | retinal binding(GO:0016918) |
| 0.2 | 1.5 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.2 | 5.7 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.2 | 2.0 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.2 | 2.2 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.2 | 6.0 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.2 | 7.0 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 1.2 | GO:0086020 | gap junction channel activity involved in SA node cell-atrial cardiac muscle cell electrical coupling(GO:0086020) gap junction channel activity involved in AV node cell-bundle of His cell electrical coupling(GO:0086077) |
| 0.2 | 2.0 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.2 | 3.2 | GO:0016594 | glycine binding(GO:0016594) |
| 0.2 | 3.7 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.2 | 3.7 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.2 | 3.5 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.2 | 2.8 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.2 | 0.6 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.2 | 7.6 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.2 | 1.5 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.2 | 2.1 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.2 | 22.5 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.2 | 2.6 | GO:0031432 | titin binding(GO:0031432) |
| 0.2 | 4.5 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.2 | 2.6 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.2 | 0.9 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.2 | 1.5 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.2 | 1.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.2 | 17.1 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.2 | 5.9 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.2 | 2.2 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.2 | 6.9 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.2 | 1.6 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.2 | 0.9 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) exodeoxyribonuclease III activity(GO:0008853) |
| 0.2 | 0.9 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.2 | 0.5 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.2 | 1.8 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.2 | 8.4 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.2 | 3.0 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.2 | 4.1 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 2.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 4.6 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 2.7 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.1 | 3.4 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 8.9 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 2.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.5 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 2.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 3.5 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.1 | 12.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 3.6 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 2.8 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.1 | 2.5 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 2.8 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 0.5 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 0.8 | GO:0019863 | IgE binding(GO:0019863) |
| 0.1 | 0.6 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 5.2 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 2.0 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 1.9 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 0.8 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 1.7 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 2.0 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 20.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 1.3 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 2.2 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 2.8 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 1.0 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.1 | 12.2 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.1 | 3.0 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 2.4 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 0.6 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 1.1 | GO:0015929 | hexosaminidase activity(GO:0015929) |
| 0.0 | 2.9 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 1.1 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.5 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.4 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 3.7 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.0 | 0.8 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 2.2 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 1.0 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.1 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 4.1 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 3.3 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.8 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.3 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 1.3 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 1.9 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.7 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.4 | GO:0008392 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.9 | GO:0001190 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 1.3 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 2.1 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 2.3 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 0.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.3 | GO:0019956 | chemokine binding(GO:0019956) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 11.0 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.4 | 15.4 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.4 | 8.6 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.4 | 4.2 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.3 | 14.1 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.3 | 14.0 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.2 | 13.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.2 | 14.2 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 2.1 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 7.7 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 5.8 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 7.1 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 3.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 1.5 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 4.1 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 2.9 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 7.6 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 4.0 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 13.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 1.4 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.1 | 7.6 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 2.9 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 3.0 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 1.1 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 3.4 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 5.7 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 12.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.5 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 2.1 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 1.5 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 1.0 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.2 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.4 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.8 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 1.9 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 7.4 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.8 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 1.1 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.5 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 32.7 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.5 | 13.2 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.4 | 8.6 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.4 | 13.9 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.4 | 13.2 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.4 | 11.0 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.4 | 5.7 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.3 | 7.5 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.3 | 13.2 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.3 | 15.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.3 | 4.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.3 | 3.0 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.3 | 19.0 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.3 | 5.7 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.2 | 5.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.2 | 6.0 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.2 | 10.0 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.2 | 2.9 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.2 | 5.0 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.2 | 8.0 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.2 | 3.3 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.2 | 1.5 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.2 | 10.8 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.2 | 3.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.2 | 2.4 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.2 | 7.1 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.2 | 2.8 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.2 | 2.1 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 5.7 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 2.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 2.9 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 4.1 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 1.9 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 2.7 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 1.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 2.6 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 2.8 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 2.9 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 3.0 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 12.2 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 17.4 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 2.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 2.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 2.4 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.1 | 2.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 1.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 3.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 3.2 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.9 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.5 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 2.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.9 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 2.0 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 1.4 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.9 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 10.9 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 1.9 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.5 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.4 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.7 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.6 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |