Project
averaged GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
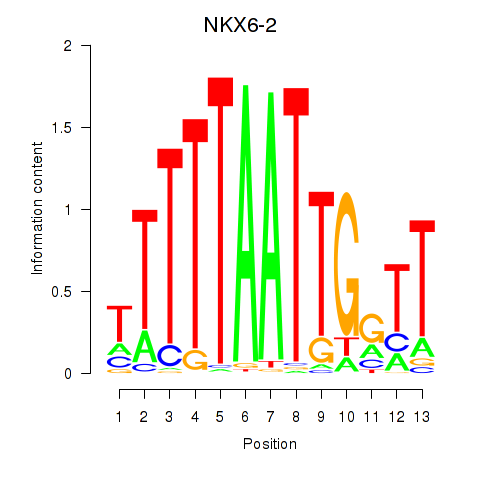
Results for NKX6-2
Z-value: 1.27
Transcription factors associated with NKX6-2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX6-2
|
ENSG00000148826.6 | NK6 homeobox 2 |
Activity profile of NKX6-2 motif
Sorted Z-values of NKX6-2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_60097524 | 35.81 |
ENST00000342503.4
|
RTN1
|
reticulon 1 |
| chr17_-_42992856 | 33.65 |
ENST00000588316.1
ENST00000435360.2 ENST00000586793.1 ENST00000588735.1 ENST00000588037.1 ENST00000592320.1 ENST00000253408.5 |
GFAP
|
glial fibrillary acidic protein |
| chr3_+_39509163 | 31.12 |
ENST00000436143.2
ENST00000441980.2 ENST00000311042.6 |
MOBP
|
myelin-associated oligodendrocyte basic protein |
| chr4_+_41258786 | 29.25 |
ENST00000503431.1
ENST00000284440.4 ENST00000508768.1 ENST00000512788.1 |
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) |
| chr3_+_39509070 | 29.22 |
ENST00000354668.4
ENST00000428261.1 ENST00000420739.1 ENST00000415443.1 ENST00000447324.1 ENST00000383754.3 |
MOBP
|
myelin-associated oligodendrocyte basic protein |
| chr14_-_60097297 | 28.58 |
ENST00000395090.1
|
RTN1
|
reticulon 1 |
| chr3_-_98241358 | 21.58 |
ENST00000503004.1
ENST00000506575.1 ENST00000513452.1 ENST00000515620.1 |
CLDND1
|
claudin domain containing 1 |
| chr2_-_175712270 | 20.58 |
ENST00000295497.7
ENST00000444394.1 |
CHN1
|
chimerin 1 |
| chr10_-_73848086 | 17.05 |
ENST00000536168.1
|
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr4_+_88896819 | 16.05 |
ENST00000237623.7
ENST00000395080.3 ENST00000508233.1 ENST00000360804.4 |
SPP1
|
secreted phosphoprotein 1 |
| chr22_+_29138013 | 13.98 |
ENST00000216027.3
ENST00000398941.2 |
HSCB
|
HscB mitochondrial iron-sulfur cluster co-chaperone |
| chr9_-_93405352 | 13.55 |
ENST00000375765.3
|
DIRAS2
|
DIRAS family, GTP-binding RAS-like 2 |
| chr7_-_37024665 | 12.83 |
ENST00000396040.2
|
ELMO1
|
engulfment and cell motility 1 |
| chr10_-_73848531 | 12.47 |
ENST00000373109.2
|
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr7_-_137028534 | 11.78 |
ENST00000348225.2
|
PTN
|
pleiotrophin |
| chr2_+_233527443 | 11.66 |
ENST00000410095.1
|
EFHD1
|
EF-hand domain family, member D1 |
| chr14_-_27066960 | 11.25 |
ENST00000539517.2
|
NOVA1
|
neuro-oncological ventral antigen 1 |
| chr7_-_137028498 | 10.76 |
ENST00000393083.2
|
PTN
|
pleiotrophin |
| chr4_-_87374283 | 10.74 |
ENST00000361569.2
|
MAPK10
|
mitogen-activated protein kinase 10 |
| chr5_+_161275320 | 10.61 |
ENST00000437025.2
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr9_+_82186872 | 10.46 |
ENST00000376544.3
ENST00000376520.4 |
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr19_+_52772821 | 9.93 |
ENST00000439461.1
|
ZNF766
|
zinc finger protein 766 |
| chr17_+_57232690 | 9.77 |
ENST00000262293.4
|
PRR11
|
proline rich 11 |
| chr7_+_77325738 | 9.74 |
ENST00000334955.8
|
RSBN1L
|
round spermatid basic protein 1-like |
| chr12_-_6233828 | 9.56 |
ENST00000572068.1
ENST00000261405.5 |
VWF
|
von Willebrand factor |
| chr2_-_73520667 | 9.15 |
ENST00000545030.1
ENST00000436467.2 |
EGR4
|
early growth response 4 |
| chrX_+_135230712 | 8.87 |
ENST00000535737.1
|
FHL1
|
four and a half LIM domains 1 |
| chr14_-_98444386 | 8.74 |
ENST00000556462.1
ENST00000556138.1 |
C14orf64
|
chromosome 14 open reading frame 64 |
| chr18_-_52989217 | 8.60 |
ENST00000570287.2
|
TCF4
|
transcription factor 4 |
| chr19_+_52772832 | 8.53 |
ENST00000593703.1
ENST00000601711.1 ENST00000599581.1 |
ZNF766
|
zinc finger protein 766 |
| chr17_-_73663168 | 8.53 |
ENST00000578201.1
ENST00000423245.2 |
RECQL5
|
RecQ protein-like 5 |
| chr16_-_4852915 | 8.26 |
ENST00000322048.7
|
ROGDI
|
rogdi homolog (Drosophila) |
| chr10_-_71169031 | 8.15 |
ENST00000373307.1
|
TACR2
|
tachykinin receptor 2 |
| chr7_+_16828866 | 7.99 |
ENST00000597084.1
|
AC073333.1
|
Uncharacterized protein |
| chr1_-_226926864 | 7.62 |
ENST00000429204.1
ENST00000366784.1 |
ITPKB
|
inositol-trisphosphate 3-kinase B |
| chr17_+_72581057 | 7.61 |
ENST00000392620.1
|
C17orf77
|
chromosome 17 open reading frame 77 |
| chr5_+_161274940 | 7.49 |
ENST00000393943.4
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr2_+_68962014 | 7.38 |
ENST00000467265.1
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr12_+_32655048 | 7.35 |
ENST00000427716.2
ENST00000266482.3 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr1_-_149858227 | 7.25 |
ENST00000369155.2
|
HIST2H2BE
|
histone cluster 2, H2be |
| chr5_+_161274685 | 7.01 |
ENST00000428797.2
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr8_+_79428539 | 6.84 |
ENST00000352966.5
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha |
| chr1_+_229440129 | 6.37 |
ENST00000366688.3
|
SPHAR
|
S-phase response (cyclin related) |
| chr1_+_115397424 | 6.19 |
ENST00000369522.3
ENST00000455987.1 |
SYCP1
|
synaptonemal complex protein 1 |
| chr17_-_73663245 | 6.16 |
ENST00000584999.1
ENST00000317905.5 ENST00000420326.2 ENST00000340830.5 |
RECQL5
|
RecQ protein-like 5 |
| chr16_+_14802801 | 5.86 |
ENST00000526520.1
ENST00000531598.2 |
NPIPA3
|
nuclear pore complex interacting protein family, member A3 |
| chr19_+_12175504 | 5.84 |
ENST00000439326.3
|
ZNF844
|
zinc finger protein 844 |
| chr5_-_20575959 | 5.73 |
ENST00000507958.1
|
CDH18
|
cadherin 18, type 2 |
| chr5_+_67586465 | 5.71 |
ENST00000336483.5
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr5_-_58295712 | 5.69 |
ENST00000317118.8
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr6_+_31582961 | 5.64 |
ENST00000376059.3
ENST00000337917.7 |
AIF1
|
allograft inflammatory factor 1 |
| chr3_+_159557637 | 5.63 |
ENST00000445224.2
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr16_+_31128978 | 5.35 |
ENST00000448516.2
ENST00000219797.4 |
KAT8
|
K(lysine) acetyltransferase 8 |
| chr14_-_27066636 | 5.30 |
ENST00000267422.7
ENST00000344429.5 ENST00000574031.1 ENST00000465357.2 ENST00000547619.1 |
NOVA1
|
neuro-oncological ventral antigen 1 |
| chr16_+_14841923 | 5.25 |
ENST00000530328.1
ENST00000529166.1 |
NPIPA2
|
nuclear pore complex interacting protein family, member A2 |
| chr5_+_161495038 | 5.06 |
ENST00000393933.4
|
GABRG2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr18_+_76829385 | 4.98 |
ENST00000426216.2
ENST00000307671.7 ENST00000586672.1 ENST00000586722.1 |
ATP9B
|
ATPase, class II, type 9B |
| chr11_-_62521614 | 4.97 |
ENST00000527994.1
ENST00000394807.3 |
ZBTB3
|
zinc finger and BTB domain containing 3 |
| chr1_+_50459990 | 4.95 |
ENST00000448346.1
|
AL645730.2
|
AL645730.2 |
| chr2_-_217559517 | 4.66 |
ENST00000449583.1
|
IGFBP5
|
insulin-like growth factor binding protein 5 |
| chr18_+_76829441 | 4.65 |
ENST00000458297.2
|
ATP9B
|
ATPase, class II, type 9B |
| chr1_+_180601139 | 4.64 |
ENST00000367590.4
ENST00000367589.3 |
XPR1
|
xenotropic and polytropic retrovirus receptor 1 |
| chrX_+_10124977 | 4.63 |
ENST00000380833.4
|
CLCN4
|
chloride channel, voltage-sensitive 4 |
| chr5_-_55412774 | 4.59 |
ENST00000434982.2
|
ANKRD55
|
ankyrin repeat domain 55 |
| chr3_-_114790179 | 4.58 |
ENST00000462705.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr18_-_52989525 | 4.50 |
ENST00000457482.3
|
TCF4
|
transcription factor 4 |
| chr6_+_26402517 | 4.49 |
ENST00000414912.2
|
BTN3A1
|
butyrophilin, subfamily 3, member A1 |
| chr10_+_26505179 | 4.38 |
ENST00000376261.3
|
GAD2
|
glutamate decarboxylase 2 (pancreatic islets and brain, 65kDa) |
| chr9_-_23826298 | 4.38 |
ENST00000380117.1
|
ELAVL2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr19_+_54641444 | 4.35 |
ENST00000221232.5
ENST00000358389.3 |
CNOT3
|
CCR4-NOT transcription complex, subunit 3 |
| chrX_-_48901012 | 4.35 |
ENST00000315869.7
|
TFE3
|
transcription factor binding to IGHM enhancer 3 |
| chr7_-_22233442 | 4.28 |
ENST00000401957.2
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr4_+_55095264 | 4.25 |
ENST00000257290.5
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr15_-_49103235 | 3.96 |
ENST00000380950.2
|
CEP152
|
centrosomal protein 152kDa |
| chr18_+_32290218 | 3.82 |
ENST00000348997.5
ENST00000588949.1 ENST00000597599.1 |
DTNA
|
dystrobrevin, alpha |
| chr7_+_138943265 | 3.71 |
ENST00000483726.1
|
UBN2
|
ubinuclein 2 |
| chr8_+_104831472 | 3.67 |
ENST00000262231.10
ENST00000507740.1 |
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chrX_+_55744166 | 3.61 |
ENST00000374941.4
ENST00000414239.1 |
RRAGB
|
Ras-related GTP binding B |
| chr6_+_158733692 | 3.61 |
ENST00000367094.2
ENST00000367097.3 |
TULP4
|
tubby like protein 4 |
| chr18_+_13218769 | 3.56 |
ENST00000399848.3
ENST00000361205.4 |
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
| chr9_-_23825956 | 3.55 |
ENST00000397312.2
|
ELAVL2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr1_+_209878182 | 3.43 |
ENST00000367027.3
|
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1 |
| chr15_-_49103184 | 3.43 |
ENST00000399334.3
ENST00000325747.5 |
CEP152
|
centrosomal protein 152kDa |
| chr7_-_140482926 | 3.29 |
ENST00000496384.2
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B |
| chr2_+_218990727 | 3.22 |
ENST00000318507.2
ENST00000454148.1 |
CXCR2
|
chemokine (C-X-C motif) receptor 2 |
| chr2_+_210444142 | 3.17 |
ENST00000360351.4
ENST00000361559.4 |
MAP2
|
microtubule-associated protein 2 |
| chr2_-_74570520 | 3.13 |
ENST00000394019.2
ENST00000346834.4 ENST00000359484.4 ENST00000423644.1 ENST00000377634.4 ENST00000436454.1 |
SLC4A5
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 5 |
| chr1_-_49242553 | 3.07 |
ENST00000371833.3
|
BEND5
|
BEN domain containing 5 |
| chr2_-_228244013 | 3.05 |
ENST00000304568.3
|
TM4SF20
|
transmembrane 4 L six family member 20 |
| chr3_-_45838011 | 3.05 |
ENST00000358525.4
ENST00000413781.1 |
SLC6A20
|
solute carrier family 6 (proline IMINO transporter), member 20 |
| chrX_+_102840408 | 3.05 |
ENST00000468024.1
ENST00000472484.1 ENST00000415568.2 ENST00000490644.1 ENST00000459722.1 ENST00000472745.1 ENST00000494801.1 ENST00000434216.2 ENST00000425011.1 |
TCEAL4
|
transcription elongation factor A (SII)-like 4 |
| chr2_-_217560248 | 3.05 |
ENST00000233813.4
|
IGFBP5
|
insulin-like growth factor binding protein 5 |
| chr5_+_7373230 | 2.99 |
ENST00000500616.2
|
CTD-2296D1.5
|
CTD-2296D1.5 |
| chr19_+_15852203 | 2.78 |
ENST00000305892.1
|
OR10H3
|
olfactory receptor, family 10, subfamily H, member 3 |
| chrX_+_68835911 | 2.76 |
ENST00000525810.1
ENST00000527388.1 ENST00000374553.2 ENST00000374552.4 ENST00000338901.3 ENST00000524573.1 |
EDA
|
ectodysplasin A |
| chrX_+_55744228 | 2.67 |
ENST00000262850.7
|
RRAGB
|
Ras-related GTP binding B |
| chr2_+_202047843 | 2.61 |
ENST00000272879.5
ENST00000374650.3 ENST00000346817.5 ENST00000313728.7 ENST00000448480.1 |
CASP10
|
caspase 10, apoptosis-related cysteine peptidase |
| chr10_-_100995540 | 2.52 |
ENST00000370546.1
ENST00000404542.1 |
HPSE2
|
heparanase 2 |
| chr9_-_110540419 | 2.48 |
ENST00000398726.3
|
AL162389.1
|
Uncharacterized protein |
| chr21_-_36421535 | 2.38 |
ENST00000416754.1
ENST00000437180.1 ENST00000455571.1 |
RUNX1
|
runt-related transcription factor 1 |
| chr16_+_31271274 | 2.34 |
ENST00000287497.8
ENST00000544665.3 |
ITGAM
|
integrin, alpha M (complement component 3 receptor 3 subunit) |
| chr14_-_101295407 | 2.28 |
ENST00000596284.1
|
AL117190.2
|
AL117190.2 |
| chr3_-_45837959 | 2.25 |
ENST00000353278.4
ENST00000456124.2 |
SLC6A20
|
solute carrier family 6 (proline IMINO transporter), member 20 |
| chrX_+_82763265 | 2.24 |
ENST00000373200.2
|
POU3F4
|
POU class 3 homeobox 4 |
| chr3_+_178276488 | 2.22 |
ENST00000432997.1
ENST00000455865.1 |
KCNMB2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
| chr6_-_26199471 | 2.22 |
ENST00000341023.1
|
HIST1H2AD
|
histone cluster 1, H2ad |
| chr10_-_100995603 | 2.20 |
ENST00000370552.3
ENST00000370549.1 |
HPSE2
|
heparanase 2 |
| chr6_-_46703430 | 2.19 |
ENST00000537365.1
|
PLA2G7
|
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) |
| chr6_+_26402465 | 2.19 |
ENST00000476549.2
ENST00000289361.6 ENST00000450085.2 ENST00000425234.2 ENST00000427334.1 ENST00000506698.1 |
BTN3A1
|
butyrophilin, subfamily 3, member A1 |
| chr8_-_82395461 | 2.15 |
ENST00000256104.4
|
FABP4
|
fatty acid binding protein 4, adipocyte |
| chr21_-_36421626 | 2.12 |
ENST00000300305.3
|
RUNX1
|
runt-related transcription factor 1 |
| chr18_-_10701979 | 2.04 |
ENST00000538948.1
ENST00000285141.4 |
PIEZO2
|
piezo-type mechanosensitive ion channel component 2 |
| chr20_-_18477862 | 2.03 |
ENST00000337227.4
|
RBBP9
|
retinoblastoma binding protein 9 |
| chr17_-_40828969 | 1.96 |
ENST00000591022.1
ENST00000587627.1 ENST00000293349.6 |
PLEKHH3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr10_-_115904361 | 1.95 |
ENST00000428953.1
ENST00000543782.1 |
C10orf118
|
chromosome 10 open reading frame 118 |
| chr5_-_146258205 | 1.94 |
ENST00000394413.3
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta |
| chr1_+_70820451 | 1.92 |
ENST00000361764.4
ENST00000359875.5 ENST00000370940.5 ENST00000531950.1 ENST00000432224.1 |
HHLA3
|
HERV-H LTR-associating 3 |
| chr12_+_9980069 | 1.89 |
ENST00000354855.3
ENST00000324214.4 ENST00000279544.3 |
KLRF1
|
killer cell lectin-like receptor subfamily F, member 1 |
| chr10_+_32873190 | 1.80 |
ENST00000375025.4
|
C10orf68
|
Homo sapiens coiled-coil domain containing 7 (CCDC7), transcript variant 5, mRNA. |
| chr17_-_40829026 | 1.69 |
ENST00000412503.1
|
PLEKHH3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr18_+_61575200 | 1.66 |
ENST00000238508.3
|
SERPINB10
|
serpin peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr2_-_136873735 | 1.63 |
ENST00000409817.1
|
CXCR4
|
chemokine (C-X-C motif) receptor 4 |
| chr8_-_91095099 | 1.62 |
ENST00000265431.3
|
CALB1
|
calbindin 1, 28kDa |
| chr9_+_82187630 | 1.54 |
ENST00000265284.6
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr2_+_68961934 | 1.52 |
ENST00000409202.3
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr21_-_27462351 | 1.50 |
ENST00000448850.1
|
APP
|
amyloid beta (A4) precursor protein |
| chr2_+_68961905 | 1.48 |
ENST00000295381.3
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr22_-_40929812 | 1.35 |
ENST00000422851.1
|
MKL1
|
megakaryoblastic leukemia (translocation) 1 |
| chr1_-_200379129 | 1.30 |
ENST00000367353.1
|
ZNF281
|
zinc finger protein 281 |
| chr1_+_196621002 | 1.29 |
ENST00000367429.4
ENST00000439155.2 |
CFH
|
complement factor H |
| chr15_+_93443419 | 1.28 |
ENST00000557381.1
ENST00000420239.2 |
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chr14_-_99737565 | 1.28 |
ENST00000357195.3
|
BCL11B
|
B-cell CLL/lymphoma 11B (zinc finger protein) |
| chr8_+_24298531 | 1.17 |
ENST00000175238.6
|
ADAM7
|
ADAM metallopeptidase domain 7 |
| chr7_-_107880508 | 1.16 |
ENST00000425651.2
|
NRCAM
|
neuronal cell adhesion molecule |
| chr7_+_143657027 | 1.16 |
ENST00000392899.1
|
OR2F1
|
olfactory receptor, family 2, subfamily F, member 1 (gene/pseudogene) |
| chr6_-_26199499 | 1.15 |
ENST00000377831.5
|
HIST1H3D
|
histone cluster 1, H3d |
| chr8_+_24298597 | 1.15 |
ENST00000380789.1
|
ADAM7
|
ADAM metallopeptidase domain 7 |
| chr3_+_130650738 | 1.13 |
ENST00000504612.1
|
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr4_+_100495864 | 1.10 |
ENST00000265517.5
ENST00000422897.2 |
MTTP
|
microsomal triglyceride transfer protein |
| chr20_+_42136308 | 0.95 |
ENST00000434666.1
ENST00000427442.2 ENST00000439769.1 ENST00000418998.1 |
L3MBTL1
|
l(3)mbt-like 1 (Drosophila) |
| chr10_+_48189612 | 0.94 |
ENST00000453919.1
|
AGAP9
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 9 |
| chr12_+_25205568 | 0.90 |
ENST00000548766.1
ENST00000556887.1 |
LRMP
|
lymphoid-restricted membrane protein |
| chr1_-_190446759 | 0.87 |
ENST00000367462.3
|
BRINP3
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr22_-_39268192 | 0.83 |
ENST00000216083.6
|
CBX6
|
chromobox homolog 6 |
| chr5_+_31193847 | 0.82 |
ENST00000514738.1
ENST00000265071.2 |
CDH6
|
cadherin 6, type 2, K-cadherin (fetal kidney) |
| chr13_-_33760216 | 0.71 |
ENST00000255486.4
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13 |
| chr19_+_4402659 | 0.70 |
ENST00000301280.5
ENST00000585854.1 |
CHAF1A
|
chromatin assembly factor 1, subunit A (p150) |
| chr5_-_111091948 | 0.65 |
ENST00000447165.2
|
NREP
|
neuronal regeneration related protein |
| chr1_+_32379174 | 0.63 |
ENST00000391369.1
|
AL136115.1
|
HCG2032337; PRO1848; Uncharacterized protein |
| chr15_-_89764929 | 0.59 |
ENST00000268125.5
|
RLBP1
|
retinaldehyde binding protein 1 |
| chr14_+_72399833 | 0.47 |
ENST00000553530.1
ENST00000556437.1 |
RGS6
|
regulator of G-protein signaling 6 |
| chr4_-_110723194 | 0.46 |
ENST00000394635.3
|
CFI
|
complement factor I |
| chr6_-_26043885 | 0.44 |
ENST00000357905.2
|
HIST1H2BB
|
histone cluster 1, H2bb |
| chr4_+_169418255 | 0.41 |
ENST00000505667.1
ENST00000511948.1 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr1_-_150849174 | 0.39 |
ENST00000515192.1
|
ARNT
|
aryl hydrocarbon receptor nuclear translocator |
| chr2_-_208030647 | 0.32 |
ENST00000309446.6
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr13_+_97928395 | 0.32 |
ENST00000445661.2
|
MBNL2
|
muscleblind-like splicing regulator 2 |
| chr6_+_25963020 | 0.30 |
ENST00000357085.3
|
TRIM38
|
tripartite motif containing 38 |
| chr21_-_36421401 | 0.26 |
ENST00000486278.2
|
RUNX1
|
runt-related transcription factor 1 |
| chr22_+_24198890 | 0.24 |
ENST00000345044.6
|
SLC2A11
|
solute carrier family 2 (facilitated glucose transporter), member 11 |
| chr6_+_41021027 | 0.21 |
ENST00000244669.2
|
APOBEC2
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 2 |
| chr19_+_852291 | 0.17 |
ENST00000263621.1
|
ELANE
|
elastase, neutrophil expressed |
| chr22_-_39268308 | 0.17 |
ENST00000407418.3
|
CBX6
|
chromobox homolog 6 |
| chr1_+_174933899 | 0.16 |
ENST00000367688.3
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr16_+_28565230 | 0.16 |
ENST00000317058.3
|
CCDC101
|
coiled-coil domain containing 101 |
| chr2_+_169757750 | 0.02 |
ENST00000375363.3
ENST00000429379.2 ENST00000421979.1 |
G6PC2
|
glucose-6-phosphatase, catalytic, 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX6-2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.8 | 29.3 | GO:0007412 | axon target recognition(GO:0007412) |
| 7.5 | 22.5 | GO:1904395 | retinal rod cell differentiation(GO:0060221) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) negative regulation of neuromuscular junction development(GO:1904397) |
| 5.3 | 16.0 | GO:0035937 | estrogen secretion(GO:0035937) estradiol secretion(GO:0035938) regulation of estrogen secretion(GO:2000861) regulation of estradiol secretion(GO:2000864) |
| 4.9 | 14.7 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 4.0 | 60.3 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 2.7 | 8.2 | GO:0014057 | positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
| 2.6 | 7.7 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 2.3 | 14.0 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 1.6 | 30.6 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 1.5 | 30.2 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 1.4 | 4.2 | GO:0072277 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 1.4 | 29.5 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 1.3 | 10.7 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 1.2 | 7.4 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 1.1 | 5.6 | GO:0014738 | regulation of muscle hyperplasia(GO:0014738) |
| 1.1 | 7.6 | GO:0033029 | regulation of neutrophil apoptotic process(GO:0033029) common myeloid progenitor cell proliferation(GO:0035726) |
| 1.1 | 3.2 | GO:0098759 | response to interleukin-8(GO:0098758) cellular response to interleukin-8(GO:0098759) |
| 1.0 | 6.3 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.8 | 5.3 | GO:0015824 | proline transport(GO:0015824) |
| 0.7 | 4.4 | GO:0006540 | glutamate decarboxylation to succinate(GO:0006540) |
| 0.7 | 6.2 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.7 | 4.6 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.7 | 3.3 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.6 | 5.7 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.6 | 3.1 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.6 | 20.6 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.5 | 2.8 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.4 | 2.2 | GO:0034441 | plasma lipoprotein particle oxidation(GO:0034441) |
| 0.4 | 6.8 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.4 | 9.6 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.4 | 4.8 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.4 | 7.3 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.4 | 9.8 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.4 | 5.7 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) |
| 0.4 | 5.4 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.3 | 12.8 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.3 | 9.6 | GO:0045332 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.3 | 1.3 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 0.3 | 1.5 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.3 | 3.6 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.3 | 6.7 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.2 | 1.6 | GO:0044800 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.2 | 11.9 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.2 | 1.9 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.2 | 1.6 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 0.2 | 4.3 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.2 | 1.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.2 | 1.4 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.2 | 1.1 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.2 | 2.2 | GO:2001054 | negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.2 | 2.3 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.2 | 8.9 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.2 | 3.4 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.1 | 0.7 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.1 | 2.1 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.1 | 2.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 1.1 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.1 | 8.3 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.1 | 4.6 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.1 | 3.8 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.1 | 2.0 | GO:0050974 | detection of mechanical stimulus involved in sensory perception(GO:0050974) |
| 0.1 | 4.6 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 3.6 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.1 | 3.9 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 4.3 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.1 | 4.6 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.1 | 24.8 | GO:0000398 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.1 | 0.2 | GO:1903238 | positive regulation of leukocyte tethering or rolling(GO:1903238) |
| 0.1 | 1.3 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 3.2 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 1.0 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 0.6 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 6.5 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 3.1 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.0 | 0.3 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 | 0.2 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 10.4 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.0 | 4.7 | GO:0043406 | positive regulation of MAP kinase activity(GO:0043406) |
| 0.0 | 0.9 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) |
| 0.0 | 2.6 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.0 | 12.0 | GO:0031175 | neuron projection development(GO:0031175) |
| 0.0 | 3.9 | GO:0007264 | small GTPase mediated signal transduction(GO:0007264) |
| 0.0 | 1.3 | GO:0032508 | DNA duplex unwinding(GO:0032508) |
| 0.0 | 7.5 | GO:0045666 | positive regulation of neuron differentiation(GO:0045666) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.4 | GO:0098536 | deuterosome(GO:0098536) |
| 1.4 | 5.7 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 1.2 | 6.2 | GO:0000801 | central element(GO:0000801) |
| 1.1 | 12.8 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 1.0 | 6.3 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.9 | 30.2 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.9 | 7.7 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.8 | 5.4 | GO:0072487 | MSL complex(GO:0072487) |
| 0.6 | 29.3 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.5 | 3.2 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.5 | 54.3 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.5 | 2.3 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.5 | 5.6 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.4 | 4.3 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.4 | 64.4 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.4 | 4.4 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.3 | 5.6 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.2 | 19.6 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.2 | 0.7 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 2.6 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.1 | 16.4 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 5.7 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 2.8 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 1.2 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 3.2 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 2.2 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.1 | 33.8 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 3.7 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 7.5 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.1 | 8.2 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 13.0 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 1.9 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 3.0 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 4.6 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 2.9 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 4.3 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 5.8 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 1.6 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 9.4 | GO:0005743 | mitochondrial inner membrane(GO:0005743) organelle inner membrane(GO:0019866) |
| 0.0 | 3.6 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 8.4 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 1.7 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 2.2 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 7.3 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.9 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 1.0 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 10.3 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.6 | GO:0044297 | cell body(GO:0044297) |
| 0.0 | 1.3 | GO:0072562 | blood microparticle(GO:0072562) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.3 | 29.3 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 5.6 | 22.5 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 5.0 | 60.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 3.1 | 25.1 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 2.4 | 14.7 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 1.8 | 10.7 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 1.6 | 8.2 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 1.4 | 4.2 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 1.3 | 7.6 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 1.2 | 29.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 1.2 | 4.7 | GO:0030305 | heparanase activity(GO:0030305) |
| 1.2 | 4.6 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 1.1 | 3.4 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 1.1 | 3.2 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 1.0 | 13.1 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.7 | 7.7 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.6 | 4.4 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.6 | 5.7 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.5 | 2.6 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.4 | 5.4 | GO:0043996 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.4 | 20.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.4 | 14.0 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.4 | 5.1 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.4 | 9.6 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.3 | 6.8 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.3 | 2.0 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.3 | 4.3 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.3 | 0.9 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.3 | 5.7 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.3 | 9.6 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.3 | 2.2 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.3 | 1.6 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.3 | 5.3 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.2 | 15.9 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.2 | 2.3 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.2 | 3.2 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.2 | 4.6 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.2 | 1.6 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.2 | 3.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.2 | 30.5 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.2 | 1.5 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.2 | 2.8 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.1 | 0.6 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.1 | 2.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 11.9 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.1 | 3.3 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.1 | 1.2 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 0.7 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.1 | 24.5 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.1 | 2.2 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 12.8 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 3.6 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 1.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 7.4 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 1.4 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 3.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 4.8 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.1 | 1.3 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 2.8 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 0.9 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 1.1 | GO:0005548 | phospholipid transporter activity(GO:0005548) |
| 0.0 | 1.3 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 1.1 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 11.4 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 11.9 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.2 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 5.6 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 2.4 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 1.2 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 16.5 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 1.9 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 1.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.0 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 7.6 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.6 | 19.1 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.5 | 29.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.5 | 29.3 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.5 | 33.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.5 | 28.9 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.4 | 16.0 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.4 | 3.2 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.2 | 4.2 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 20.1 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 2.6 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 17.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 2.2 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 2.5 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 4.8 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 1.5 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 1.6 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 3.2 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.9 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 8.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 3.3 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 1.4 | PID RHOA PATHWAY | RhoA signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 30.2 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.6 | 9.6 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.5 | 10.7 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.5 | 33.6 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.3 | 1.6 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.3 | 5.7 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.3 | 7.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.3 | 12.8 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.2 | 12.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.2 | 5.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.2 | 39.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.2 | 13.1 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.2 | 10.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.2 | 11.9 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.2 | 3.3 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.2 | 6.2 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.2 | 2.6 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.2 | 18.4 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 2.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 4.4 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 3.3 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 4.3 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 2.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 5.7 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 3.0 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 1.5 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 3.9 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 1.8 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 1.1 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 2.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 3.2 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 4.2 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 2.3 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 1.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 4.2 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 3.1 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |