Project
averaged GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
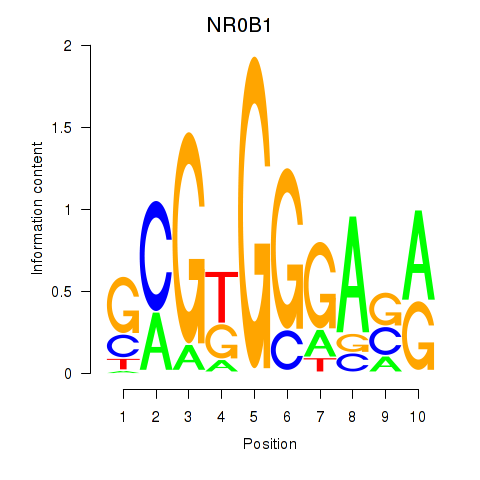
Results for NR0B1
Z-value: 1.03
Transcription factors associated with NR0B1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR0B1
|
ENSG00000169297.6 | nuclear receptor subfamily 0 group B member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR0B1 | hg19_v2_chrX_-_30327495_30327509, hg19_v2_chrX_-_30326445_30326605 | 0.06 | 3.6e-01 | Click! |
Activity profile of NR0B1 motif
Sorted Z-values of NR0B1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_56954867 | 10.33 |
ENST00000370708.4
ENST00000370702.1 |
ZNF451
|
zinc finger protein 451 |
| chr8_-_101734170 | 9.06 |
ENST00000522387.1
ENST00000518196.1 |
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr8_-_101734308 | 8.80 |
ENST00000519004.1
ENST00000519363.1 ENST00000520142.1 |
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr19_-_2050852 | 8.53 |
ENST00000541165.1
ENST00000591601.1 |
MKNK2
|
MAP kinase interacting serine/threonine kinase 2 |
| chr2_+_30454390 | 7.64 |
ENST00000395323.3
ENST00000406087.1 ENST00000404397.1 |
LBH
|
limb bud and heart development |
| chr20_+_35807449 | 7.59 |
ENST00000237530.6
|
RPN2
|
ribophorin II |
| chr20_+_35807512 | 7.49 |
ENST00000373622.5
|
RPN2
|
ribophorin II |
| chr13_-_52027134 | 7.16 |
ENST00000311234.4
ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6
|
integrator complex subunit 6 |
| chr2_+_105471969 | 6.76 |
ENST00000361360.2
|
POU3F3
|
POU class 3 homeobox 3 |
| chr1_+_211433275 | 6.69 |
ENST00000367005.4
|
RCOR3
|
REST corepressor 3 |
| chr9_+_128509624 | 6.58 |
ENST00000342287.5
ENST00000373487.4 |
PBX3
|
pre-B-cell leukemia homeobox 3 |
| chr13_+_25670268 | 6.36 |
ENST00000281589.3
|
PABPC3
|
poly(A) binding protein, cytoplasmic 3 |
| chr2_-_60780607 | 6.16 |
ENST00000537768.1
ENST00000335712.6 ENST00000356842.4 |
BCL11A
|
B-cell CLL/lymphoma 11A (zinc finger protein) |
| chr5_-_172198190 | 6.13 |
ENST00000239223.3
|
DUSP1
|
dual specificity phosphatase 1 |
| chr19_-_2051223 | 6.08 |
ENST00000309340.7
ENST00000589534.1 ENST00000250896.3 ENST00000589509.1 |
MKNK2
|
MAP kinase interacting serine/threonine kinase 2 |
| chr9_-_110251836 | 5.68 |
ENST00000374672.4
|
KLF4
|
Kruppel-like factor 4 (gut) |
| chr2_+_232573208 | 5.28 |
ENST00000409115.3
|
PTMA
|
prothymosin, alpha |
| chr14_+_57857262 | 4.83 |
ENST00000555166.1
ENST00000556492.1 ENST00000554703.1 |
NAA30
|
N(alpha)-acetyltransferase 30, NatC catalytic subunit |
| chr5_-_180237445 | 4.79 |
ENST00000393340.3
|
MGAT1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr15_+_74422585 | 4.71 |
ENST00000561740.1
ENST00000435464.1 ENST00000453268.2 |
ISLR2
|
immunoglobulin superfamily containing leucine-rich repeat 2 |
| chr7_+_26438187 | 4.53 |
ENST00000439120.1
ENST00000430548.1 ENST00000421862.1 ENST00000449537.1 ENST00000420774.1 ENST00000418758.2 |
AC004540.5
|
AC004540.5 |
| chr18_-_29522989 | 4.47 |
ENST00000582539.1
ENST00000283351.4 ENST00000582513.1 |
TRAPPC8
|
trafficking protein particle complex 8 |
| chr11_-_64512273 | 4.43 |
ENST00000377497.3
ENST00000377487.1 ENST00000430645.1 |
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr12_+_50355647 | 4.41 |
ENST00000293599.6
|
AQP5
|
aquaporin 5 |
| chr11_-_64512469 | 4.32 |
ENST00000377485.1
|
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr1_+_43148625 | 4.20 |
ENST00000436427.1
|
YBX1
|
Y box binding protein 1 |
| chr3_+_133292851 | 4.18 |
ENST00000503932.1
|
CDV3
|
CDV3 homolog (mouse) |
| chr1_+_226250379 | 4.18 |
ENST00000366815.3
ENST00000366814.3 |
H3F3A
|
H3 histone, family 3A |
| chr6_+_7541808 | 4.08 |
ENST00000379802.3
|
DSP
|
desmoplakin |
| chr6_+_7541845 | 4.04 |
ENST00000418664.2
|
DSP
|
desmoplakin |
| chr10_+_70715884 | 3.93 |
ENST00000354185.4
|
DDX21
|
DEAD (Asp-Glu-Ala-Asp) box helicase 21 |
| chr1_+_211432593 | 3.91 |
ENST00000367006.4
|
RCOR3
|
REST corepressor 3 |
| chr4_-_151936865 | 3.90 |
ENST00000535741.1
|
LRBA
|
LPS-responsive vesicle trafficking, beach and anchor containing |
| chr8_+_125486939 | 3.87 |
ENST00000303545.3
|
RNF139
|
ring finger protein 139 |
| chr8_-_125486755 | 3.72 |
ENST00000499418.2
ENST00000530778.1 |
RNF139-AS1
|
RNF139 antisense RNA 1 (head to head) |
| chr20_+_46130601 | 3.70 |
ENST00000341724.6
|
NCOA3
|
nuclear receptor coactivator 3 |
| chr17_-_42277203 | 3.52 |
ENST00000587097.1
|
ATXN7L3
|
ataxin 7-like 3 |
| chr17_+_76165213 | 3.38 |
ENST00000590201.1
|
SYNGR2
|
synaptogyrin 2 |
| chr8_-_144679532 | 3.34 |
ENST00000534380.1
ENST00000533494.1 ENST00000531218.1 ENST00000526340.1 ENST00000533204.1 ENST00000532400.1 ENST00000529516.1 ENST00000534377.1 ENST00000531621.1 ENST00000530191.1 ENST00000524900.1 ENST00000526838.1 ENST00000531931.1 ENST00000534475.1 ENST00000442189.2 ENST00000524624.1 ENST00000532596.1 ENST00000529832.1 ENST00000530306.1 ENST00000530545.1 ENST00000525261.1 ENST00000534804.1 ENST00000528303.1 ENST00000528610.1 |
EEF1D
|
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| chr22_-_39639021 | 3.32 |
ENST00000455790.1
|
PDGFB
|
platelet-derived growth factor beta polypeptide |
| chr5_+_121647386 | 3.20 |
ENST00000542191.1
ENST00000506272.1 ENST00000508681.1 ENST00000509154.2 |
SNCAIP
|
synuclein, alpha interacting protein |
| chr1_-_235491462 | 3.17 |
ENST00000418304.1
ENST00000264183.3 ENST00000349213.3 |
ARID4B
|
AT rich interactive domain 4B (RBP1-like) |
| chr1_-_40041925 | 3.13 |
ENST00000372862.3
|
PABPC4
|
poly(A) binding protein, cytoplasmic 4 (inducible form) |
| chr6_+_126070726 | 3.13 |
ENST00000368364.3
|
HEY2
|
hes-related family bHLH transcription factor with YRPW motif 2 |
| chr9_+_128509663 | 3.09 |
ENST00000373489.5
ENST00000373483.2 |
PBX3
|
pre-B-cell leukemia homeobox 3 |
| chr18_+_29672573 | 3.09 |
ENST00000578107.1
ENST00000257190.5 ENST00000580499.1 |
RNF138
|
ring finger protein 138, E3 ubiquitin protein ligase |
| chr1_+_235491714 | 3.08 |
ENST00000471812.1
ENST00000358966.2 ENST00000282841.5 ENST00000391855.2 |
GGPS1
|
geranylgeranyl diphosphate synthase 1 |
| chr20_-_18038521 | 3.06 |
ENST00000278780.6
|
OVOL2
|
ovo-like zinc finger 2 |
| chr11_-_119993979 | 3.06 |
ENST00000524816.3
ENST00000525327.1 |
TRIM29
|
tripartite motif containing 29 |
| chr4_+_38665810 | 3.05 |
ENST00000261438.5
ENST00000514033.1 |
KLF3
|
Kruppel-like factor 3 (basic) |
| chr17_-_36413133 | 3.04 |
ENST00000523089.1
ENST00000312412.4 ENST00000520237.1 |
RP11-1407O15.2
|
TBC1 domain family member 3 |
| chr2_+_232573222 | 2.99 |
ENST00000341369.7
ENST00000409683.1 |
PTMA
|
prothymosin, alpha |
| chr9_-_35691017 | 2.97 |
ENST00000378292.3
|
TPM2
|
tropomyosin 2 (beta) |
| chr19_-_49576198 | 2.90 |
ENST00000221444.1
|
KCNA7
|
potassium voltage-gated channel, shaker-related subfamily, member 7 |
| chr1_-_23810664 | 2.86 |
ENST00000336689.3
ENST00000437606.2 |
ASAP3
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 3 |
| chr11_-_2160180 | 2.86 |
ENST00000381406.4
|
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chr10_+_38299546 | 2.83 |
ENST00000374618.3
ENST00000432900.2 ENST00000458705.2 ENST00000469037.2 |
ZNF33A
|
zinc finger protein 33A |
| chr11_-_64511789 | 2.79 |
ENST00000419843.1
ENST00000394430.1 |
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr9_+_71320596 | 2.77 |
ENST00000265382.3
|
PIP5K1B
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta |
| chr1_-_29508321 | 2.76 |
ENST00000546138.1
|
SRSF4
|
serine/arginine-rich splicing factor 4 |
| chr8_-_144679296 | 2.73 |
ENST00000317198.6
|
EEF1D
|
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| chr4_-_151936416 | 2.72 |
ENST00000510413.1
ENST00000507224.1 |
LRBA
|
LPS-responsive vesicle trafficking, beach and anchor containing |
| chr1_-_40098672 | 2.71 |
ENST00000535435.1
|
HEYL
|
hes-related family bHLH transcription factor with YRPW motif-like |
| chr21_+_17102311 | 2.70 |
ENST00000285679.6
ENST00000351097.5 ENST00000285681.2 ENST00000400183.2 |
USP25
|
ubiquitin specific peptidase 25 |
| chr12_-_58240470 | 2.65 |
ENST00000548823.1
ENST00000398073.2 |
CTDSP2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr4_-_83351005 | 2.59 |
ENST00000295470.5
|
HNRNPDL
|
heterogeneous nuclear ribonucleoprotein D-like |
| chr11_+_64863587 | 2.57 |
ENST00000530773.1
ENST00000279281.3 ENST00000529180.1 |
VPS51
|
vacuolar protein sorting 51 homolog (S. cerevisiae) |
| chr6_+_391739 | 2.56 |
ENST00000380956.4
|
IRF4
|
interferon regulatory factor 4 |
| chr5_-_42811986 | 2.55 |
ENST00000511224.1
ENST00000507920.1 ENST00000510965.1 |
SEPP1
|
selenoprotein P, plasma, 1 |
| chr19_-_45909585 | 2.54 |
ENST00000593226.1
ENST00000418234.2 |
PPP1R13L
|
protein phosphatase 1, regulatory subunit 13 like |
| chr11_+_64794875 | 2.50 |
ENST00000377244.3
ENST00000534637.1 ENST00000524831.1 |
SNX15
|
sorting nexin 15 |
| chr1_-_48937682 | 2.48 |
ENST00000371843.3
|
SPATA6
|
spermatogenesis associated 6 |
| chr17_+_29421987 | 2.47 |
ENST00000431387.4
|
NF1
|
neurofibromin 1 |
| chr8_+_128748308 | 2.47 |
ENST00000377970.2
|
MYC
|
v-myc avian myelocytomatosis viral oncogene homolog |
| chr6_+_7107999 | 2.43 |
ENST00000491191.1
ENST00000379938.2 ENST00000471433.1 |
RREB1
|
ras responsive element binding protein 1 |
| chr17_+_78075361 | 2.42 |
ENST00000577106.1
ENST00000390015.3 |
GAA
|
glucosidase, alpha; acid |
| chr5_+_43121698 | 2.33 |
ENST00000505606.2
ENST00000509634.1 ENST00000509341.1 |
ZNF131
|
zinc finger protein 131 |
| chr9_+_137218362 | 2.29 |
ENST00000481739.1
|
RXRA
|
retinoid X receptor, alpha |
| chr19_-_44143939 | 2.29 |
ENST00000222374.2
|
CADM4
|
cell adhesion molecule 4 |
| chr8_+_95653302 | 2.27 |
ENST00000423620.2
ENST00000433389.2 |
ESRP1
|
epithelial splicing regulatory protein 1 |
| chr2_-_60780702 | 2.23 |
ENST00000359629.5
|
BCL11A
|
B-cell CLL/lymphoma 11A (zinc finger protein) |
| chr5_+_121647764 | 2.19 |
ENST00000261368.8
ENST00000379533.2 ENST00000379536.2 ENST00000379538.3 |
SNCAIP
|
synuclein, alpha interacting protein |
| chr7_+_142960505 | 2.18 |
ENST00000409500.3
ENST00000443571.2 ENST00000358406.5 ENST00000479303.1 |
GSTK1
|
glutathione S-transferase kappa 1 |
| chr12_-_6484715 | 2.18 |
ENST00000228916.2
|
SCNN1A
|
sodium channel, non-voltage-gated 1 alpha subunit |
| chr19_+_17970693 | 2.18 |
ENST00000600147.1
ENST00000599898.1 |
RPL18A
|
ribosomal protein L18a |
| chr12_+_69864129 | 2.17 |
ENST00000547219.1
ENST00000299293.2 ENST00000549921.1 ENST00000550316.1 ENST00000548154.1 ENST00000547414.1 ENST00000550389.1 ENST00000550937.1 ENST00000549092.1 ENST00000550169.1 |
FRS2
|
fibroblast growth factor receptor substrate 2 |
| chr17_+_7211280 | 2.16 |
ENST00000419711.2
ENST00000571955.1 ENST00000573714.1 |
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr20_-_3996165 | 2.15 |
ENST00000545616.2
ENST00000358395.6 |
RNF24
|
ring finger protein 24 |
| chr10_-_69834973 | 2.15 |
ENST00000395187.2
|
HERC4
|
HECT and RLD domain containing E3 ubiquitin protein ligase 4 |
| chr22_-_50217981 | 2.13 |
ENST00000457780.2
|
BRD1
|
bromodomain containing 1 |
| chr18_+_56338750 | 2.13 |
ENST00000345724.3
|
MALT1
|
mucosa associated lymphoid tissue lymphoma translocation gene 1 |
| chr16_-_2014804 | 2.11 |
ENST00000526522.1
ENST00000527302.1 ENST00000529806.1 ENST00000563194.1 ENST00000343262.4 |
RPS2
|
ribosomal protein S2 |
| chr4_-_2758015 | 2.09 |
ENST00000510267.1
ENST00000503235.1 ENST00000315423.7 |
TNIP2
|
TNFAIP3 interacting protein 2 |
| chr7_+_2559399 | 2.07 |
ENST00000222725.5
ENST00000359574.3 |
LFNG
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr15_+_74833518 | 2.06 |
ENST00000346246.5
|
ARID3B
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr7_-_8301768 | 2.04 |
ENST00000265577.7
|
ICA1
|
islet cell autoantigen 1, 69kDa |
| chr4_-_109090106 | 2.03 |
ENST00000379951.2
|
LEF1
|
lymphoid enhancer-binding factor 1 |
| chr1_+_43148059 | 2.03 |
ENST00000321358.7
ENST00000332220.6 |
YBX1
|
Y box binding protein 1 |
| chr5_+_110559603 | 2.02 |
ENST00000512453.1
|
CAMK4
|
calcium/calmodulin-dependent protein kinase IV |
| chr16_-_28937027 | 2.01 |
ENST00000358201.4
|
RABEP2
|
rabaptin, RAB GTPase binding effector protein 2 |
| chr1_+_244816237 | 2.01 |
ENST00000302550.11
|
DESI2
|
desumoylating isopeptidase 2 |
| chr17_+_4736627 | 2.00 |
ENST00000355280.6
ENST00000347992.7 |
MINK1
|
misshapen-like kinase 1 |
| chr19_-_49622348 | 2.00 |
ENST00000408991.2
|
C19orf73
|
chromosome 19 open reading frame 73 |
| chr19_-_4065730 | 1.99 |
ENST00000601588.1
|
ZBTB7A
|
zinc finger and BTB domain containing 7A |
| chr2_-_165698521 | 1.96 |
ENST00000409184.3
ENST00000392717.2 ENST00000456693.1 |
COBLL1
|
cordon-bleu WH2 repeat protein-like 1 |
| chr11_+_64053311 | 1.94 |
ENST00000540370.1
|
GPR137
|
G protein-coupled receptor 137 |
| chr1_+_153700518 | 1.93 |
ENST00000318967.2
ENST00000456435.1 ENST00000435409.2 |
INTS3
|
integrator complex subunit 3 |
| chr3_+_73045936 | 1.91 |
ENST00000356692.5
ENST00000488810.1 ENST00000394284.3 ENST00000295862.9 ENST00000495566.1 |
PPP4R2
|
protein phosphatase 4, regulatory subunit 2 |
| chr19_+_17970677 | 1.90 |
ENST00000222247.5
|
RPL18A
|
ribosomal protein L18a |
| chr20_+_35201857 | 1.89 |
ENST00000373874.2
|
TGIF2
|
TGFB-induced factor homeobox 2 |
| chr9_+_71320557 | 1.88 |
ENST00000541509.1
|
PIP5K1B
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta |
| chr1_+_212458834 | 1.87 |
ENST00000261461.2
|
PPP2R5A
|
protein phosphatase 2, regulatory subunit B', alpha |
| chr19_+_1205740 | 1.87 |
ENST00000326873.7
|
STK11
|
serine/threonine kinase 11 |
| chr1_-_48937838 | 1.86 |
ENST00000371847.3
|
SPATA6
|
spermatogenesis associated 6 |
| chr20_+_49348081 | 1.86 |
ENST00000371610.2
|
PARD6B
|
par-6 family cell polarity regulator beta |
| chr14_+_57046500 | 1.85 |
ENST00000261556.6
|
TMEM260
|
transmembrane protein 260 |
| chr12_+_12764773 | 1.82 |
ENST00000228865.2
|
CREBL2
|
cAMP responsive element binding protein-like 2 |
| chr18_-_32924372 | 1.81 |
ENST00000261332.6
ENST00000399061.3 |
ZNF24
|
zinc finger protein 24 |
| chr8_+_38644778 | 1.80 |
ENST00000276520.8
|
TACC1
|
transforming, acidic coiled-coil containing protein 1 |
| chrX_-_130423386 | 1.80 |
ENST00000370903.3
|
IGSF1
|
immunoglobulin superfamily, member 1 |
| chr2_-_43823093 | 1.78 |
ENST00000405006.4
|
THADA
|
thyroid adenoma associated |
| chr2_-_43823119 | 1.74 |
ENST00000403856.1
ENST00000404790.1 ENST00000405975.2 ENST00000415080.2 |
THADA
|
thyroid adenoma associated |
| chr20_+_57466629 | 1.74 |
ENST00000371081.1
ENST00000338783.6 |
GNAS
|
GNAS complex locus |
| chr7_-_8301682 | 1.73 |
ENST00000396675.3
ENST00000430867.1 |
ICA1
|
islet cell autoantigen 1, 69kDa |
| chr10_-_69835099 | 1.72 |
ENST00000373700.4
|
HERC4
|
HECT and RLD domain containing E3 ubiquitin protein ligase 4 |
| chr22_-_50218452 | 1.71 |
ENST00000216267.8
|
BRD1
|
bromodomain containing 1 |
| chr14_+_57046530 | 1.70 |
ENST00000536419.1
ENST00000538838.1 |
TMEM260
|
transmembrane protein 260 |
| chr12_-_54020107 | 1.70 |
ENST00000588232.1
ENST00000548446.2 ENST00000420353.2 ENST00000591397.1 ENST00000415113.1 |
ATF7
|
activating transcription factor 7 |
| chr10_-_21463116 | 1.69 |
ENST00000417816.2
|
NEBL
|
nebulette |
| chr1_-_19229014 | 1.68 |
ENST00000538839.1
ENST00000290597.5 |
ALDH4A1
|
aldehyde dehydrogenase 4 family, member A1 |
| chr10_+_112257596 | 1.68 |
ENST00000369583.3
|
DUSP5
|
dual specificity phosphatase 5 |
| chr14_-_21566731 | 1.65 |
ENST00000360947.3
|
ZNF219
|
zinc finger protein 219 |
| chr18_-_51750948 | 1.61 |
ENST00000583046.1
ENST00000398398.2 |
MBD2
|
methyl-CpG binding domain protein 2 |
| chr1_+_235492300 | 1.60 |
ENST00000476121.1
ENST00000497327.1 |
GGPS1
|
geranylgeranyl diphosphate synthase 1 |
| chr1_-_236030216 | 1.60 |
ENST00000389794.3
ENST00000389793.2 |
LYST
|
lysosomal trafficking regulator |
| chr16_+_3014217 | 1.59 |
ENST00000572045.1
|
KREMEN2
|
kringle containing transmembrane protein 2 |
| chr10_+_70661014 | 1.58 |
ENST00000373585.3
|
DDX50
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 50 |
| chr14_+_73525229 | 1.57 |
ENST00000527432.1
ENST00000531500.1 ENST00000525321.1 ENST00000526754.1 |
RBM25
|
RNA binding motif protein 25 |
| chr1_+_222817629 | 1.55 |
ENST00000340535.7
|
MIA3
|
melanoma inhibitory activity family, member 3 |
| chrX_-_153363125 | 1.54 |
ENST00000407218.1
ENST00000453960.2 |
MECP2
|
methyl CpG binding protein 2 (Rett syndrome) |
| chr14_+_73525144 | 1.53 |
ENST00000261973.7
ENST00000540173.1 |
RBM25
|
RNA binding motif protein 25 |
| chr7_-_155604967 | 1.53 |
ENST00000297261.2
|
SHH
|
sonic hedgehog |
| chr4_-_146101304 | 1.52 |
ENST00000447906.2
|
OTUD4
|
OTU domain containing 4 |
| chr12_+_49372251 | 1.50 |
ENST00000293549.3
|
WNT1
|
wingless-type MMTV integration site family, member 1 |
| chr2_+_97203082 | 1.48 |
ENST00000454558.2
|
ARID5A
|
AT rich interactive domain 5A (MRF1-like) |
| chr8_+_100025476 | 1.48 |
ENST00000355155.1
ENST00000357162.2 ENST00000358544.2 ENST00000395996.1 ENST00000441350.2 |
VPS13B
|
vacuolar protein sorting 13 homolog B (yeast) |
| chr12_-_133338379 | 1.48 |
ENST00000539605.1
|
ANKLE2
|
ankyrin repeat and LEM domain containing 2 |
| chr8_+_38644715 | 1.47 |
ENST00000317827.4
ENST00000379931.3 |
TACC1
|
transforming, acidic coiled-coil containing protein 1 |
| chr8_+_95653427 | 1.44 |
ENST00000454170.2
|
ESRP1
|
epithelial splicing regulatory protein 1 |
| chr1_+_110754094 | 1.43 |
ENST00000369787.3
ENST00000413138.3 ENST00000438661.2 |
KCNC4
|
potassium voltage-gated channel, Shaw-related subfamily, member 4 |
| chr14_-_89021077 | 1.42 |
ENST00000556564.1
|
PTPN21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr17_+_45608614 | 1.41 |
ENST00000544660.1
|
NPEPPS
|
aminopeptidase puromycin sensitive |
| chr1_-_16302565 | 1.41 |
ENST00000537142.1
ENST00000448462.2 |
ZBTB17
|
zinc finger and BTB domain containing 17 |
| chr2_+_191513789 | 1.40 |
ENST00000409581.1
|
NAB1
|
NGFI-A binding protein 1 (EGR1 binding protein 1) |
| chr1_+_197170592 | 1.38 |
ENST00000535699.1
|
CRB1
|
crumbs homolog 1 (Drosophila) |
| chr12_-_96794143 | 1.38 |
ENST00000543119.2
|
CDK17
|
cyclin-dependent kinase 17 |
| chr8_+_22022800 | 1.38 |
ENST00000397814.3
|
BMP1
|
bone morphogenetic protein 1 |
| chr6_-_29595779 | 1.37 |
ENST00000355973.3
ENST00000377012.4 |
GABBR1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr15_-_83953466 | 1.36 |
ENST00000345382.2
|
BNC1
|
basonuclin 1 |
| chr3_+_170075436 | 1.36 |
ENST00000476188.1
ENST00000259119.4 ENST00000426052.2 |
SKIL
|
SKI-like oncogene |
| chr12_-_133338426 | 1.35 |
ENST00000337516.5
ENST00000357997.5 |
ANKLE2
|
ankyrin repeat and LEM domain containing 2 |
| chrX_-_130423240 | 1.33 |
ENST00000370910.1
ENST00000370901.4 |
IGSF1
|
immunoglobulin superfamily, member 1 |
| chr4_+_159593418 | 1.33 |
ENST00000507475.1
ENST00000307738.5 |
ETFDH
|
electron-transferring-flavoprotein dehydrogenase |
| chr1_-_171711177 | 1.31 |
ENST00000415773.1
ENST00000367740.2 |
VAMP4
|
vesicle-associated membrane protein 4 |
| chr12_-_95467267 | 1.31 |
ENST00000330677.7
|
NR2C1
|
nuclear receptor subfamily 2, group C, member 1 |
| chr2_-_105946491 | 1.30 |
ENST00000393359.2
|
TGFBRAP1
|
transforming growth factor, beta receptor associated protein 1 |
| chr5_+_175792459 | 1.30 |
ENST00000310389.5
|
ARL10
|
ADP-ribosylation factor-like 10 |
| chr8_-_145559943 | 1.30 |
ENST00000332135.4
|
SCRT1
|
scratch family zinc finger 1 |
| chr17_-_2614927 | 1.29 |
ENST00000435359.1
|
CLUH
|
clustered mitochondria (cluA/CLU1) homolog |
| chr7_-_8302207 | 1.29 |
ENST00000407906.1
|
ICA1
|
islet cell autoantigen 1, 69kDa |
| chr14_+_96968707 | 1.28 |
ENST00000216277.8
ENST00000557320.1 ENST00000557471.1 |
PAPOLA
|
poly(A) polymerase alpha |
| chr2_-_219925189 | 1.27 |
ENST00000295731.6
|
IHH
|
indian hedgehog |
| chr17_-_7155802 | 1.26 |
ENST00000572043.1
|
CTDNEP1
|
CTD nuclear envelope phosphatase 1 |
| chr22_-_31885727 | 1.26 |
ENST00000330125.5
ENST00000344710.5 ENST00000397518.1 |
EIF4ENIF1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr1_-_183604794 | 1.26 |
ENST00000367534.1
ENST00000359856.6 ENST00000294742.6 |
ARPC5
|
actin related protein 2/3 complex, subunit 5, 16kDa |
| chr19_+_49661037 | 1.26 |
ENST00000427978.2
|
TRPM4
|
transient receptor potential cation channel, subfamily M, member 4 |
| chr22_+_32870661 | 1.25 |
ENST00000266087.7
|
FBXO7
|
F-box protein 7 |
| chr5_+_175298573 | 1.25 |
ENST00000512824.1
|
CPLX2
|
complexin 2 |
| chr7_-_8302164 | 1.25 |
ENST00000447326.1
ENST00000406470.2 |
ICA1
|
islet cell autoantigen 1, 69kDa |
| chr5_+_159343688 | 1.24 |
ENST00000306675.3
|
ADRA1B
|
adrenoceptor alpha 1B |
| chr1_-_16302608 | 1.24 |
ENST00000375743.4
ENST00000375733.2 |
ZBTB17
|
zinc finger and BTB domain containing 17 |
| chr16_-_18937726 | 1.23 |
ENST00000389467.3
ENST00000446231.2 |
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr7_-_8301869 | 1.22 |
ENST00000402384.3
|
ICA1
|
islet cell autoantigen 1, 69kDa |
| chr17_+_77751931 | 1.22 |
ENST00000310942.4
ENST00000269399.5 |
CBX2
|
chromobox homolog 2 |
| chrX_-_119445306 | 1.22 |
ENST00000371369.4
ENST00000440464.1 ENST00000519908.1 |
TMEM255A
|
transmembrane protein 255A |
| chr17_-_65241281 | 1.21 |
ENST00000358691.5
ENST00000580168.1 |
HELZ
|
helicase with zinc finger |
| chr17_-_40761375 | 1.20 |
ENST00000543197.1
ENST00000309428.5 |
FAM134C
|
family with sequence similarity 134, member C |
| chr19_-_38720294 | 1.20 |
ENST00000412732.1
ENST00000456296.1 |
DPF1
|
D4, zinc and double PHD fingers family 1 |
| chr6_+_7107830 | 1.19 |
ENST00000379933.3
|
RREB1
|
ras responsive element binding protein 1 |
| chr12_-_99288536 | 1.19 |
ENST00000549797.1
ENST00000333732.7 ENST00000341752.7 |
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr11_+_76092353 | 1.19 |
ENST00000530460.1
ENST00000321844.4 |
RP11-111M22.2
|
Homo sapiens putative uncharacterized protein FLJ37770-like (LOC100506127), mRNA. |
| chr1_-_171711387 | 1.15 |
ENST00000236192.7
|
VAMP4
|
vesicle-associated membrane protein 4 |
| chr11_+_67159416 | 1.15 |
ENST00000307980.2
ENST00000544620.1 |
RAD9A
|
RAD9 homolog A (S. pombe) |
| chr5_+_79331164 | 1.13 |
ENST00000350881.2
|
THBS4
|
thrombospondin 4 |
| chr19_-_18392422 | 1.13 |
ENST00000252818.3
|
JUND
|
jun D proto-oncogene |
| chr5_-_81046922 | 1.12 |
ENST00000514493.1
ENST00000320672.4 |
SSBP2
|
single-stranded DNA binding protein 2 |
| chr8_+_22022653 | 1.12 |
ENST00000354870.5
ENST00000397816.3 ENST00000306349.8 |
BMP1
|
bone morphogenetic protein 1 |
| chr6_+_43149903 | 1.10 |
ENST00000252050.4
ENST00000354495.3 ENST00000372647.2 |
CUL9
|
cullin 9 |
| chr14_+_96968802 | 1.09 |
ENST00000556619.1
ENST00000392990.2 |
PAPOLA
|
poly(A) polymerase alpha |
| chr12_-_53715328 | 1.09 |
ENST00000547757.1
ENST00000394384.3 ENST00000209873.4 |
AAAS
|
achalasia, adrenocortical insufficiency, alacrimia |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR0B1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 8.8 | GO:1904800 | negative regulation of dendrite extension(GO:1903860) regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 2.2 | 2.2 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 2.0 | 17.9 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 2.0 | 11.7 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 1.9 | 5.7 | GO:0071409 | negative regulation of muscle hyperplasia(GO:0014740) cellular response to cycloheximide(GO:0071409) |
| 1.6 | 4.7 | GO:0033386 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 1.5 | 7.6 | GO:2000035 | positive regulation of somatic stem cell population maintenance(GO:1904674) regulation of stem cell division(GO:2000035) |
| 1.5 | 13.5 | GO:0060633 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) |
| 1.4 | 6.8 | GO:0072240 | thick ascending limb development(GO:0072023) DCT cell differentiation(GO:0072069) metanephric thick ascending limb development(GO:0072233) metanephric DCT cell differentiation(GO:0072240) |
| 1.2 | 4.8 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 1.2 | 3.6 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 1.1 | 4.5 | GO:0030242 | pexophagy(GO:0030242) |
| 1.0 | 4.2 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 1.0 | 7.3 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.9 | 3.6 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.9 | 2.7 | GO:2000824 | negative regulation of androgen receptor activity(GO:2000824) |
| 0.9 | 6.2 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.9 | 4.4 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.9 | 3.5 | GO:1901895 | negative regulation of calcium-transporting ATPase activity(GO:1901895) |
| 0.9 | 2.6 | GO:0045082 | interleukin-4 biosynthetic process(GO:0042097) positive regulation of interleukin-10 biosynthetic process(GO:0045082) regulation of interleukin-4 biosynthetic process(GO:0045402) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.8 | 3.3 | GO:1900241 | metanephric glomerular mesangial cell development(GO:0072255) reversible differentiation(GO:0090677) cell dedifferentiation involved in phenotypic switching(GO:0090678) positive regulation of phenotypic switching(GO:1900241) regulation of vascular smooth muscle cell dedifferentiation(GO:1905174) positive regulation of vascular smooth muscle cell dedifferentiation(GO:1905176) vascular smooth muscle cell dedifferentiation(GO:1990936) |
| 0.8 | 4.8 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.8 | 3.2 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.8 | 3.1 | GO:0060214 | endocardium formation(GO:0060214) |
| 0.8 | 2.3 | GO:0045994 | positive regulation of translational initiation by iron(GO:0045994) |
| 0.7 | 2.1 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 0.7 | 14.6 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.6 | 1.9 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.6 | 2.4 | GO:0000023 | maltose metabolic process(GO:0000023) diaphragm contraction(GO:0002086) |
| 0.6 | 1.8 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.6 | 2.3 | GO:1990736 | positive regulation of cardiac conduction(GO:1903781) positive regulation of atrial cardiac muscle cell action potential(GO:1903949) positive regulation of regulation of vascular smooth muscle cell membrane depolarization(GO:1904199) regulation of vascular smooth muscle cell membrane depolarization(GO:1990736) |
| 0.5 | 1.6 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.5 | 3.7 | GO:0035624 | receptor transactivation(GO:0035624) |
| 0.5 | 1.5 | GO:1900114 | positive regulation of histone H3-K9 trimethylation(GO:1900114) |
| 0.5 | 1.5 | GO:2000729 | bud outgrowth involved in lung branching(GO:0060447) positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.5 | 2.5 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.5 | 1.5 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.5 | 7.8 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.5 | 9.1 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.4 | 3.1 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.4 | 1.7 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 0.4 | 2.5 | GO:0072133 | kidney mesenchyme morphogenesis(GO:0072131) metanephric mesenchyme morphogenesis(GO:0072133) |
| 0.4 | 2.0 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.4 | 1.2 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.4 | 1.9 | GO:1901090 | regulation of protein tetramerization(GO:1901090) negative regulation of protein tetramerization(GO:1901091) regulation of protein homotetramerization(GO:1901093) negative regulation of protein homotetramerization(GO:1901094) |
| 0.3 | 1.3 | GO:0048073 | regulation of eye pigmentation(GO:0048073) |
| 0.3 | 2.9 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.3 | 1.2 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.3 | 2.7 | GO:1904293 | negative regulation of ERAD pathway(GO:1904293) |
| 0.3 | 2.2 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.3 | 2.1 | GO:0023035 | CD40 signaling pathway(GO:0023035) |
| 0.3 | 13.9 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.2 | 1.9 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.2 | 1.1 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.2 | 0.6 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.2 | 0.8 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.2 | 0.8 | GO:0051899 | membrane depolarization(GO:0051899) |
| 0.2 | 2.0 | GO:0002371 | dendritic cell cytokine production(GO:0002371) |
| 0.2 | 3.2 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.2 | 0.4 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.2 | 0.8 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.2 | 0.8 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.2 | 0.6 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.2 | 0.9 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.2 | 3.9 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.2 | 4.3 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.2 | 2.1 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.2 | 0.6 | GO:1902949 | positive regulation of tau-protein kinase activity(GO:1902949) |
| 0.2 | 0.6 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.2 | 1.1 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.1 | 2.8 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.1 | 1.3 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 | 2.5 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.1 | 0.7 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.1 | 1.3 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 0.8 | GO:0071321 | cellular response to cGMP(GO:0071321) |
| 0.1 | 3.5 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.1 | 0.5 | GO:1902723 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.1 | 1.3 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 2.5 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.1 | 2.1 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.1 | 3.9 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.1 | 1.7 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.1 | 10.8 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.1 | 0.9 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 2.6 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.1 | 2.5 | GO:0034380 | high-density lipoprotein particle assembly(GO:0034380) |
| 0.1 | 1.0 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.1 | 0.8 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.1 | 2.0 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 | 5.5 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 1.0 | GO:0072178 | nephric duct morphogenesis(GO:0072178) |
| 0.1 | 0.3 | GO:0071529 | cementum mineralization(GO:0071529) |
| 0.1 | 1.0 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.1 | 1.0 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.1 | 1.6 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.1 | 3.8 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.1 | 0.6 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.1 | 1.3 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 2.6 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.1 | 0.3 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.1 | 0.9 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.1 | 2.8 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.1 | 0.8 | GO:0098719 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 0.4 | GO:0051138 | positive regulation of NK T cell differentiation(GO:0051138) |
| 0.1 | 1.1 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.1 | 0.3 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.1 | 1.4 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.1 | 1.5 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.1 | 1.3 | GO:1903599 | positive regulation of mitophagy(GO:1903599) |
| 0.1 | 1.6 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 3.3 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 0.6 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.1 | 1.9 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.1 | 0.8 | GO:0034465 | response to carbon monoxide(GO:0034465) negative regulation of cell volume(GO:0045794) |
| 0.1 | 0.7 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.1 | 0.6 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.1 | 2.2 | GO:1901687 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.1 | 0.9 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 0.5 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.1 | 6.1 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.1 | 1.0 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.1 | 0.2 | GO:0010700 | negative regulation of norepinephrine secretion(GO:0010700) |
| 0.1 | 0.4 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.1 | 0.5 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.1 | 2.4 | GO:0031440 | regulation of mRNA 3'-end processing(GO:0031440) |
| 0.1 | 0.9 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.9 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 | 0.1 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 1.9 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 3.2 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 1.0 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 4.2 | GO:0008283 | cell proliferation(GO:0008283) |
| 0.0 | 2.3 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 1.8 | GO:0061098 | positive regulation of protein tyrosine kinase activity(GO:0061098) |
| 0.0 | 1.4 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 7.4 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 3.0 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 1.3 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.0 | 2.2 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 3.1 | GO:1900181 | negative regulation of protein localization to nucleus(GO:1900181) |
| 0.0 | 0.5 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 1.4 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.9 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.0 | 1.8 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 2.7 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.0 | 3.4 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 3.5 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 1.4 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.7 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 2.9 | GO:0051492 | regulation of stress fiber assembly(GO:0051492) |
| 0.0 | 1.4 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.5 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.0 | 5.5 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.0 | 1.6 | GO:0030279 | negative regulation of ossification(GO:0030279) |
| 0.0 | 0.4 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.1 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) regulation of NFAT protein import into nucleus(GO:0051532) positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 0.4 | GO:0046320 | regulation of fatty acid oxidation(GO:0046320) |
| 0.0 | 1.4 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.3 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.1 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.1 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.0 | 4.7 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.4 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 1.6 | GO:0006401 | RNA catabolic process(GO:0006401) |
| 0.0 | 1.0 | GO:0001657 | ureteric bud development(GO:0001657) mesonephric epithelium development(GO:0072163) mesonephric tubule development(GO:0072164) |
| 0.0 | 0.7 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.7 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.0 | 0.7 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 15.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 1.2 | 4.8 | GO:0031417 | NatC complex(GO:0031417) |
| 1.1 | 4.3 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 1.0 | 9.1 | GO:0032039 | integrator complex(GO:0032039) |
| 0.9 | 17.9 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.8 | 6.2 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.7 | 4.2 | GO:0001740 | Barr body(GO:0001740) |
| 0.6 | 2.6 | GO:1990745 | EARP complex(GO:1990745) |
| 0.6 | 1.9 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.6 | 6.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.5 | 8.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.5 | 1.4 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.4 | 2.1 | GO:0032449 | CBM complex(GO:0032449) |
| 0.4 | 3.8 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.4 | 1.1 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.4 | 1.9 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.4 | 4.5 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.4 | 2.5 | GO:0070554 | synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.4 | 1.8 | GO:0071546 | perinucleolar chromocenter(GO:0010370) pi-body(GO:0071546) |
| 0.4 | 3.9 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.3 | 1.0 | GO:0016935 | glycine-gated chloride channel complex(GO:0016935) |
| 0.3 | 1.1 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.3 | 1.3 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.2 | 3.5 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.2 | 2.0 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.2 | 4.6 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.2 | 1.5 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.2 | 1.3 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.2 | 28.5 | GO:0016605 | PML body(GO:0016605) |
| 0.2 | 3.6 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.2 | 2.6 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.2 | 3.7 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.2 | 2.2 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.2 | 1.1 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.2 | 6.9 | GO:0000791 | euchromatin(GO:0000791) |
| 0.2 | 3.0 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 0.9 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 3.6 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.1 | 2.5 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 13.7 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 0.5 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 1.0 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 8.9 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 1.4 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.1 | 4.4 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 1.8 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.1 | 11.0 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 0.5 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 0.9 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 0.3 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.1 | 1.6 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.1 | 3.9 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 9.8 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 1.1 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.1 | 5.4 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 1.1 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 5.8 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 1.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 1.3 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 1.0 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 1.2 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 4.8 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.4 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 4.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 2.1 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 2.4 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 4.7 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 3.2 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.8 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.7 | GO:0031082 | BLOC complex(GO:0031082) |
| 0.0 | 2.1 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.7 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.8 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 10.7 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 1.4 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 3.4 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 6.6 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 0.7 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.4 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 1.4 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 1.0 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.4 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.1 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 1.7 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 3.0 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 1.0 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.1 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 4.5 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 1.1 | GO:0031461 | cullin-RING ubiquitin ligase complex(GO:0031461) |
| 0.0 | 1.0 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.0 | 0.2 | GO:0098589 | membrane region(GO:0098589) |
| 0.0 | 0.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.2 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.9 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 34.2 | GO:0005654 | nucleoplasm(GO:0005654) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 10.3 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 2.0 | 6.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 1.5 | 4.6 | GO:0052810 | 1-phosphatidylinositol-5-kinase activity(GO:0052810) |
| 1.5 | 15.1 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 1.5 | 5.8 | GO:0035939 | microsatellite binding(GO:0035939) |
| 1.2 | 4.8 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 1.1 | 5.7 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 1.1 | 17.9 | GO:0008494 | translation activator activity(GO:0008494) |
| 1.0 | 3.1 | GO:0017130 | poly(C) RNA binding(GO:0017130) |
| 0.9 | 4.7 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.9 | 8.1 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.9 | 2.7 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.9 | 16.6 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.8 | 2.4 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.6 | 11.5 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.6 | 1.8 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.6 | 2.3 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.5 | 2.7 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.5 | 2.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.5 | 3.9 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.5 | 1.4 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.4 | 3.0 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.4 | 1.2 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.4 | 3.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.4 | 1.6 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.4 | 1.9 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.4 | 4.8 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.4 | 4.3 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.3 | 3.3 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.3 | 3.9 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.3 | 8.2 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.3 | 2.4 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.3 | 6.4 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.3 | 6.8 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.3 | 3.6 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.3 | 1.6 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.3 | 11.7 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.3 | 4.4 | GO:0015250 | water channel activity(GO:0015250) |
| 0.2 | 0.7 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.2 | 1.4 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.2 | 0.8 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.2 | 1.0 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.2 | 2.2 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.2 | 2.8 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.2 | 0.6 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.2 | 1.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.2 | 5.4 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.2 | 1.7 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.2 | 0.5 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.2 | 0.9 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.2 | 1.0 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.2 | 1.5 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.1 | 3.8 | GO:0042393 | histone binding(GO:0042393) |
| 0.1 | 2.6 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 1.0 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.1 | 1.7 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 2.0 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.1 | 10.7 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.1 | 0.8 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.1 | 3.1 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 2.7 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.1 | 2.0 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 0.7 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.1 | 1.3 | GO:0005113 | patched binding(GO:0005113) |
| 0.1 | 4.2 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 5.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 3.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 1.3 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 2.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.8 | GO:0015386 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 2.2 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.1 | 1.7 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 0.9 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.1 | 0.8 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 3.2 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 1.5 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.1 | 2.8 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.1 | 0.7 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.1 | 0.9 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.1 | 0.9 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 2.0 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 2.3 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.1 | 1.7 | GO:0016646 | aldehyde dehydrogenase (NAD) activity(GO:0004029) oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.1 | 1.8 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 3.5 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 0.5 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.1 | 1.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 14.4 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 0.3 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.1 | 1.1 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.1 | 2.5 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 2.0 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 4.3 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.1 | 1.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.2 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 0.2 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 2.1 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 1.7 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.2 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.6 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 5.6 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 6.1 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.3 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 1.2 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 1.8 | GO:0016278 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 0.4 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 3.1 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.9 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 1.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 3.0 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 2.1 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 1.2 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.3 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 5.9 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.7 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.0 | 6.0 | GO:0061659 | ubiquitin-like protein ligase activity(GO:0061659) |
| 0.0 | 1.6 | GO:0008186 | ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 1.4 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 1.3 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 1.5 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.5 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.4 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 2.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.3 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 2.2 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 4.1 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 1.2 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 3.4 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 1.7 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 1.1 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.4 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 2.1 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 1.3 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.1 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.0 | 0.5 | GO:0035064 | methylated histone binding(GO:0035064) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 20.7 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.3 | 15.1 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.2 | 6.5 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.2 | 3.7 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 10.5 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 0.3 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 4.0 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 2.1 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 3.3 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.1 | 2.6 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 5.6 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.1 | 5.5 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 1.8 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 6.2 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 2.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 1.4 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 2.3 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 3.3 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 7.8 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 2.1 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.1 | 6.3 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 2.2 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.1 | 2.8 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 1.2 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 1.8 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 2.6 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.0 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 2.2 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 1.9 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.7 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 1.6 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.9 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.3 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.1 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.5 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.6 | PID ILK PATHWAY | Integrin-linked kinase signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 17.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.4 | 8.1 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.3 | 5.7 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.3 | 4.4 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.3 | 10.8 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.2 | 6.0 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.2 | 19.3 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.2 | 4.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 0.9 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 2.4 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 2.9 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 7.0 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 2.8 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.1 | 2.5 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 5.8 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 2.3 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 1.9 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.1 | 3.7 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 1.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 5.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 1.1 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 1.2 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.1 | 1.4 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 1.5 | REACTOME PROLONGED ERK ACTIVATION EVENTS | Genes involved in Prolonged ERK activation events |
| 0.1 | 2.2 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.1 | 2.0 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 12.3 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.1 | 5.9 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.1 | 0.8 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 2.0 | REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | Genes involved in Post NMDA receptor activation events |
| 0.0 | 1.9 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 3.0 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 1.8 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 2.1 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 4.0 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 0.9 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 2.0 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.2 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.7 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.6 | REACTOME GLUCAGON SIGNALING IN METABOLIC REGULATION | Genes involved in Glucagon signaling in metabolic regulation |
| 0.0 | 2.1 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 1.0 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 1.2 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.0 | 0.6 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.8 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 0.8 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 1.4 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.4 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.0 | 0.5 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 3.5 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |