Project
averaged GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
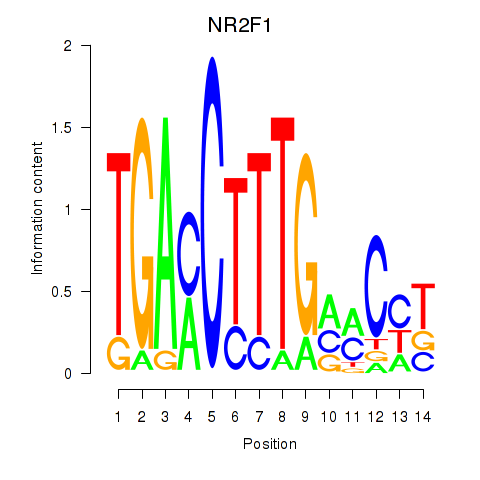
Results for NR2F1
Z-value: 0.74
Transcription factors associated with NR2F1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR2F1
|
ENSG00000175745.7 | nuclear receptor subfamily 2 group F member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR2F1 | hg19_v2_chr5_+_92919043_92919082 | 0.08 | 2.3e-01 | Click! |
Activity profile of NR2F1 motif
Sorted Z-values of NR2F1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_222846 | 13.93 |
ENST00000251595.6
ENST00000397806.1 |
HBA2
|
hemoglobin, alpha 2 |
| chr9_+_139873264 | 7.48 |
ENST00000446677.1
|
PTGDS
|
prostaglandin D2 synthase 21kDa (brain) |
| chr11_-_111794446 | 6.62 |
ENST00000527950.1
|
CRYAB
|
crystallin, alpha B |
| chr19_+_1205740 | 5.46 |
ENST00000326873.7
|
STK11
|
serine/threonine kinase 11 |
| chr4_+_159593418 | 5.46 |
ENST00000507475.1
ENST00000307738.5 |
ETFDH
|
electron-transferring-flavoprotein dehydrogenase |
| chr19_-_39390440 | 5.34 |
ENST00000249396.7
ENST00000414941.1 ENST00000392081.2 |
SIRT2
|
sirtuin 2 |
| chr14_-_21492251 | 5.12 |
ENST00000554398.1
|
NDRG2
|
NDRG family member 2 |
| chr14_-_21492113 | 4.62 |
ENST00000554094.1
|
NDRG2
|
NDRG family member 2 |
| chr19_-_1174226 | 4.48 |
ENST00000587024.1
ENST00000361757.3 |
SBNO2
|
strawberry notch homolog 2 (Drosophila) |
| chr8_-_56685859 | 4.46 |
ENST00000523423.1
ENST00000523073.1 ENST00000519784.1 ENST00000434581.2 ENST00000519780.1 ENST00000521229.1 ENST00000522576.1 ENST00000523180.1 ENST00000522090.1 |
TMEM68
|
transmembrane protein 68 |
| chr13_+_52586517 | 4.43 |
ENST00000523764.1
ENST00000521508.1 |
ALG11
|
ALG11, alpha-1,2-mannosyltransferase |
| chr5_-_42812143 | 4.42 |
ENST00000514985.1
|
SEPP1
|
selenoprotein P, plasma, 1 |
| chr5_-_42811986 | 4.42 |
ENST00000511224.1
ENST00000507920.1 ENST00000510965.1 |
SEPP1
|
selenoprotein P, plasma, 1 |
| chr14_-_23285069 | 4.37 |
ENST00000554758.1
ENST00000397528.4 |
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr4_+_159593271 | 4.31 |
ENST00000512251.1
ENST00000511912.1 |
ETFDH
|
electron-transferring-flavoprotein dehydrogenase |
| chr2_+_113763031 | 4.27 |
ENST00000259211.6
|
IL36A
|
interleukin 36, alpha |
| chr8_-_56685966 | 4.26 |
ENST00000334667.2
|
TMEM68
|
transmembrane protein 68 |
| chr20_+_56136136 | 4.18 |
ENST00000319441.4
ENST00000543666.1 |
PCK1
|
phosphoenolpyruvate carboxykinase 1 (soluble) |
| chr16_+_30710462 | 4.14 |
ENST00000262518.4
ENST00000395059.2 ENST00000344771.4 |
SRCAP
|
Snf2-related CREBBP activator protein |
| chr14_-_23285011 | 4.13 |
ENST00000397532.3
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr1_+_160085501 | 4.12 |
ENST00000361216.3
|
ATP1A2
|
ATPase, Na+/K+ transporting, alpha 2 polypeptide |
| chr17_-_42992856 | 4.05 |
ENST00000588316.1
ENST00000435360.2 ENST00000586793.1 ENST00000588735.1 ENST00000588037.1 ENST00000592320.1 ENST00000253408.5 |
GFAP
|
glial fibrillary acidic protein |
| chr8_+_144816303 | 4.03 |
ENST00000533004.1
|
FAM83H-AS1
|
FAM83H antisense RNA 1 (head to head) |
| chr17_+_7531281 | 3.92 |
ENST00000575729.1
ENST00000340624.5 |
SHBG
|
sex hormone-binding globulin |
| chr1_-_36235559 | 3.89 |
ENST00000251195.5
|
CLSPN
|
claspin |
| chr18_+_54318566 | 3.88 |
ENST00000589935.1
ENST00000357574.3 |
WDR7
|
WD repeat domain 7 |
| chr2_+_220491973 | 3.80 |
ENST00000358055.3
|
SLC4A3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr12_-_7125770 | 3.78 |
ENST00000261407.4
|
LPCAT3
|
lysophosphatidylcholine acyltransferase 3 |
| chr4_-_681114 | 3.66 |
ENST00000503156.1
|
MFSD7
|
major facilitator superfamily domain containing 7 |
| chr4_-_120243545 | 3.57 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr11_-_45939565 | 3.55 |
ENST00000525192.1
ENST00000378750.5 |
PEX16
|
peroxisomal biogenesis factor 16 |
| chr11_-_45939374 | 3.50 |
ENST00000533151.1
ENST00000241041.3 |
PEX16
|
peroxisomal biogenesis factor 16 |
| chr7_-_100844193 | 3.49 |
ENST00000440203.2
ENST00000379423.3 ENST00000223114.4 |
MOGAT3
|
monoacylglycerol O-acyltransferase 3 |
| chr7_+_150264365 | 3.46 |
ENST00000255945.2
ENST00000461940.1 |
GIMAP4
|
GTPase, IMAP family member 4 |
| chrX_-_43741594 | 3.42 |
ENST00000536181.1
ENST00000378069.4 |
MAOB
|
monoamine oxidase B |
| chr10_+_114133773 | 3.40 |
ENST00000354655.4
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5 |
| chr17_-_34329084 | 3.36 |
ENST00000354059.4
ENST00000536149.1 |
CCL15
CCL14
|
chemokine (C-C motif) ligand 15 chemokine (C-C motif) ligand 14 |
| chr6_+_7108210 | 3.36 |
ENST00000467782.1
ENST00000334984.6 ENST00000349384.6 |
RREB1
|
ras responsive element binding protein 1 |
| chr18_+_54318616 | 3.24 |
ENST00000254442.3
|
WDR7
|
WD repeat domain 7 |
| chr4_+_2965307 | 3.19 |
ENST00000398051.4
ENST00000503518.2 ENST00000398052.4 ENST00000345167.6 ENST00000504933.1 ENST00000442472.2 |
GRK4
|
G protein-coupled receptor kinase 4 |
| chr10_+_96443204 | 3.12 |
ENST00000339022.5
|
CYP2C18
|
cytochrome P450, family 2, subfamily C, polypeptide 18 |
| chr10_+_96443378 | 3.07 |
ENST00000285979.6
|
CYP2C18
|
cytochrome P450, family 2, subfamily C, polypeptide 18 |
| chr2_+_220492116 | 3.04 |
ENST00000373760.2
|
SLC4A3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr1_+_156123359 | 3.03 |
ENST00000368284.1
ENST00000368286.2 ENST00000438830.1 |
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr19_+_11877838 | 3.00 |
ENST00000357901.4
ENST00000454339.2 |
ZNF441
|
zinc finger protein 441 |
| chr19_+_58790314 | 2.98 |
ENST00000196548.5
ENST00000608843.1 |
ZNF8
ZNF8
|
Zinc finger protein 8 zinc finger protein 8 |
| chr17_-_16472483 | 2.96 |
ENST00000395824.1
ENST00000448349.2 ENST00000395825.3 |
ZNF287
|
zinc finger protein 287 |
| chr19_-_633576 | 2.93 |
ENST00000588649.2
|
POLRMT
|
polymerase (RNA) mitochondrial (DNA directed) |
| chr18_+_29769978 | 2.92 |
ENST00000269202.6
ENST00000581447.1 |
MEP1B
|
meprin A, beta |
| chr7_-_115608304 | 2.91 |
ENST00000457268.1
|
TFEC
|
transcription factor EC |
| chr1_+_156123318 | 2.89 |
ENST00000368285.3
|
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr22_+_47158518 | 2.87 |
ENST00000337137.4
ENST00000380995.1 ENST00000407381.3 |
TBC1D22A
|
TBC1 domain family, member 22A |
| chr2_+_220492373 | 2.85 |
ENST00000317151.3
|
SLC4A3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr2_+_162016827 | 2.82 |
ENST00000429217.1
ENST00000406287.1 ENST00000402568.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr4_-_11431188 | 2.82 |
ENST00000510712.1
|
HS3ST1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 1 |
| chr12_+_113659234 | 2.81 |
ENST00000551096.1
ENST00000551099.1 ENST00000335509.6 ENST00000552897.1 ENST00000550785.1 ENST00000549279.1 |
TPCN1
|
two pore segment channel 1 |
| chr12_-_6665200 | 2.71 |
ENST00000336604.4
ENST00000396840.2 ENST00000356896.4 |
IFFO1
|
intermediate filament family orphan 1 |
| chr6_+_7107830 | 2.70 |
ENST00000379933.3
|
RREB1
|
ras responsive element binding protein 1 |
| chr17_-_2614927 | 2.69 |
ENST00000435359.1
|
CLUH
|
clustered mitochondria (cluA/CLU1) homolog |
| chr2_-_28113217 | 2.67 |
ENST00000444339.2
|
RBKS
|
ribokinase |
| chr3_+_37284824 | 2.67 |
ENST00000431105.1
|
GOLGA4
|
golgin A4 |
| chr19_-_41256207 | 2.64 |
ENST00000598485.2
ENST00000470681.1 ENST00000339153.3 ENST00000598729.1 |
C19orf54
|
chromosome 19 open reading frame 54 |
| chr15_+_75105170 | 2.62 |
ENST00000379709.3
|
LMAN1L
|
lectin, mannose-binding, 1 like |
| chr9_+_34458771 | 2.59 |
ENST00000437363.1
ENST00000242317.4 |
DNAI1
|
dynein, axonemal, intermediate chain 1 |
| chr1_+_158149737 | 2.57 |
ENST00000368171.3
|
CD1D
|
CD1d molecule |
| chr10_-_103815874 | 2.57 |
ENST00000370033.4
ENST00000311122.5 |
C10orf76
|
chromosome 10 open reading frame 76 |
| chr10_-_17171817 | 2.55 |
ENST00000377833.4
|
CUBN
|
cubilin (intrinsic factor-cobalamin receptor) |
| chr1_-_173886491 | 2.55 |
ENST00000367698.3
|
SERPINC1
|
serpin peptidase inhibitor, clade C (antithrombin), member 1 |
| chr1_-_169555779 | 2.54 |
ENST00000367797.3
ENST00000367796.3 |
F5
|
coagulation factor V (proaccelerin, labile factor) |
| chr12_-_56727487 | 2.49 |
ENST00000548043.1
ENST00000425394.2 |
PAN2
|
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr2_+_219433281 | 2.49 |
ENST00000273064.6
ENST00000509807.2 ENST00000542068.1 |
RQCD1
|
RCD1 required for cell differentiation1 homolog (S. pombe) |
| chr22_+_47158578 | 2.47 |
ENST00000355704.3
|
TBC1D22A
|
TBC1 domain family, member 22A |
| chr19_+_49713991 | 2.39 |
ENST00000597316.1
|
TRPM4
|
transient receptor potential cation channel, subfamily M, member 4 |
| chr20_+_57875658 | 2.34 |
ENST00000371025.3
|
EDN3
|
endothelin 3 |
| chr19_+_47813110 | 2.34 |
ENST00000355085.3
|
C5AR1
|
complement component 5a receptor 1 |
| chr2_+_68592305 | 2.32 |
ENST00000234313.7
|
PLEK
|
pleckstrin |
| chr6_+_142468361 | 2.32 |
ENST00000367630.4
|
VTA1
|
vesicle (multivesicular body) trafficking 1 |
| chr19_-_8408139 | 2.31 |
ENST00000330915.3
ENST00000593649.1 ENST00000595639.1 |
KANK3
|
KN motif and ankyrin repeat domains 3 |
| chr2_+_220492287 | 2.31 |
ENST00000273063.6
ENST00000373762.3 |
SLC4A3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr22_-_29784519 | 2.31 |
ENST00000357586.2
ENST00000356015.2 ENST00000432560.2 ENST00000317368.7 |
AP1B1
|
adaptor-related protein complex 1, beta 1 subunit |
| chr22_-_51021397 | 2.30 |
ENST00000406938.2
|
CHKB
|
choline kinase beta |
| chr4_+_667686 | 2.24 |
ENST00000505477.1
|
MYL5
|
myosin, light chain 5, regulatory |
| chr10_-_75634219 | 2.23 |
ENST00000305762.7
|
CAMK2G
|
calcium/calmodulin-dependent protein kinase II gamma |
| chr1_-_23520755 | 2.23 |
ENST00000314113.3
|
HTR1D
|
5-hydroxytryptamine (serotonin) receptor 1D, G protein-coupled |
| chr14_+_101299520 | 2.21 |
ENST00000455531.1
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr19_-_36304201 | 2.20 |
ENST00000301175.3
|
PRODH2
|
proline dehydrogenase (oxidase) 2 |
| chr17_+_1646130 | 2.19 |
ENST00000453066.1
ENST00000324015.3 ENST00000450523.2 ENST00000453723.1 ENST00000382061.4 |
SERPINF2
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2 |
| chr12_-_56727676 | 2.18 |
ENST00000547572.1
ENST00000257931.5 ENST00000440411.3 |
PAN2
|
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr12_+_58013693 | 2.17 |
ENST00000320442.4
ENST00000379218.2 |
SLC26A10
|
solute carrier family 26, member 10 |
| chr1_+_145438469 | 2.17 |
ENST00000369317.4
|
TXNIP
|
thioredoxin interacting protein |
| chr1_-_109618566 | 2.16 |
ENST00000338366.5
|
TAF13
|
TAF13 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 18kDa |
| chr19_-_1021113 | 2.15 |
ENST00000333175.5
ENST00000356663.3 |
TMEM259
|
transmembrane protein 259 |
| chr20_+_57875457 | 2.13 |
ENST00000337938.2
ENST00000311585.7 ENST00000371028.2 |
EDN3
|
endothelin 3 |
| chr19_+_39616410 | 2.12 |
ENST00000602004.1
ENST00000599470.1 ENST00000321944.4 ENST00000593480.1 ENST00000358301.3 ENST00000593690.1 ENST00000599386.1 |
PAK4
|
p21 protein (Cdc42/Rac)-activated kinase 4 |
| chr17_+_42264395 | 2.09 |
ENST00000587989.1
ENST00000590235.1 |
TMUB2
|
transmembrane and ubiquitin-like domain containing 2 |
| chr16_+_83932684 | 2.08 |
ENST00000262430.4
|
MLYCD
|
malonyl-CoA decarboxylase |
| chr1_+_205473720 | 2.07 |
ENST00000429964.2
ENST00000506784.1 ENST00000360066.2 |
CDK18
|
cyclin-dependent kinase 18 |
| chr11_-_73720276 | 2.06 |
ENST00000348534.4
|
UCP3
|
uncoupling protein 3 (mitochondrial, proton carrier) |
| chr1_+_196857144 | 2.06 |
ENST00000367416.2
ENST00000251424.4 ENST00000367418.2 |
CFHR4
|
complement factor H-related 4 |
| chr12_-_14996355 | 2.06 |
ENST00000228936.4
|
ART4
|
ADP-ribosyltransferase 4 (Dombrock blood group) |
| chr2_+_219646462 | 2.04 |
ENST00000258415.4
|
CYP27A1
|
cytochrome P450, family 27, subfamily A, polypeptide 1 |
| chr2_+_162016916 | 2.00 |
ENST00000405852.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr1_-_23521222 | 1.98 |
ENST00000374619.1
|
HTR1D
|
5-hydroxytryptamine (serotonin) receptor 1D, G protein-coupled |
| chr22_+_45898712 | 1.93 |
ENST00000455233.1
ENST00000348697.2 ENST00000402984.3 ENST00000262722.7 ENST00000327858.6 ENST00000442170.2 ENST00000340923.5 ENST00000439835.1 |
FBLN1
|
fibulin 1 |
| chr14_+_58754751 | 1.88 |
ENST00000598233.1
|
AL132989.1
|
AL132989.1 |
| chr1_-_229569834 | 1.85 |
ENST00000366684.3
ENST00000366683.2 |
ACTA1
|
actin, alpha 1, skeletal muscle |
| chr4_+_69962185 | 1.83 |
ENST00000305231.7
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr4_+_69962212 | 1.82 |
ENST00000508661.1
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr1_+_43855560 | 1.80 |
ENST00000562955.1
|
SZT2
|
seizure threshold 2 homolog (mouse) |
| chr11_-_124767693 | 1.80 |
ENST00000533054.1
|
ROBO4
|
roundabout, axon guidance receptor, homolog 4 (Drosophila) |
| chr6_+_31926857 | 1.79 |
ENST00000375394.2
ENST00000544581.1 |
SKIV2L
|
superkiller viralicidic activity 2-like (S. cerevisiae) |
| chr13_+_100741269 | 1.78 |
ENST00000376286.4
ENST00000376279.3 ENST00000376285.1 |
PCCA
|
propionyl CoA carboxylase, alpha polypeptide |
| chrX_+_138612889 | 1.75 |
ENST00000218099.2
ENST00000394090.2 |
F9
|
coagulation factor IX |
| chr19_+_589893 | 1.72 |
ENST00000251287.2
|
HCN2
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 2 |
| chr10_+_96522361 | 1.71 |
ENST00000371321.3
|
CYP2C19
|
cytochrome P450, family 2, subfamily C, polypeptide 19 |
| chr4_-_11431389 | 1.71 |
ENST00000002596.5
|
HS3ST1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 1 |
| chr16_-_31439735 | 1.69 |
ENST00000287490.4
|
COX6A2
|
cytochrome c oxidase subunit VIa polypeptide 2 |
| chr14_+_24590560 | 1.67 |
ENST00000558325.1
|
RP11-468E2.6
|
RP11-468E2.6 |
| chr9_+_6716478 | 1.67 |
ENST00000452643.1
|
RP11-390F4.3
|
RP11-390F4.3 |
| chr20_+_53092123 | 1.63 |
ENST00000262593.5
|
DOK5
|
docking protein 5 |
| chr1_-_11107280 | 1.62 |
ENST00000400897.3
ENST00000400898.3 |
MASP2
|
mannan-binding lectin serine peptidase 2 |
| chr9_+_91933407 | 1.61 |
ENST00000375807.3
ENST00000339901.4 |
SECISBP2
|
SECIS binding protein 2 |
| chr2_+_162016804 | 1.60 |
ENST00000392749.2
ENST00000440506.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr19_-_38806560 | 1.58 |
ENST00000591755.1
ENST00000337679.8 ENST00000339413.6 |
YIF1B
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr6_-_33771757 | 1.57 |
ENST00000507738.1
ENST00000266003.5 ENST00000430124.2 |
MLN
|
motilin |
| chr19_-_51568324 | 1.56 |
ENST00000595547.1
ENST00000335422.3 ENST00000595793.1 ENST00000596955.1 |
KLK13
|
kallikrein-related peptidase 13 |
| chr19_+_36195467 | 1.55 |
ENST00000426659.2
|
ZBTB32
|
zinc finger and BTB domain containing 32 |
| chr9_+_34989638 | 1.54 |
ENST00000453597.3
ENST00000335998.3 ENST00000312316.5 |
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5 |
| chr3_+_186560476 | 1.53 |
ENST00000320741.2
ENST00000444204.2 |
ADIPOQ
|
adiponectin, C1Q and collagen domain containing |
| chr6_+_30594619 | 1.53 |
ENST00000318999.7
ENST00000376485.4 ENST00000376478.2 ENST00000319027.5 ENST00000376483.4 ENST00000329992.8 ENST00000330083.5 |
ATAT1
|
alpha tubulin acetyltransferase 1 |
| chr3_+_186560462 | 1.52 |
ENST00000412955.2
|
ADIPOQ
|
adiponectin, C1Q and collagen domain containing |
| chr22_+_25003626 | 1.50 |
ENST00000451366.1
ENST00000406383.2 ENST00000428855.1 |
GGT1
|
gamma-glutamyltransferase 1 |
| chr10_-_74114714 | 1.49 |
ENST00000338820.3
ENST00000394903.2 ENST00000444643.2 |
DNAJB12
|
DnaJ (Hsp40) homolog, subfamily B, member 12 |
| chr19_+_41594377 | 1.49 |
ENST00000330436.3
|
CYP2A13
|
cytochrome P450, family 2, subfamily A, polypeptide 13 |
| chr19_+_10197463 | 1.49 |
ENST00000590378.1
ENST00000397881.3 |
C19orf66
|
chromosome 19 open reading frame 66 |
| chr4_+_100495864 | 1.49 |
ENST00000265517.5
ENST00000422897.2 |
MTTP
|
microsomal triglyceride transfer protein |
| chr16_-_4292071 | 1.48 |
ENST00000399609.3
|
SRL
|
sarcalumenin |
| chr4_+_76649797 | 1.47 |
ENST00000538159.1
ENST00000514213.2 |
USO1
|
USO1 vesicle transport factor |
| chr1_+_156611704 | 1.45 |
ENST00000329117.5
|
BCAN
|
brevican |
| chr19_-_16045619 | 1.45 |
ENST00000402119.4
|
CYP4F11
|
cytochrome P450, family 4, subfamily F, polypeptide 11 |
| chr15_-_75660919 | 1.44 |
ENST00000569482.1
ENST00000565683.1 ENST00000561615.1 ENST00000563622.1 ENST00000568374.1 ENST00000566256.1 ENST00000267978.5 |
MAN2C1
|
mannosidase, alpha, class 2C, member 1 |
| chr12_-_53074182 | 1.41 |
ENST00000252244.3
|
KRT1
|
keratin 1 |
| chr19_-_42133420 | 1.40 |
ENST00000221954.2
ENST00000600925.1 |
CEACAM4
|
carcinoembryonic antigen-related cell adhesion molecule 4 |
| chr18_-_44336754 | 1.40 |
ENST00000538168.1
ENST00000536490.1 |
ST8SIA5
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 5 |
| chr15_-_41120896 | 1.40 |
ENST00000299174.5
ENST00000427255.2 |
PPP1R14D
|
protein phosphatase 1, regulatory (inhibitor) subunit 14D |
| chr7_-_37956409 | 1.37 |
ENST00000436072.2
|
SFRP4
|
secreted frizzled-related protein 4 |
| chr3_+_113616317 | 1.37 |
ENST00000440446.2
ENST00000488680.1 |
GRAMD1C
|
GRAM domain containing 1C |
| chr6_+_33172407 | 1.35 |
ENST00000374662.3
|
HSD17B8
|
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr2_-_222436988 | 1.34 |
ENST00000409854.1
ENST00000281821.2 ENST00000392071.4 ENST00000443796.1 |
EPHA4
|
EPH receptor A4 |
| chr19_-_16045665 | 1.33 |
ENST00000248041.8
|
CYP4F11
|
cytochrome P450, family 4, subfamily F, polypeptide 11 |
| chr11_-_82746587 | 1.29 |
ENST00000528379.1
ENST00000534103.1 |
RAB30
|
RAB30, member RAS oncogene family |
| chr11_+_72929402 | 1.28 |
ENST00000393596.2
|
P2RY2
|
purinergic receptor P2Y, G-protein coupled, 2 |
| chr2_-_169887827 | 1.27 |
ENST00000263817.6
|
ABCB11
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11 |
| chr19_-_51472222 | 1.25 |
ENST00000376851.3
|
KLK6
|
kallikrein-related peptidase 6 |
| chr2_-_70780770 | 1.25 |
ENST00000444975.1
ENST00000445399.1 ENST00000418333.2 |
TGFA
|
transforming growth factor, alpha |
| chr2_+_219283815 | 1.24 |
ENST00000248444.5
ENST00000454069.1 ENST00000392114.2 |
VIL1
|
villin 1 |
| chr2_-_152382500 | 1.23 |
ENST00000434685.1
|
NEB
|
nebulin |
| chr1_+_110162448 | 1.22 |
ENST00000342115.4
ENST00000469039.2 ENST00000474459.1 ENST00000528667.1 |
AMPD2
|
adenosine monophosphate deaminase 2 |
| chr12_-_53594227 | 1.21 |
ENST00000550743.2
|
ITGB7
|
integrin, beta 7 |
| chr19_+_54606145 | 1.21 |
ENST00000485876.1
ENST00000391762.1 ENST00000471292.1 ENST00000391763.3 ENST00000391764.3 ENST00000303553.5 |
NDUFA3
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 3, 9kDa |
| chr1_+_181003067 | 1.20 |
ENST00000434571.2
ENST00000367579.3 ENST00000282990.6 ENST00000367580.5 |
MR1
|
major histocompatibility complex, class I-related |
| chr2_-_165698662 | 1.17 |
ENST00000194871.6
ENST00000445474.2 |
COBLL1
|
cordon-bleu WH2 repeat protein-like 1 |
| chr2_+_44502597 | 1.16 |
ENST00000260649.6
ENST00000409387.1 |
SLC3A1
|
solute carrier family 3 (amino acid transporter heavy chain), member 1 |
| chr6_-_31864977 | 1.15 |
ENST00000395728.3
ENST00000375528.4 |
EHMT2
|
euchromatic histone-lysine N-methyltransferase 2 |
| chr11_+_72929319 | 1.14 |
ENST00000393597.2
ENST00000311131.2 |
P2RY2
|
