Project
averaged GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
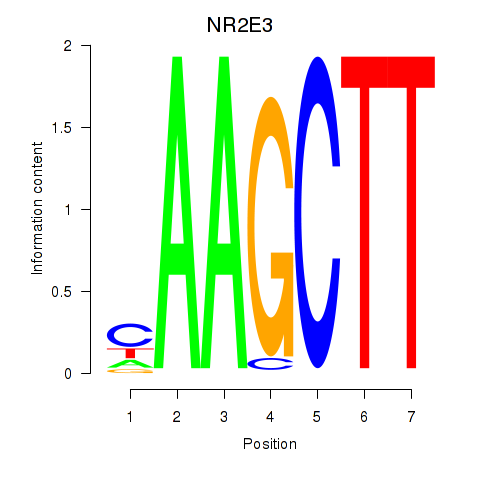
Results for NR2E3
Z-value: 1.47
Transcription factors associated with NR2E3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR2E3
|
ENSG00000031544.10 | nuclear receptor subfamily 2 group E member 3 |
Activity profile of NR2E3 motif
Sorted Z-values of NR2E3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_60337684 | 45.14 |
ENST00000267484.5
|
RTN1
|
reticulon 1 |
| chr3_-_195310802 | 32.50 |
ENST00000421243.1
ENST00000453131.1 |
APOD
|
apolipoprotein D |
| chr5_-_138210977 | 27.78 |
ENST00000274711.6
ENST00000521094.2 |
LRRTM2
|
leucine rich repeat transmembrane neuronal 2 |
| chrX_-_13835147 | 25.83 |
ENST00000493677.1
ENST00000355135.2 |
GPM6B
|
glycoprotein M6B |
| chr12_+_79258444 | 25.44 |
ENST00000261205.4
|
SYT1
|
synaptotagmin I |
| chr22_+_41865109 | 25.10 |
ENST00000216254.4
ENST00000396512.3 |
ACO2
|
aconitase 2, mitochondrial |
| chr2_-_175712270 | 24.70 |
ENST00000295497.7
ENST00000444394.1 |
CHN1
|
chimerin 1 |
| chr9_-_93405352 | 24.54 |
ENST00000375765.3
|
DIRAS2
|
DIRAS family, GTP-binding RAS-like 2 |
| chr3_-_58563094 | 23.95 |
ENST00000464064.1
|
FAM107A
|
family with sequence similarity 107, member A |
| chr12_+_79258547 | 22.53 |
ENST00000457153.2
|
SYT1
|
synaptotagmin I |
| chr4_+_88896819 | 21.88 |
ENST00000237623.7
ENST00000395080.3 ENST00000508233.1 ENST00000360804.4 |
SPP1
|
secreted phosphoprotein 1 |
| chr16_+_222846 | 20.00 |
ENST00000251595.6
ENST00000397806.1 |
HBA2
|
hemoglobin, alpha 2 |
| chr12_-_45269430 | 15.71 |
ENST00000395487.2
|
NELL2
|
NEL-like 2 (chicken) |
| chr1_+_65775204 | 15.69 |
ENST00000371069.4
|
DNAJC6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr11_-_5271122 | 13.29 |
ENST00000330597.3
|
HBG1
|
hemoglobin, gamma A |
| chr16_+_2039946 | 12.62 |
ENST00000248121.2
ENST00000568896.1 |
SYNGR3
|
synaptogyrin 3 |
| chr15_+_42696954 | 12.02 |
ENST00000337571.4
ENST00000569136.1 |
CAPN3
|
calpain 3, (p94) |
| chr7_-_137028534 | 11.92 |
ENST00000348225.2
|
PTN
|
pleiotrophin |
| chr7_-_137028498 | 11.90 |
ENST00000393083.2
|
PTN
|
pleiotrophin |
| chr15_+_42697018 | 11.70 |
ENST00000397204.4
|
CAPN3
|
calpain 3, (p94) |
| chr15_+_42696992 | 11.70 |
ENST00000561817.1
|
CAPN3
|
calpain 3, (p94) |
| chr13_-_45010939 | 11.57 |
ENST00000261489.2
|
TSC22D1
|
TSC22 domain family, member 1 |
| chr1_+_10271674 | 11.31 |
ENST00000377086.1
|
KIF1B
|
kinesin family member 1B |
| chr4_-_87374283 | 11.30 |
ENST00000361569.2
|
MAPK10
|
mitogen-activated protein kinase 10 |
| chr4_+_71587669 | 10.90 |
ENST00000381006.3
ENST00000226328.4 |
RUFY3
|
RUN and FYVE domain containing 3 |
| chr3_-_33700933 | 10.88 |
ENST00000480013.1
|
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr5_+_140868717 | 10.82 |
ENST00000252087.1
|
PCDHGC5
|
protocadherin gamma subfamily C, 5 |
| chr2_-_216257849 | 10.68 |
ENST00000456923.1
|
FN1
|
fibronectin 1 |
| chr5_-_11589131 | 10.55 |
ENST00000511377.1
|
CTNND2
|
catenin (cadherin-associated protein), delta 2 |
| chr3_-_33686743 | 10.20 |
ENST00000333778.6
ENST00000539981.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr3_+_69915385 | 10.00 |
ENST00000314589.5
|
MITF
|
microphthalmia-associated transcription factor |
| chr14_+_101297740 | 9.91 |
ENST00000555928.1
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr8_-_22089533 | 9.84 |
ENST00000321613.3
|
PHYHIP
|
phytanoyl-CoA 2-hydroxylase interacting protein |
| chr8_-_22089845 | 9.75 |
ENST00000454243.2
|
PHYHIP
|
phytanoyl-CoA 2-hydroxylase interacting protein |
| chr2_-_224467093 | 9.64 |
ENST00000305409.2
|
SCG2
|
secretogranin II |
| chr9_-_116102530 | 9.56 |
ENST00000374195.3
ENST00000341761.4 |
WDR31
|
WD repeat domain 31 |
| chr15_+_42697065 | 9.44 |
ENST00000565559.1
|
CAPN3
|
calpain 3, (p94) |
| chr1_-_153599426 | 9.40 |
ENST00000392622.1
|
S100A13
|
S100 calcium binding protein A13 |
| chr6_+_69942298 | 9.35 |
ENST00000238918.8
|
BAI3
|
brain-specific angiogenesis inhibitor 3 |
| chr1_-_144995074 | 9.34 |
ENST00000534536.1
|
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr11_-_5276008 | 9.33 |
ENST00000336906.4
|
HBG2
|
hemoglobin, gamma G |
| chr5_+_140734570 | 9.13 |
ENST00000571252.1
|
PCDHGA4
|
protocadherin gamma subfamily A, 4 |
| chr9_-_116102562 | 9.11 |
ENST00000374193.4
ENST00000465979.1 |
WDR31
|
WD repeat domain 31 |
| chr19_+_33182823 | 8.72 |
ENST00000397061.3
|
NUDT19
|
nudix (nucleoside diphosphate linked moiety X)-type motif 19 |
| chr1_-_153599732 | 8.71 |
ENST00000392623.1
|
S100A13
|
S100 calcium binding protein A13 |
| chr18_-_5544241 | 8.43 |
ENST00000341928.2
ENST00000540638.2 |
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3 |
| chr16_-_66785497 | 8.27 |
ENST00000440564.2
ENST00000379482.2 ENST00000443351.2 ENST00000566150.1 |
DYNC1LI2
|
dynein, cytoplasmic 1, light intermediate chain 2 |
| chr2_+_187454749 | 8.18 |
ENST00000261023.3
ENST00000374907.3 |
ITGAV
|
integrin, alpha V |
| chr2_+_166095898 | 7.83 |
ENST00000424833.1
ENST00000375437.2 ENST00000357398.3 |
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit |
| chr21_-_27542972 | 7.81 |
ENST00000346798.3
ENST00000439274.2 ENST00000354192.3 ENST00000348990.5 ENST00000357903.3 ENST00000358918.3 ENST00000359726.3 |
APP
|
amyloid beta (A4) precursor protein |
| chr10_-_93392811 | 7.64 |
ENST00000238994.5
|
PPP1R3C
|
protein phosphatase 1, regulatory subunit 3C |
| chr1_-_95391315 | 7.59 |
ENST00000545882.1
ENST00000415017.1 |
CNN3
|
calponin 3, acidic |
| chr14_+_78870030 | 7.44 |
ENST00000553631.1
ENST00000554719.1 |
NRXN3
|
neurexin 3 |
| chr7_-_130080818 | 7.23 |
ENST00000343969.5
ENST00000541543.1 ENST00000489512.1 |
CEP41
|
centrosomal protein 41kDa |
| chr7_-_79082867 | 7.12 |
ENST00000419488.1
ENST00000354212.4 |
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr19_-_13617037 | 7.11 |
ENST00000360228.5
|
CACNA1A
|
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit |
| chr16_+_7382745 | 7.00 |
ENST00000436368.2
ENST00000311745.5 ENST00000355637.4 ENST00000340209.4 |
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr17_+_12692774 | 6.96 |
ENST00000379672.5
ENST00000340825.3 |
ARHGAP44
|
Rho GTPase activating protein 44 |
| chr1_+_87797351 | 6.86 |
ENST00000370542.1
|
LMO4
|
LIM domain only 4 |
| chr15_-_83240507 | 6.71 |
ENST00000564522.1
ENST00000398592.2 |
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr21_+_40752170 | 6.64 |
ENST00000333781.5
ENST00000541890.1 |
WRB
|
tryptophan rich basic protein |
| chr1_+_89829610 | 6.64 |
ENST00000370456.4
ENST00000535065.1 |
GBP6
|
guanylate binding protein family, member 6 |
| chr5_+_161274940 | 6.60 |
ENST00000393943.4
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr4_+_128554081 | 6.57 |
ENST00000335251.6
ENST00000296461.5 |
INTU
|
inturned planar cell polarity protein |
| chr1_+_22979676 | 6.51 |
ENST00000432749.2
ENST00000314933.6 |
C1QB
|
complement component 1, q subcomponent, B chain |
| chr15_-_83240553 | 6.34 |
ENST00000423133.2
ENST00000398591.2 |
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chrX_+_102883887 | 6.29 |
ENST00000372625.3
ENST00000372624.3 |
TCEAL1
|
transcription elongation factor A (SII)-like 1 |
| chr21_-_27543425 | 6.29 |
ENST00000448388.2
|
APP
|
amyloid beta (A4) precursor protein |
| chr15_+_43803143 | 6.26 |
ENST00000382031.1
|
MAP1A
|
microtubule-associated protein 1A |
| chr12_+_48516357 | 6.16 |
ENST00000549022.1
ENST00000547587.1 ENST00000312352.7 |
PFKM
|
phosphofructokinase, muscle |
| chrX_-_48858667 | 6.16 |
ENST00000376423.4
ENST00000376441.1 |
GRIPAP1
|
GRIP1 associated protein 1 |
| chr8_-_67341208 | 6.12 |
ENST00000499642.1
|
RP11-346I3.4
|
RP11-346I3.4 |
| chr5_+_161274685 | 6.10 |
ENST00000428797.2
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr1_-_144995002 | 5.90 |
ENST00000369356.4
|
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr7_+_29519486 | 5.87 |
ENST00000409041.4
|
CHN2
|
chimerin 2 |
| chrM_+_12331 | 5.66 |
ENST00000361567.2
|
MT-ND5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr12_+_20848486 | 5.66 |
ENST00000545102.1
|
SLCO1C1
|
solute carrier organic anion transporter family, member 1C1 |
| chr4_-_153332886 | 5.66 |
ENST00000603841.1
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr1_+_28099683 | 5.66 |
ENST00000373943.4
|
STX12
|
syntaxin 12 |
| chr3_-_33700589 | 5.55 |
ENST00000461133.3
ENST00000496954.2 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr5_+_161275320 | 5.52 |
ENST00000437025.2
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr12_-_91539918 | 5.50 |
ENST00000548218.1
|
DCN
|
decorin |
| chr11_-_71639670 | 5.40 |
ENST00000533047.1
ENST00000529844.1 |
RP11-849H4.2
|
Putative short transient receptor potential channel 2-like protein |
| chr12_+_53443963 | 5.34 |
ENST00000546602.1
ENST00000552570.1 ENST00000549700.1 |
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2) |
| chr12_+_53773944 | 5.32 |
ENST00000551969.1
ENST00000327443.4 |
SP1
|
Sp1 transcription factor |
| chr9_+_72002837 | 5.26 |
ENST00000377216.3
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chr4_+_114214125 | 5.23 |
ENST00000509550.1
|
ANK2
|
ankyrin 2, neuronal |
| chr12_+_56477093 | 5.20 |
ENST00000549672.1
ENST00000415288.2 |
ERBB3
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr3_-_185826855 | 5.12 |
ENST00000306376.5
|
ETV5
|
ets variant 5 |
| chr11_-_71639480 | 5.04 |
ENST00000529513.1
|
RP11-849H4.2
|
Putative short transient receptor potential channel 2-like protein |
| chr1_+_84630053 | 5.01 |
ENST00000394838.2
ENST00000370682.3 ENST00000432111.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr8_+_125954281 | 4.98 |
ENST00000510897.2
ENST00000533286.1 |
LINC00964
|
long intergenic non-protein coding RNA 964 |
| chr2_-_50201327 | 4.96 |
ENST00000412315.1
|
NRXN1
|
neurexin 1 |
| chr12_+_20848282 | 4.93 |
ENST00000545604.1
|
SLCO1C1
|
solute carrier organic anion transporter family, member 1C1 |
| chr2_+_173600565 | 4.88 |
ENST00000397081.3
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr2_+_173600514 | 4.81 |
ENST00000264111.6
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr12_-_71182695 | 4.80 |
ENST00000342084.4
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr19_+_47759716 | 4.73 |
ENST00000221922.6
|
CCDC9
|
coiled-coil domain containing 9 |
| chr5_+_140718396 | 4.72 |
ENST00000394576.2
|
PCDHGA2
|
protocadherin gamma subfamily A, 2 |
| chr13_-_48575443 | 4.71 |
ENST00000378654.3
|
SUCLA2
|
succinate-CoA ligase, ADP-forming, beta subunit |
| chr7_-_100860851 | 4.68 |
ENST00000223127.3
|
PLOD3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 |
| chr17_+_57232690 | 4.68 |
ENST00000262293.4
|
PRR11
|
proline rich 11 |
| chr2_+_173686303 | 4.63 |
ENST00000397087.3
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr1_+_164528866 | 4.63 |
ENST00000420696.2
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr1_+_233765353 | 4.61 |
ENST00000366620.1
|
KCNK1
|
potassium channel, subfamily K, member 1 |
| chr5_+_137774706 | 4.58 |
ENST00000378339.2
ENST00000254901.5 ENST00000506158.1 |
REEP2
|
receptor accessory protein 2 |
| chr19_-_11669960 | 4.53 |
ENST00000589171.1
ENST00000590700.1 ENST00000586683.1 ENST00000593077.1 ENST00000252445.3 |
ELOF1
|
elongation factor 1 homolog (S. cerevisiae) |
| chr2_+_172543919 | 4.50 |
ENST00000452242.1
ENST00000340296.4 |
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2 |
| chr2_-_145277640 | 4.39 |
ENST00000539609.3
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr17_+_39845134 | 4.36 |
ENST00000591776.1
ENST00000469257.1 |
EIF1
|
eukaryotic translation initiation factor 1 |
| chr1_+_84630645 | 4.33 |
ENST00000394839.2
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr19_-_40596767 | 4.31 |
ENST00000599972.1
ENST00000450241.2 ENST00000595687.2 |
ZNF780A
|
zinc finger protein 780A |
| chr11_-_6426635 | 4.27 |
ENST00000608645.1
ENST00000608394.1 ENST00000529519.1 |
APBB1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr3_-_73673991 | 4.27 |
ENST00000308537.4
ENST00000263666.4 |
PDZRN3
|
PDZ domain containing ring finger 3 |
| chr12_+_20848377 | 4.25 |
ENST00000540354.1
ENST00000266509.2 ENST00000381552.1 |
SLCO1C1
|
solute carrier organic anion transporter family, member 1C1 |
| chr11_+_88910620 | 4.13 |
ENST00000263321.5
|
TYR
|
tyrosinase |
| chr18_+_7754957 | 4.12 |
ENST00000400053.4
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M |
| chr8_-_141810634 | 4.12 |
ENST00000521986.1
ENST00000523539.1 ENST00000538769.1 |
PTK2
|
protein tyrosine kinase 2 |
| chr19_+_2977444 | 4.01 |
ENST00000246112.4
ENST00000453329.1 ENST00000482627.1 ENST00000452088.1 |
TLE6
|
transducin-like enhancer of split 6 (E(sp1) homolog, Drosophila) |
| chr19_-_36822595 | 3.99 |
ENST00000585356.1
ENST00000438368.2 ENST00000590622.1 |
LINC00665
|
long intergenic non-protein coding RNA 665 |
| chr17_+_68165657 | 3.96 |
ENST00000243457.3
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2 |
| chrX_-_148586804 | 3.92 |
ENST00000428056.2
ENST00000340855.6 ENST00000370441.4 ENST00000370443.4 |
IDS
|
iduronate 2-sulfatase |
| chr14_+_74004051 | 3.90 |
ENST00000557556.1
|
ACOT1
|
acyl-CoA thioesterase 1 |
| chr3_+_186501336 | 3.90 |
ENST00000323963.5
ENST00000440191.2 ENST00000356531.5 |
EIF4A2
|
eukaryotic translation initiation factor 4A2 |
| chr3_-_165555200 | 3.76 |
ENST00000479451.1
ENST00000540653.1 ENST00000488954.1 ENST00000264381.3 |
BCHE
|
butyrylcholinesterase |
| chr1_-_154150651 | 3.74 |
ENST00000302206.5
|
TPM3
|
tropomyosin 3 |
| chr10_+_82168240 | 3.74 |
ENST00000372187.5
ENST00000372185.1 |
FAM213A
|
family with sequence similarity 213, member A |
| chr5_+_102201722 | 3.60 |
ENST00000274392.9
ENST00000455264.2 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr4_-_186877806 | 3.56 |
ENST00000355634.5
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr3_-_129407535 | 3.56 |
ENST00000432054.2
|
TMCC1
|
transmembrane and coiled-coil domain family 1 |
| chr11_+_65479702 | 3.55 |
ENST00000530446.1
ENST00000534104.1 ENST00000530605.1 ENST00000528198.1 ENST00000531880.1 ENST00000534650.1 |
KAT5
|
K(lysine) acetyltransferase 5 |
| chr15_-_77197620 | 3.50 |
ENST00000565970.1
ENST00000563290.1 ENST00000565372.1 ENST00000564177.1 ENST00000568382.1 ENST00000563919.1 |
SCAPER
|
S-phase cyclin A-associated protein in the ER |
| chr3_-_149470229 | 3.41 |
ENST00000473414.1
|
COMMD2
|
COMM domain containing 2 |
| chr1_+_22963158 | 3.39 |
ENST00000438241.1
|
C1QA
|
complement component 1, q subcomponent, A chain |
| chr18_-_53069419 | 3.36 |
ENST00000570177.2
|
TCF4
|
transcription factor 4 |
| chr9_+_71939488 | 3.33 |
ENST00000455972.1
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chr5_+_140710061 | 3.32 |
ENST00000517417.1
ENST00000378105.3 |
PCDHGA1
|
protocadherin gamma subfamily A, 1 |
| chr15_+_73976715 | 3.32 |
ENST00000558689.1
ENST00000560786.2 ENST00000561213.1 ENST00000563584.1 ENST00000561416.1 |
CD276
|
CD276 molecule |
| chr6_-_91296737 | 3.31 |
ENST00000369332.3
ENST00000369329.3 |
MAP3K7
|
mitogen-activated protein kinase kinase kinase 7 |
| chr4_+_100737954 | 3.31 |
ENST00000296414.7
ENST00000512369.1 |
DAPP1
|
dual adaptor of phosphotyrosine and 3-phosphoinositides |
| chr2_+_130737223 | 3.31 |
ENST00000410061.2
|
RAB6C
|
RAB6C, member RAS oncogene family |
| chr13_-_53422640 | 3.25 |
ENST00000338862.4
ENST00000377942.3 |
PCDH8
|
protocadherin 8 |
| chr2_-_145277569 | 3.25 |
ENST00000303660.4
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr1_+_24742264 | 3.24 |
ENST00000374399.4
ENST00000003912.3 ENST00000358028.4 ENST00000339255.2 |
NIPAL3
|
NIPA-like domain containing 3 |
| chr6_+_22146851 | 3.21 |
ENST00000606197.1
|
CASC15
|
cancer susceptibility candidate 15 (non-protein coding) |
| chr9_+_71944241 | 3.16 |
ENST00000257515.8
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chr5_+_102201509 | 3.14 |
ENST00000348126.2
ENST00000379787.4 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr16_-_75569068 | 3.11 |
ENST00000336257.3
ENST00000565039.1 |
CHST5
|
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 5 |
| chr12_+_122064398 | 3.09 |
ENST00000330079.7
|
ORAI1
|
ORAI calcium release-activated calcium modulator 1 |
| chrX_+_65384052 | 3.05 |
ENST00000336279.5
ENST00000458621.1 |
HEPH
|
hephaestin |
| chr5_+_102201687 | 3.04 |
ENST00000304400.7
|
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr17_-_41977964 | 3.04 |
ENST00000377184.3
|
MPP2
|
membrane protein, palmitoylated 2 (MAGUK p55 subfamily member 2) |
| chr20_-_9819674 | 2.96 |
ENST00000378429.3
|
PAK7
|
p21 protein (Cdc42/Rac)-activated kinase 7 |
| chr13_+_98086445 | 2.89 |
ENST00000245304.4
|
RAP2A
|
RAP2A, member of RAS oncogene family |
| chr7_+_110731062 | 2.88 |
ENST00000308478.5
ENST00000451085.1 ENST00000422987.3 ENST00000421101.1 |
LRRN3
|
leucine rich repeat neuronal 3 |
| chr19_-_54693521 | 2.84 |
ENST00000391754.1
ENST00000245615.1 ENST00000431666.2 |
MBOAT7
|
membrane bound O-acyltransferase domain containing 7 |
| chr18_-_52989217 | 2.82 |
ENST00000570287.2
|
TCF4
|
transcription factor 4 |
| chr11_-_108408895 | 2.81 |
ENST00000443411.1
ENST00000533052.1 |
EXPH5
|
exophilin 5 |
| chr4_+_71588372 | 2.80 |
ENST00000536664.1
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chrX_-_10588595 | 2.74 |
ENST00000423614.1
ENST00000317552.4 |
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr12_-_25150373 | 2.73 |
ENST00000549828.1
|
C12orf77
|
chromosome 12 open reading frame 77 |
| chrX_+_54835493 | 2.73 |
ENST00000396224.1
|
MAGED2
|
melanoma antigen family D, 2 |
| chr13_-_103053946 | 2.73 |
ENST00000376131.4
|
FGF14
|
fibroblast growth factor 14 |
| chr12_+_58138800 | 2.70 |
ENST00000547992.1
ENST00000552816.1 ENST00000547472.1 |
TSPAN31
|
tetraspanin 31 |
| chr2_+_171036635 | 2.69 |
ENST00000484338.2
ENST00000334231.6 |
MYO3B
|
myosin IIIB |
| chr21_-_30257669 | 2.65 |
ENST00000303775.5
ENST00000351429.3 |
N6AMT1
|
N-6 adenine-specific DNA methyltransferase 1 (putative) |
| chrX_-_10588459 | 2.62 |
ENST00000380782.2
|
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr8_+_54764346 | 2.57 |
ENST00000297313.3
ENST00000344277.6 |
RGS20
|
regulator of G-protein signaling 20 |
| chr15_+_75639296 | 2.56 |
ENST00000564500.1
ENST00000355059.4 ENST00000566752.1 |
NEIL1
|
nei endonuclease VIII-like 1 (E. coli) |
| chr14_+_74035763 | 2.48 |
ENST00000238651.5
|
ACOT2
|
acyl-CoA thioesterase 2 |
| chr13_-_48575401 | 2.48 |
ENST00000433022.1
ENST00000544100.1 |
SUCLA2
|
succinate-CoA ligase, ADP-forming, beta subunit |
| chr19_+_41509851 | 2.48 |
ENST00000593831.1
ENST00000330446.5 |
CYP2B6
|
cytochrome P450, family 2, subfamily B, polypeptide 6 |
| chrX_+_65384182 | 2.48 |
ENST00000441993.2
ENST00000419594.1 |
HEPH
|
hephaestin |
| chr6_-_28220002 | 2.47 |
ENST00000377294.2
|
ZKSCAN4
|
zinc finger with KRAB and SCAN domains 4 |
| chr19_-_12886327 | 2.44 |
ENST00000397668.3
ENST00000587178.1 ENST00000264827.5 |
HOOK2
|
hook microtubule-tethering protein 2 |
| chr7_+_90338712 | 2.43 |
ENST00000265741.3
ENST00000406263.1 |
CDK14
|
cyclin-dependent kinase 14 |
| chr8_+_132952112 | 2.42 |
ENST00000520362.1
ENST00000519656.1 |
EFR3A
|
EFR3 homolog A (S. cerevisiae) |
| chr6_+_45296391 | 2.41 |
ENST00000371436.6
ENST00000576263.1 |
RUNX2
|
runt-related transcription factor 2 |
| chr3_+_181429704 | 2.41 |
ENST00000431565.2
ENST00000325404.1 |
SOX2
|
SRY (sex determining region Y)-box 2 |
| chr9_-_90589402 | 2.36 |
ENST00000375871.4
ENST00000605159.1 ENST00000336654.5 |
CDK20
|
cyclin-dependent kinase 20 |
| chr10_-_45474237 | 2.35 |
ENST00000448778.1
ENST00000298295.3 |
C10orf10
|
chromosome 10 open reading frame 10 |
| chr7_+_92158083 | 2.32 |
ENST00000265732.5
ENST00000481551.1 ENST00000496410.1 |
RBM48
|
RNA binding motif protein 48 |
| chr1_-_193155729 | 2.32 |
ENST00000367434.4
|
B3GALT2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
| chr15_+_79165222 | 2.28 |
ENST00000559930.1
|
MORF4L1
|
mortality factor 4 like 1 |
| chrX_+_107683096 | 2.28 |
ENST00000328300.6
ENST00000361603.2 |
COL4A5
|
collagen, type IV, alpha 5 |
| chr9_-_14180778 | 2.27 |
ENST00000380924.1
ENST00000543693.1 |
NFIB
|
nuclear factor I/B |
| chr16_+_66878282 | 2.25 |
ENST00000338437.2
|
CA7
|
carbonic anhydrase VII |
| chr5_+_102455853 | 2.24 |
ENST00000515845.1
ENST00000321521.9 ENST00000507921.1 |
PPIP5K2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr8_+_67579807 | 2.20 |
ENST00000519289.1
ENST00000519561.1 ENST00000521889.1 |
C8orf44-SGK3
C8orf44
|
C8orf44-SGK3 readthrough chromosome 8 open reading frame 44 |
| chr4_-_130692631 | 2.20 |
ENST00000500092.2
ENST00000509105.1 |
RP11-519M16.1
|
RP11-519M16.1 |
| chr8_+_38677850 | 2.18 |
ENST00000518809.1
ENST00000520611.1 |
TACC1
|
transforming, acidic coiled-coil containing protein 1 |
| chr1_+_203595689 | 2.12 |
ENST00000357681.5
|
ATP2B4
|
ATPase, Ca++ transporting, plasma membrane 4 |
| chr5_+_102201430 | 2.12 |
ENST00000438793.3
ENST00000346918.2 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr2_+_237994519 | 2.07 |
ENST00000392008.2
ENST00000409334.1 ENST00000409629.1 |
COPS8
|
COP9 signalosome subunit 8 |
| chr17_-_76870126 | 2.05 |
ENST00000586057.1
|
TIMP2
|
TIMP metallopeptidase inhibitor 2 |
| chr12_+_81110684 | 2.04 |
ENST00000228644.3
|
MYF5
|
myogenic factor 5 |
| chr12_+_32655048 | 2.04 |
ENST00000427716.2
ENST00000266482.3 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr14_+_74003818 | 2.02 |
ENST00000311148.4
|
ACOT1
|
acyl-CoA thioesterase 1 |
| chr8_+_9953061 | 1.99 |
ENST00000522907.1
ENST00000528246.1 |
MSRA
|
methionine sulfoxide reductase A |
| chrX_+_103810874 | 1.97 |
ENST00000372582.1
|
IL1RAPL2
|
interleukin 1 receptor accessory protein-like 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR2E3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 16.0 | 48.0 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 10.8 | 32.5 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 7.9 | 23.8 | GO:1904395 | retinal rod cell differentiation(GO:0060221) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) negative regulation of neuromuscular junction development(GO:1904397) |
| 7.5 | 44.9 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 7.3 | 21.9 | GO:2000864 | estrogen secretion(GO:0035937) estradiol secretion(GO:0035938) regulation of estrogen secretion(GO:2000861) regulation of estradiol secretion(GO:2000864) |
| 5.0 | 25.1 | GO:0035900 | response to isolation stress(GO:0035900) |
| 3.8 | 11.3 | GO:1904647 | response to rotenone(GO:1904647) |
| 3.6 | 10.7 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 3.3 | 26.6 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 3.0 | 11.9 | GO:0001519 | peptide amidation(GO:0001519) protein amidation(GO:0018032) peptide modification(GO:0031179) |
| 2.8 | 14.1 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 2.6 | 25.8 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 2.3 | 9.3 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 2.3 | 27.8 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 2.1 | 6.2 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 2.0 | 8.2 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 1.9 | 9.3 | GO:0097338 | response to clozapine(GO:0097338) |
| 1.8 | 7.2 | GO:0006780 | uroporphyrinogen III biosynthetic process(GO:0006780) uroporphyrinogen III metabolic process(GO:0046502) |
| 1.7 | 15.7 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 1.7 | 5.2 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 1.6 | 13.0 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 1.6 | 24.4 | GO:0015671 | oxygen transport(GO:0015671) |
| 1.6 | 4.7 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 1.5 | 4.6 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 1.4 | 4.3 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 1.4 | 7.1 | GO:1901631 | planar cell polarity pathway involved in axis elongation(GO:0003402) positive regulation of presynaptic membrane organization(GO:1901631) positive regulation of synaptic vesicle clustering(GO:2000809) |
| 1.4 | 11.3 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 1.4 | 15.2 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 1.3 | 3.8 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 1.2 | 6.2 | GO:1905244 | regulation of modification of synaptic structure(GO:1905244) |
| 1.2 | 7.2 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 1.2 | 8.4 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 1.2 | 3.6 | GO:1903249 | regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) negative regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901205) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 1.1 | 5.7 | GO:2000639 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 1.1 | 15.7 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 1.1 | 6.6 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 1.1 | 14.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 1.1 | 8.6 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 1.0 | 4.1 | GO:0043474 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) |
| 1.0 | 5.1 | GO:0098728 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 1.0 | 7.0 | GO:0099563 | negative regulation of Rac protein signal transduction(GO:0035021) modification of synaptic structure(GO:0099563) |
| 0.9 | 18.2 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.9 | 5.3 | GO:1900239 | phenotypic switching(GO:0036166) regulation of phenotypic switching(GO:1900239) |
| 0.9 | 6.9 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.8 | 4.8 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.8 | 2.3 | GO:0060151 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.7 | 0.7 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.7 | 9.6 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.7 | 12.4 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.7 | 24.7 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.7 | 4.0 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.6 | 1.9 | GO:0019474 | L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.6 | 5.5 | GO:0051964 | netrin-activated signaling pathway(GO:0038007) negative regulation of synapse assembly(GO:0051964) |
| 0.6 | 7.8 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.6 | 3.4 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.5 | 1.6 | GO:0008050 | female courtship behavior(GO:0008050) positive regulation of female receptivity(GO:0045925) |
| 0.5 | 1.6 | GO:1902997 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.5 | 2.0 | GO:0072166 | posterior mesonephric tubule development(GO:0072166) negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) |
| 0.5 | 5.4 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.5 | 1.9 | GO:0060084 | regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) synaptic transmission involved in micturition(GO:0060084) |
| 0.5 | 3.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.5 | 5.5 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.5 | 1.8 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.4 | 2.2 | GO:0032849 | positive regulation of cellular pH reduction(GO:0032849) |
| 0.4 | 3.6 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.4 | 3.5 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.4 | 6.6 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.4 | 4.4 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.4 | 1.7 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.4 | 0.4 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.4 | 1.7 | GO:0003095 | pressure natriuresis(GO:0003095) |
| 0.4 | 2.1 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.4 | 4.5 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.4 | 2.4 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.4 | 3.5 | GO:0030091 | protein repair(GO:0030091) |
| 0.4 | 2.7 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.4 | 2.7 | GO:1903421 | regulation of synaptic vesicle recycling(GO:1903421) |
| 0.4 | 1.9 | GO:0030070 | insulin processing(GO:0030070) |
| 0.4 | 3.8 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.4 | 4.5 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.4 | 2.9 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.4 | 11.6 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.3 | 3.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.3 | 4.4 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.3 | 1.3 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.3 | 7.6 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.3 | 2.3 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.3 | 1.6 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.3 | 1.8 | GO:0002865 | negative regulation of acute inflammatory response to antigenic stimulus(GO:0002865) |
| 0.3 | 2.4 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.3 | 2.3 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.3 | 0.9 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.3 | 3.1 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.3 | 1.9 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.3 | 0.5 | GO:0070054 | mRNA splicing via endonucleolytic cleavage and ligation involved in unfolded protein response(GO:0030969) mRNA splicing, via endonucleolytic cleavage and ligation(GO:0070054) mRNA endonucleolytic cleavage involved in unfolded protein response(GO:0070055) |
| 0.3 | 8.3 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.2 | 8.4 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.2 | 0.9 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.2 | 0.5 | GO:0010767 | regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010767) |
| 0.2 | 0.7 | GO:0051673 | pore formation in membrane of other organism(GO:0035915) membrane disruption in other organism(GO:0051673) |
| 0.2 | 0.6 | GO:0098758 | response to interleukin-8(GO:0098758) cellular response to interleukin-8(GO:0098759) |
| 0.2 | 5.5 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.2 | 10.5 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.2 | 3.3 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.2 | 0.8 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.2 | 5.2 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.2 | 10.0 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.2 | 1.0 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.2 | 1.3 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.2 | 4.1 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.2 | 3.9 | GO:0030207 | chondroitin sulfate catabolic process(GO:0030207) |
| 0.2 | 2.0 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.2 | 2.3 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.2 | 0.7 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.2 | 0.5 | GO:1902723 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.2 | 3.9 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.2 | 0.5 | GO:1903413 | cellular response to bile acid(GO:1903413) |
| 0.2 | 0.6 | GO:0072180 | mesonephric duct morphogenesis(GO:0072180) |
| 0.2 | 12.9 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.1 | 1.2 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.1 | 3.6 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.1 | 0.7 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 3.6 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.1 | 2.1 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 0.9 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.1 | 2.0 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 1.4 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.1 | 2.5 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.1 | 19.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 0.9 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 0.7 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.1 | 0.5 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 2.3 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 0.7 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.1 | 0.9 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.1 | 0.9 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.1 | 5.1 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.1 | 8.0 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.1 | 3.3 | GO:0010824 | regulation of centrosome duplication(GO:0010824) |
| 0.1 | 2.8 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 1.8 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.1 | 3.9 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.1 | 1.4 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.1 | 3.4 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.1 | 1.1 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.1 | 1.1 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.1 | 0.3 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.1 | 3.9 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 6.1 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.1 | 3.2 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.1 | 5.7 | GO:0048278 | vesicle docking(GO:0048278) |
| 0.1 | 2.4 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.1 | 0.4 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.1 | 0.4 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) secretory granule localization(GO:0032252) |
| 0.1 | 0.3 | GO:1904845 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.1 | 0.4 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.1 | 0.6 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.1 | 0.1 | GO:0050955 | thermoception(GO:0050955) detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.1 | 1.6 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.1 | 0.7 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.1 | 0.3 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) positive regulation of autophagosome maturation(GO:1901098) |
| 0.1 | 1.0 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 1.3 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 1.5 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.1 | GO:0042713 | sperm ejaculation(GO:0042713) |
| 0.0 | 0.2 | GO:0032053 | ciliary basal body organization(GO:0032053) positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.3 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.0 | 0.6 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 9.3 | GO:0043406 | positive regulation of MAP kinase activity(GO:0043406) |
| 0.0 | 1.4 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 4.0 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 1.5 | GO:0060338 | regulation of type I interferon-mediated signaling pathway(GO:0060338) |
| 0.0 | 1.9 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 1.4 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.3 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.0 | 4.8 | GO:0071346 | cellular response to interferon-gamma(GO:0071346) |
| 0.0 | 0.2 | GO:0045007 | depurination(GO:0045007) |
| 0.0 | 0.6 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.0 | 0.8 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.4 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.2 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.0 | 8.0 | GO:0001558 | regulation of cell growth(GO:0001558) |
| 0.0 | 0.9 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.0 | 0.9 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.8 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.0 | 0.9 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.7 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 1.5 | GO:0006633 | fatty acid biosynthetic process(GO:0006633) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.6 | 48.0 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 2.7 | 8.2 | GO:0034686 | integrin alphav-beta6 complex(GO:0034685) integrin alphav-beta8 complex(GO:0034686) |
| 2.7 | 26.6 | GO:0045180 | basal cortex(GO:0045180) |
| 2.0 | 18.4 | GO:1990812 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 2.0 | 9.9 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 1.6 | 22.6 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 1.2 | 6.2 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 1.2 | 6.2 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 1.1 | 3.4 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 1.0 | 16.3 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.9 | 4.6 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.9 | 7.1 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.9 | 3.6 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.8 | 10.7 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.8 | 5.7 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.8 | 9.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.8 | 13.0 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.7 | 56.7 | GO:0030315 | T-tubule(GO:0030315) |
| 0.7 | 2.0 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.6 | 2.4 | GO:0070695 | FHF complex(GO:0070695) |
| 0.6 | 9.6 | GO:0031045 | dense core granule(GO:0031045) |
| 0.5 | 13.7 | GO:0071437 | invadopodium(GO:0071437) |
| 0.5 | 43.0 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.5 | 5.3 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.5 | 16.0 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.4 | 16.2 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.4 | 12.6 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.4 | 5.5 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.3 | 5.7 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.3 | 45.7 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.3 | 11.3 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.3 | 1.6 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.3 | 3.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.2 | 4.7 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.2 | 3.7 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.2 | 0.9 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.2 | 2.3 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.2 | 22.2 | GO:0043204 | perikaryon(GO:0043204) |
| 0.2 | 0.9 | GO:0071012 | catalytic step 1 spliceosome(GO:0071012) |
| 0.2 | 1.2 | GO:0000235 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.2 | 51.4 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.1 | 1.8 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 1.9 | GO:0097060 | synaptic membrane(GO:0097060) |
| 0.1 | 3.0 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 0.9 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 23.0 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.1 | 27.7 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 7.1 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 2.8 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 4.2 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 0.8 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.1 | 0.5 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.1 | 1.9 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 5.5 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 1.6 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.1 | 3.1 | GO:0031228 | intrinsic component of Golgi membrane(GO:0031228) |
| 0.1 | 4.5 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 0.3 | GO:0030430 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.1 | 7.4 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 27.8 | GO:0031252 | cell leading edge(GO:0031252) |
| 0.1 | 3.3 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 4.1 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.1 | 2.9 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 2.9 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 1.6 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 5.3 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 0.4 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 5.8 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 1.6 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 10.6 | GO:0045121 | membrane raft(GO:0045121) |
| 0.0 | 7.1 | GO:0030016 | myofibril(GO:0030016) |
| 0.0 | 22.2 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 0.4 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 4.0 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 2.7 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.5 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 2.3 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 1.1 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 1.3 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.2 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.3 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.9 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 0.9 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 1.3 | GO:0098793 | presynapse(GO:0098793) |
| 0.0 | 0.7 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 2.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.6 | 48.0 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 8.4 | 25.1 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 6.0 | 23.8 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 3.2 | 44.9 | GO:0031432 | titin binding(GO:0031432) |
| 3.0 | 11.9 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 2.4 | 7.2 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 2.4 | 7.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 2.3 | 18.2 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 2.0 | 27.8 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 1.9 | 11.3 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 1.9 | 24.4 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 1.8 | 14.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 1.6 | 8.2 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 1.6 | 4.7 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) procollagen galactosyltransferase activity(GO:0050211) |
| 1.4 | 14.1 | GO:0051425 | PTB domain binding(GO:0051425) |
| 1.4 | 4.1 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 1.2 | 3.6 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 1.1 | 27.5 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 1.1 | 17.1 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 1.0 | 6.2 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.9 | 3.8 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.8 | 13.0 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.8 | 12.4 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.8 | 2.3 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.8 | 6.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.7 | 15.0 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.7 | 3.5 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.6 | 12.8 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.6 | 5.5 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.6 | 1.8 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.6 | 9.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.6 | 31.3 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.6 | 4.4 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.5 | 1.5 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.5 | 6.2 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.5 | 23.7 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.4 | 1.3 | GO:0008267 | poly-glutamine tract binding(GO:0008267) |
| 0.4 | 2.2 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.4 | 3.1 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.4 | 5.2 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.4 | 1.7 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.4 | 18.1 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.4 | 2.0 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.4 | 24.8 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.4 | 2.0 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.4 | 8.9 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.4 | 7.1 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.3 | 7.8 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.3 | 3.3 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.3 | 11.1 | GO:1990939 | ATP-dependent microtubule motor activity(GO:1990939) |
| 0.3 | 9.6 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.3 | 7.6 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.3 | 4.3 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.2 | 11.1 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.2 | 3.8 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.2 | 1.0 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.2 | 0.9 | GO:0035651 | AP-1 adaptor complex binding(GO:0035650) AP-3 adaptor complex binding(GO:0035651) |
| 0.2 | 3.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.2 | 1.4 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.2 | 5.3 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.2 | 9.1 | GO:0070888 | E-box binding(GO:0070888) |
| 0.2 | 3.9 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.2 | 0.6 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.2 | 2.4 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.2 | 3.9 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.2 | 0.7 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.2 | 7.6 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.2 | 2.5 | GO:0008391 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.2 | 7.1 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.2 | 1.6 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.2 | 4.5 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.2 | 0.6 | GO:0060422 | peptidyl-dipeptidase inhibitor activity(GO:0060422) |
| 0.2 | 3.6 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.7 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.1 | 3.3 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 2.4 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 0.4 | GO:0000035 | acyl binding(GO:0000035) |
| 0.1 | 0.4 | GO:0047223 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,3-N-acetylglucosaminyltransferase activity(GO:0047223) |
| 0.1 | 13.7 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 1.8 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 2.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 1.6 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 1.9 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 0.9 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.1 | 1.3 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.1 | 4.8 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 0.5 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 3.6 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 1.6 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 3.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 7.0 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 1.3 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.1 | 2.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 1.6 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 1.5 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 9.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 1.9 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.1 | 3.8 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.1 | 3.4 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 1.0 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.1 | 0.9 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.1 | 7.5 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.1 | 0.5 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.1 | 5.4 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.1 | 1.8 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 6.9 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 22.7 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 0.1 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.1 | 3.4 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 0.4 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.1 | 0.7 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 1.1 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 3.1 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.1 | 0.7 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 3.5 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 0.2 | GO:0032408 | MutLbeta complex binding(GO:0032406) MutSbeta complex binding(GO:0032408) |
| 0.1 | 3.7 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.5 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.2 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.0 | 3.5 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.8 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 2.7 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 0.9 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 2.4 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 2.8 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 1.6 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) |
| 0.0 | 2.0 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 3.4 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.4 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) acidic amino acid transmembrane transporter activity(GO:0015172) |
| 0.0 | 0.3 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.8 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 1.9 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.2 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.9 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 1.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.7 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.9 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 8.5 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.8 | GO:0005496 | steroid binding(GO:0005496) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 59.3 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.8 | 33.8 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.8 | 23.8 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.5 | 17.8 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.4 | 30.6 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.3 | 14.1 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.3 | 6.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.3 | 14.6 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.2 | 11.0 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.2 | 4.9 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.2 | 10.5 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.2 | 10.5 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 5.1 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 2.3 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 3.3 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.1 | 4.8 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 4.5 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 2.6 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 0.7 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.1 | 0.9 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 1.3 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 1.7 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.1 | 8.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.1 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 2.0 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 4.3 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.9 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 3.0 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 8.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.4 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.9 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 1.3 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.5 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.5 | PID RHOA PATHWAY | RhoA signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 48.0 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 1.2 | 28.6 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.9 | 9.9 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.7 | 14.6 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.6 | 16.2 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.6 | 14.1 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.6 | 11.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.6 | 18.2 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.5 | 8.2 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.5 | 21.7 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.5 | 3.3 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.4 | 14.6 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.3 | 9.4 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.3 | 3.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.3 | 4.8 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.3 | 7.1 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.3 | 7.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 41.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.2 | 5.7 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.2 | 4.4 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.2 | 6.8 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.2 | 5.5 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.2 | 3.8 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.2 | 5.7 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 2.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 13.1 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 0.4 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.1 | 2.5 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.1 | 6.1 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 3.9 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 1.9 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.1 | 3.1 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.1 | 11.2 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 1.5 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.1 | 4.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 10.7 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 7.1 | REACTOME REGULATION OF INSULIN SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.1 | 1.6 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 4.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 4.8 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 2.3 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 0.9 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 2.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 1.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 3.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 1.1 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 1.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 1.4 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 1.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.4 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.7 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.0 | 0.6 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.9 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.0 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.0 | 0.4 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.9 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.0 | 0.4 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.9 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.8 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.3 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.0 | 0.2 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.5 | REACTOME SIGNALING BY ERBB4 | Genes involved in Signaling by ERBB4 |