purinergic receptor P2Y, G-protein coupled, 2 |
| chr20_-_62130474 | 1.14 |
ENST00000217182.3
|
EEF1A2
|
eukaryotic translation elongation factor 1 alpha 2 |
| chr11_+_63137251 | 1.13 |
ENST00000310969.4
ENST00000279178.3 |
SLC22A9
|
solute carrier family 22 (organic anion transporter), member 9 |
| chr2_+_138721850 | 1.10 |
ENST00000329366.4
ENST00000280097.3 |
HNMT
|
histamine N-methyltransferase |
| chr12_+_121416489 | 1.10 |
ENST00000541395.1
ENST00000544413.1 |
HNF1A
|
HNF1 homeobox A |
| chr4_-_668108 | 1.10 |
ENST00000304312.4
|
ATP5I
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit E |
| chr1_+_196946680 | 1.09 |
ENST00000256785.4
|
CFHR5
|
complement factor H-related 5 |
| chr13_+_113777105 | 1.08 |
ENST00000409306.1
ENST00000375551.3 ENST00000375559.3 |
F10
|
coagulation factor X |
| chr20_+_57875758 | 1.07 |
ENST00000395654.3
|
EDN3
|
endothelin 3 |
| chr7_+_121513143 | 1.07 |
ENST00000393386.2
|
PTPRZ1
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1 |
| chr2_+_48796120 | 1.07 |
ENST00000394754.1
|
STON1-GTF2A1L
|
STON1-GTF2A1L readthrough |
| chr11_-_116708302 | 1.05 |
ENST00000375320.1
ENST00000359492.2 ENST00000375329.2 ENST00000375323.1 |
APOA1
|
apolipoprotein A-I |
| chrX_+_66764375 | 1.02 |
ENST00000374690.3
|
AR
|
androgen receptor |
| chr7_-_99332719 | 1.00 |
ENST00000336374.2
|
CYP3A7
|
cytochrome P450, family 3, subfamily A, polypeptide 7 |
| chr6_-_30080876 | 0.99 |
ENST00000376734.3
|
TRIM31
|
tripartite motif containing 31 |
| chr17_-_10372875 | 0.99 |
ENST00000255381.2
|
MYH4
|
myosin, heavy chain 4, skeletal muscle |
| chr19_+_10196981 | 0.99 |
ENST00000591813.1
|
C19orf66
|
chromosome 19 open reading frame 66 |
| chr1_+_62439037 | 0.95 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr10_+_104503727 | 0.94 |
ENST00000448841.1
|
WBP1L
|
WW domain binding protein 1-like |
| chr12_+_6494285 | 0.94 |
ENST00000541102.1
|
LTBR
|
lymphotoxin beta receptor (TNFR superfamily, member 3) |
| chr19_+_5823813 | 0.93 |
ENST00000303212.2
|
NRTN
|
neurturin |
| chr10_-_104211294 | 0.93 |
ENST00000239125.1
|
C10orf95
|
chromosome 10 open reading frame 95 |
| chr1_+_101185290 | 0.92 |
ENST00000370119.4
ENST00000347652.2 ENST00000294728.2 ENST00000370115.1 |
VCAM1
|
vascular cell adhesion molecule 1 |
| chr12_+_121416340 | 0.91 |
ENST00000257555.6
ENST00000400024.2 |
HNF1A
|
HNF1 homeobox A |
| chr7_-_99277610 | 0.91 |
ENST00000343703.5
ENST00000222982.4 ENST00000439761.1 ENST00000339843.2 |
CYP3A5
|
cytochrome P450, family 3, subfamily A, polypeptide 5 |
| chr1_+_196946664 | 0.89 |
ENST00000367414.5
|
CFHR5
|
complement factor H-related 5 |
| chr5_+_140593509 | 0.88 |
ENST00000341948.4
|
PCDHB13
|
protocadherin beta 13 |
| chr7_+_94536898 | 0.88 |
ENST00000433360.1
ENST00000340694.4 ENST00000424654.1 |
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A |
| chr11_+_67798090 | 0.87 |
ENST00000313468.5
|
NDUFS8
|
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr7_-_74489609 | 0.86 |
ENST00000329959.4
ENST00000503250.2 ENST00000543840.1 |
WBSCR16
|
Williams-Beuren syndrome chromosome region 16 |
| chr16_+_21312170 | 0.86 |
ENST00000338573.5
ENST00000561968.1 |
CRYM-AS1
|
CRYM antisense RNA 1 |
| chr1_-_161102421 | 0.85 |
ENST00000490843.2
ENST00000368006.3 ENST00000392188.1 ENST00000545495.1 |
DEDD
|
death effector domain containing |
| chr11_-_73720122 | 0.85 |
ENST00000426995.2
|
UCP3
|
uncoupling protein 3 (mitochondrial, proton carrier) |
| chr9_-_95896550 | 0.84 |
ENST00000375446.4
|
NINJ1
|
ninjurin 1 |
| chr11_+_113848216 | 0.84 |
ENST00000299961.5
|
HTR3A
|
5-hydroxytryptamine (serotonin) receptor 3A, ionotropic |
| chr11_-_66445219 | 0.83 |
ENST00000525754.1
ENST00000531969.1 ENST00000524637.1 ENST00000531036.2 ENST00000310046.4 |
RBM4B
|
RNA binding motif protein 4B |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR2F1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 7.0 | GO:0032581 | ER-dependent peroxisome organization(GO:0032581) |
| 1.8 | 5.3 | GO:2000777 | positive regulation of oocyte maturation(GO:1900195) negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 1.6 | 9.7 | GO:1990822 | basic amino acid transmembrane transport(GO:1990822) |
| 1.4 | 5.6 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 1.4 | 5.5 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 1.1 | 6.4 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 1.0 | 4.2 | GO:0006114 | glycerol biosynthetic process(GO:0006114) response to methionine(GO:1904640) |
| 1.0 | 3.1 | GO:0097018 | renal albumin absorption(GO:0097018) regulation of renal albumin absorption(GO:2000532) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.9 | 7.3 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.9 | 2.7 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.8 | 4.1 | GO:0051946 | regulation of amino acid uptake involved in synaptic transmission(GO:0051941) regulation of glutamate uptake involved in transmission of nerve impulse(GO:0051946) regulation of L-glutamate import(GO:1900920) |
| 0.8 | 2.3 | GO:0038178 | complement component C5a signaling pathway(GO:0038178) |
| 0.8 | 3.9 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.7 | 4.5 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.7 | 2.9 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.7 | 2.8 | GO:0042361 | menaquinone catabolic process(GO:0042361) vitamin K catabolic process(GO:0042377) |
| 0.7 | 9.8 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.6 | 2.5 | GO:0032571 | response to vitamin K(GO:0032571) |
| 0.6 | 9.7 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.6 | 2.4 | GO:1990736 | positive regulation of cardiac conduction(GO:1903781) positive regulation of atrial cardiac muscle cell action potential(GO:1903949) positive regulation of regulation of vascular smooth muscle cell membrane depolarization(GO:1904199) regulation of vascular smooth muscle cell membrane depolarization(GO:1990736) |
| 0.6 | 1.8 | GO:0035565 | regulation of pronephros size(GO:0035565) renal glucose absorption(GO:0035623) |
| 0.6 | 1.8 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.5 | 4.2 | GO:0014827 | intestine smooth muscle contraction(GO:0014827) |
| 0.5 | 3.4 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.5 | 1.4 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.4 | 1.3 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.4 | 7.5 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.4 | 10.1 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.4 | 1.7 | GO:0010903 | negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) |
| 0.4 | 2.1 | GO:2001293 | malonyl-CoA metabolic process(GO:2001293) |
| 0.4 | 2.8 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.4 | 2.8 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.4 | 2.3 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.4 | 1.9 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.4 | 6.6 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.4 | 2.2 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.3 | 1.0 | GO:0060599 | lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) |
| 0.3 | 2.6 | GO:0016045 | detection of bacterium(GO:0016045) |
| 0.3 | 2.2 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.3 | 1.8 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.3 | 1.2 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.3 | 1.2 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.3 | 2.7 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.3 | 5.2 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.3 | 1.7 | GO:0071321 | cellular response to cGMP(GO:0071321) |
| 0.3 | 3.5 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.3 | 2.5 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.2 | 1.2 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.2 | 4.4 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.2 | 2.4 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.2 | 12.0 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.2 | 3.0 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.2 | 1.4 | GO:0006909 | phagocytosis(GO:0006909) |
| 0.2 | 0.9 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.2 | 1.8 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.2 | 0.9 | GO:1904048 | regulation of spontaneous neurotransmitter secretion(GO:1904048) |
| 0.2 | 2.6 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.2 | 1.5 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.2 | 1.5 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.2 | 1.8 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.2 | 1.0 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.2 | 0.6 | GO:2001045 | closure of optic fissure(GO:0061386) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.2 | 2.3 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.2 | 4.5 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.2 | 1.1 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.2 | 1.4 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.2 | 1.6 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.2 | 1.1 | GO:0070444 | oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.2 | 2.3 | GO:1904903 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.2 | 1.5 | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.2 | 3.7 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.2 | 2.0 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.2 | 5.9 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.2 | 3.8 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.2 | 0.8 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.2 | 0.7 | GO:1902358 | sulfate transmembrane transport(GO:1902358) |
| 0.2 | 0.8 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.2 | 2.9 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.2 | 3.2 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.2 | 2.2 | GO:0002347 | response to tumor cell(GO:0002347) |
| 0.2 | 2.2 | GO:1904294 | positive regulation of ERAD pathway(GO:1904294) |
| 0.1 | 3.4 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.1 | 0.7 | GO:0002442 | serotonin production involved in inflammatory response(GO:0002351) serotonin secretion involved in inflammatory response(GO:0002442) serotonin secretion by platelet(GO:0002554) |
| 0.1 | 0.8 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 0.6 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.1 | 1.1 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 1.5 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 0.6 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.1 | 0.6 | GO:0031337 | cellular response to phosphate starvation(GO:0016036) positive regulation of sulfur amino acid metabolic process(GO:0031337) negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) positive regulation of homocysteine metabolic process(GO:0050668) |
| 0.1 | 2.2 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.1 | 0.9 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.1 | 3.9 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.1 | 4.0 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.1 | 0.6 | GO:0051458 | corticotropin secretion(GO:0051458) |
| 0.1 | 1.6 | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.1 | 0.3 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.1 | 1.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 2.3 | GO:0006525 | arginine metabolic process(GO:0006525) |
| 0.1 | 1.8 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.1 | 1.4 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 1.7 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.6 | GO:0046618 | drug export(GO:0046618) |
| 0.1 | 2.3 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 1.1 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.1 | 3.0 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.1 | 0.6 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.1 | 1.1 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.1 | 0.7 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.1 | 4.3 | GO:0032755 | positive regulation of interleukin-6 production(GO:0032755) |
| 0.1 | 1.0 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.1 | 0.7 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 0.8 | GO:0060033 | anatomical structure regression(GO:0060033) |
| 0.1 | 4.9 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.1 | 1.4 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.1 | 1.4 | GO:0097503 | sialylation(GO:0097503) |
| 0.1 | 1.3 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.1 | 0.5 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.0 | 0.4 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.0 | 0.5 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.0 | 1.2 | GO:0032620 | interleukin-17 production(GO:0032620) |
| 0.0 | 0.9 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 3.9 | GO:0006024 | aminoglycan biosynthetic process(GO:0006023) glycosaminoglycan biosynthetic process(GO:0006024) |
| 0.0 | 2.2 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.9 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 0.3 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.8 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 1.5 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.9 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 2.3 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.0 | 2.2 | GO:0042795 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.2 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.0 | 2.8 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 2.6 | GO:0030509 | BMP signaling pathway(GO:0030509) |
| 0.0 | 0.6 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 0.6 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.0 | 0.1 | GO:0009441 | fatty acid alpha-oxidation(GO:0001561) glycolate metabolic process(GO:0009441) |
| 0.0 | 0.8 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 1.1 | GO:0030279 | negative regulation of ossification(GO:0030279) |
| 0.0 | 1.0 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.8 | GO:0042220 | response to cocaine(GO:0042220) |
| 0.0 | 1.6 | GO:0060996 | dendritic spine development(GO:0060996) |
| 0.0 | 2.1 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.0 | 0.7 | GO:0006306 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 1.4 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.0 | 1.7 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.6 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) cellular polysaccharide biosynthetic process(GO:0033692) |
| 0.0 | 0.5 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.0 | 0.1 | GO:0042791 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.0 | 0.6 | GO:0002228 | natural killer cell mediated immunity(GO:0002228) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.7 | GO:0031251 | PAN complex(GO:0031251) |
| 1.1 | 5.5 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.6 | 5.3 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.4 | 1.8 | GO:0055087 | Ski complex(GO:0055087) |
| 0.4 | 2.5 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.4 | 6.6 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.4 | 1.1 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.3 | 1.0 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.3 | 9.8 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.3 | 2.6 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.3 | 4.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.3 | 0.9 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.3 | 1.2 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.3 | 1.8 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.3 | 7.0 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.2 | 4.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.2 | 2.6 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.2 | 0.9 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.2 | 1.5 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.1 | 1.0 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 1.5 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 2.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 0.6 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.1 | 1.7 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 3.2 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.1 | 1.2 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 0.5 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.1 | 2.4 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.1 | 1.1 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.1 | 6.7 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 2.4 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 8.8 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 1.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 1.2 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 3.1 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 5.8 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.1 | 3.0 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 2.9 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 2.2 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 1.0 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.3 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 2.2 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 4.5 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 6.0 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 1.8 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 7.1 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 12.3 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 3.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.7 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 1.8 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 4.5 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 5.6 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.9 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.2 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 1.8 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.6 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 2.3 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 2.3 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.5 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 1.7 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.6 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 2.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 13.5 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 26.1 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.5 | GO:0031226 | intrinsic component of plasma membrane(GO:0031226) |
| 0.0 | 0.4 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 30.1 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 0.9 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.4 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 1.2 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.8 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.6 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.9 | GO:0030424 | axon(GO:0030424) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 7.5 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 1.8 | 5.3 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 1.5 | 9.2 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 1.5 | 4.4 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 1.4 | 4.2 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 1.2 | 3.5 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 1.1 | 5.5 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 1.1 | 6.4 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.9 | 9.7 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.8 | 2.3 | GO:0004878 | complement component C5a receptor activity(GO:0004878) |
| 0.8 | 2.3 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.8 | 3.8 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.8 | 4.5 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.7 | 12.0 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.6 | 3.9 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.6 | 3.2 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.6 | 4.9 | GO:0005497 | androgen binding(GO:0005497) |
| 0.6 | 10.2 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.6 | 2.2 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.5 | 4.3 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.5 | 1.9 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.5 | 2.8 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.5 | 1.4 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.4 | 1.7 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.4 | 1.7 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.4 | 5.1 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.4 | 2.9 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.4 | 2.1 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.4 | 1.6 | GO:0001855 | complement component C4b binding(GO:0001855) |
| 0.3 | 5.9 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.3 | 0.7 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.3 | 1.5 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.3 | 4.1 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.3 | 0.8 | GO:0061513 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.3 | 1.4 | GO:0070404 | NADH binding(GO:0070404) |
| 0.3 | 1.6 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.3 | 4.7 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.3 | 2.6 | GO:0030882 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.2 | 2.5 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.2 | 1.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.2 | 3.1 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.2 | 4.3 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.2 | 6.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.2 | 5.5 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.2 | 0.8 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.2 | 0.6 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.2 | 3.4 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.2 | 1.8 | GO:0009374 | biotin carboxylase activity(GO:0004075) biotin binding(GO:0009374) |
| 0.2 | 0.3 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.2 | 3.4 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.2 | 0.8 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.2 | 0.6 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.2 | 2.2 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.2 | 1.6 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.2 | 3.8 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.2 | 0.8 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.1 | 7.6 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 4.6 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 0.5 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.1 | 1.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) PH domain binding(GO:0042731) |
| 0.1 | 2.6 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 0.6 | GO:0015307 | drug:proton antiporter activity(GO:0015307) |
| 0.1 | 0.6 | GO:0015056 | corticotrophin-releasing factor receptor activity(GO:0015056) |
| 0.1 | 0.6 | GO:0015087 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.1 | 2.8 | GO:0099604 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.1 | 1.3 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.1 | 0.6 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.1 | 0.6 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.1 | 1.4 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 2.3 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 3.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 1.3 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.1 | 0.7 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.1 | 1.2 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.1 | 0.7 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 0.6 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.1 | 1.4 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.1 | 2.1 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 2.7 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.1 | 3.0 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 0.7 | GO:0001614 | purinergic nucleotide receptor activity(GO:0001614) nucleotide receptor activity(GO:0016502) |
| 0.1 | 2.4 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.1 | 1.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.5 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 0.9 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 1.7 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 1.3 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 0.5 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.1 | 0.6 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.1 | 5.6 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.1 | 0.9 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 1.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 1.2 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 4.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 2.5 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.3 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.0 | 1.1 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 1.8 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 1.6 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 6.7 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 2.9 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 1.9 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 3.5 | GO:0005319 | lipid transporter activity(GO:0005319) |
| 0.0 | 1.0 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 2.1 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 9.7 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 1.1 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.9 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.3 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 4.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.2 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 2.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.3 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 4.5 | GO:0016746 | transferase activity, transferring acyl groups(GO:0016746) |
| 0.0 | 2.5 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.4 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.8 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 3.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.3 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 1.6 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.2 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 1.1 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.0 | 0.4 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 3.6 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 2.6 | GO:0030246 | carbohydrate binding(GO:0030246) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 9.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 7.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 2.2 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 5.3 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 4.8 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 3.8 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 3.4 | ST ADRENERGIC | Adrenergic Pathway |
| 0.1 | 9.7 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 2.1 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 5.3 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 4.1 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 1.1 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 2.2 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 1.5 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 1.3 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 8.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.8 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 9.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 2.1 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 1.4 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.6 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 1.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.7 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.8 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 4.6 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.1 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.6 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 1.7 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.4 | PID P73PATHWAY | p73 transcription factor network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 12.0 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.3 | 5.4 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.3 | 4.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.3 | 4.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.2 | 5.9 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.2 | 3.8 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.2 | 9.7 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.2 | 5.5 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.2 | 2.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.2 | 1.7 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.2 | 3.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.2 | 3.6 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.2 | 3.4 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.2 | 6.4 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.2 | 12.6 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.2 | 2.0 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.1 | 1.6 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 2.5 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 4.5 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 3.7 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 2.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 1.5 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 2.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 2.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 1.5 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 6.7 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.1 | 2.3 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 1.8 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 8.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.1 | 1.9 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 2.6 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 4.1 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 2.5 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 3.1 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 1.4 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.1 | 0.7 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 4.0 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.1 | 2.0 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 1.5 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.7 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.8 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.6 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.9 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.6 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 2.2 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 7.1 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.6 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 0.9 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 1.0 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 2.1 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.8 | REACTOME ION CHANNEL TRANSPORT | Genes involved in Ion channel transport |
| 0.0 | 0.6 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 1.7 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.2 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.6 | REACTOME BIOLOGICAL OXIDATIONS | Genes involved in Biological oxidations |