Project
averaged GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
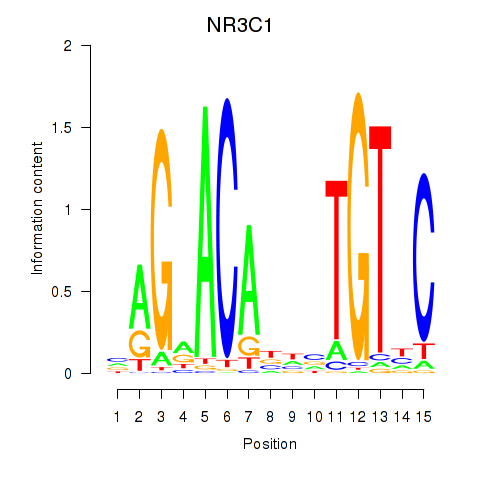
Results for NR3C1
Z-value: 2.68
Transcription factors associated with NR3C1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR3C1
|
ENSG00000113580.10 | nuclear receptor subfamily 3 group C member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR3C1 | hg19_v2_chr5_-_142780280_142780417 | 0.10 | 1.5e-01 | Click! |
Activity profile of NR3C1 motif
Sorted Z-values of NR3C1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_106054659 | 116.72 |
ENST00000390539.2
|
IGHA2
|
immunoglobulin heavy constant alpha 2 (A2m marker) |
| chr22_+_23237555 | 88.89 |
ENST00000390321.2
|
IGLC1
|
immunoglobulin lambda constant 1 (Mcg marker) |
| chr22_+_23248512 | 83.99 |
ENST00000390325.2
|
IGLC3
|
immunoglobulin lambda constant 3 (Kern-Oz+ marker) |
| chr14_-_106174960 | 74.59 |
ENST00000390547.2
|
IGHA1
|
immunoglobulin heavy constant alpha 1 |
| chr22_+_23243156 | 73.46 |
ENST00000390323.2
|
IGLC2
|
immunoglobulin lambda constant 2 (Kern-Oz- marker) |
| chr14_-_106692191 | 56.68 |
ENST00000390607.2
|
IGHV3-21
|
immunoglobulin heavy variable 3-21 |
| chr22_+_23241661 | 54.04 |
ENST00000390322.2
|
IGLJ2
|
immunoglobulin lambda joining 2 |
| chr6_+_33048222 | 52.13 |
ENST00000428835.1
|
HLA-DPB1
|
major histocompatibility complex, class II, DP beta 1 |
| chr14_-_106642049 | 50.09 |
ENST00000390605.2
|
IGHV1-18
|
immunoglobulin heavy variable 1-18 |
| chr2_-_89513402 | 49.77 |
ENST00000498435.1
|
IGKV1-27
|
immunoglobulin kappa variable 1-27 |
| chr2_-_136875712 | 45.98 |
ENST00000241393.3
|
CXCR4
|
chemokine (C-X-C motif) receptor 4 |
| chr5_-_149792295 | 43.85 |
ENST00000518797.1
ENST00000524315.1 ENST00000009530.7 ENST00000377795.3 |
CD74
|
CD74 molecule, major histocompatibility complex, class II invariant chain |
| chr22_+_23264766 | 41.92 |
ENST00000390331.2
|
IGLC7
|
immunoglobulin lambda constant 7 |
| chr6_-_33048483 | 41.71 |
ENST00000419277.1
|
HLA-DPA1
|
major histocompatibility complex, class II, DP alpha 1 |
| chr22_-_17680472 | 37.29 |
ENST00000330232.4
|
CECR1
|
cat eye syndrome chromosome region, candidate 1 |
| chr15_-_22448819 | 34.17 |
ENST00000604066.1
|
IGHV1OR15-1
|
immunoglobulin heavy variable 1/OR15-1 (non-functional) |
| chr14_-_106994333 | 34.12 |
ENST00000390624.2
|
IGHV3-48
|
immunoglobulin heavy variable 3-48 |
| chr19_-_10445399 | 34.04 |
ENST00000592945.1
|
ICAM3
|
intercellular adhesion molecule 3 |
| chr2_-_89278535 | 33.85 |
ENST00000390247.2
|
IGKV3-7
|
immunoglobulin kappa variable 3-7 (non-functional) |
| chr14_-_106330458 | 33.75 |
ENST00000461719.1
|
IGHJ4
|
immunoglobulin heavy joining 4 |
| chr14_-_106781017 | 33.32 |
ENST00000390612.2
|
IGHV4-28
|
immunoglobulin heavy variable 4-28 |
| chr14_-_107114267 | 32.05 |
ENST00000454421.2
|
IGHV3-64
|
immunoglobulin heavy variable 3-64 |
| chr22_+_22712087 | 31.63 |
ENST00000390294.2
|
IGLV1-47
|
immunoglobulin lambda variable 1-47 |
| chr10_+_70847852 | 31.09 |
ENST00000242465.3
|
SRGN
|
serglycin |
| chr16_+_32077386 | 31.09 |
ENST00000354689.6
|
IGHV3OR16-9
|
immunoglobulin heavy variable 3/OR16-9 (non-functional) |
| chr14_-_106866934 | 30.39 |
ENST00000390618.2
|
IGHV3-38
|
immunoglobulin heavy variable 3-38 (non-functional) |
| chr2_-_89340242 | 29.95 |
ENST00000480492.1
|
IGKV1-12
|
immunoglobulin kappa variable 1-12 |
| chr14_-_106805716 | 29.83 |
ENST00000438142.2
|
IGHV4-31
|
immunoglobulin heavy variable 4-31 |
| chr2_-_89292422 | 29.16 |
ENST00000495489.1
|
IGKV1-8
|
immunoglobulin kappa variable 1-8 |
| chr14_-_106453155 | 28.24 |
ENST00000390594.2
|
IGHV1-2
|
immunoglobulin heavy variable 1-2 |
| chr2_-_89545079 | 28.21 |
ENST00000468494.1
|
IGKV2-30
|
immunoglobulin kappa variable 2-30 |
| chr14_-_106725723 | 27.41 |
ENST00000390609.2
|
IGHV3-23
|
immunoglobulin heavy variable 3-23 |
| chr6_+_32605195 | 26.35 |
ENST00000374949.2
|
HLA-DQA1
|
major histocompatibility complex, class II, DQ alpha 1 |
| chr22_+_22681656 | 26.05 |
ENST00000390291.2
|
IGLV1-50
|
immunoglobulin lambda variable 1-50 (non-functional) |
| chr15_+_31658349 | 25.17 |
ENST00000558844.1
|
KLF13
|
Kruppel-like factor 13 |
| chr2_+_90192768 | 25.07 |
ENST00000390275.2
|
IGKV1D-13
|
immunoglobulin kappa variable 1D-13 |
| chr2_+_89975669 | 24.79 |
ENST00000474213.1
|
IGKV2D-30
|
immunoglobulin kappa variable 2D-30 |
| chr14_-_107219365 | 24.70 |
ENST00000424969.2
|
IGHV3-74
|
immunoglobulin heavy variable 3-74 |
| chr14_-_107049312 | 24.08 |
ENST00000390627.2
|
IGHV3-53
|
immunoglobulin heavy variable 3-53 |
| chr14_-_106518922 | 22.97 |
ENST00000390598.2
|
IGHV3-7
|
immunoglobulin heavy variable 3-7 |
| chr3_+_122044084 | 22.07 |
ENST00000264474.3
ENST00000479204.1 |
CSTA
|
cystatin A (stefin A) |
| chr2_+_89999259 | 21.60 |
ENST00000558026.1
|
IGKV2D-28
|
immunoglobulin kappa variable 2D-28 |
| chr2_+_89998789 | 21.58 |
ENST00000453166.2
|
IGKV2D-28
|
immunoglobulin kappa variable 2D-28 |
| chr11_+_57365150 | 21.38 |
ENST00000457869.1
ENST00000340687.6 ENST00000378323.4 ENST00000378324.2 ENST00000403558.1 |
SERPING1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 |
| chr12_-_15114492 | 21.03 |
ENST00000541546.1
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr5_-_172198190 | 20.58 |
ENST00000239223.3
|
DUSP1
|
dual specificity phosphatase 1 |
| chr15_-_20193370 | 20.14 |
ENST00000558565.2
|
IGHV3OR15-7
|
immunoglobulin heavy variable 3/OR15-7 (pseudogene) |
| chr2_+_90248739 | 19.67 |
ENST00000468879.1
|
IGKV1D-43
|
immunoglobulin kappa variable 1D-43 |
| chr11_+_117857063 | 19.28 |
ENST00000227752.3
ENST00000541785.1 ENST00000545409.1 |
IL10RA
|
interleukin 10 receptor, alpha |
| chr2_-_89521942 | 19.23 |
ENST00000482769.1
|
IGKV2-28
|
immunoglobulin kappa variable 2-28 |
| chr19_-_36399149 | 19.17 |
ENST00000585901.2
ENST00000544690.2 ENST00000424586.3 ENST00000262629.4 ENST00000589517.1 |
TYROBP
|
TYRO protein tyrosine kinase binding protein |
| chr2_+_90198535 | 19.06 |
ENST00000390276.2
|
IGKV1D-12
|
immunoglobulin kappa variable 1D-12 |
| chr22_+_23040274 | 18.98 |
ENST00000390306.2
|
IGLV2-23
|
immunoglobulin lambda variable 2-23 |
| chr14_-_107095662 | 18.80 |
ENST00000390630.2
|
IGHV4-61
|
immunoglobulin heavy variable 4-61 |
| chr16_+_226658 | 18.25 |
ENST00000320868.5
ENST00000397797.1 |
HBA1
|
hemoglobin, alpha 1 |
| chr14_-_106845789 | 18.17 |
ENST00000390617.2
|
IGHV3-35
|
immunoglobulin heavy variable 3-35 (non-functional) |
| chr12_-_92539614 | 18.13 |
ENST00000256015.3
|
BTG1
|
B-cell translocation gene 1, anti-proliferative |
| chr14_-_106552755 | 17.53 |
ENST00000390600.2
|
IGHV3-9
|
immunoglobulin heavy variable 3-9 |
| chr14_-_106926724 | 17.45 |
ENST00000434710.1
|
IGHV3-43
|
immunoglobulin heavy variable 3-43 |
| chr22_+_22676808 | 16.87 |
ENST00000390290.2
|
IGLV1-51
|
immunoglobulin lambda variable 1-51 |
| chr21_-_46340770 | 16.82 |
ENST00000397854.3
|
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chr21_-_46340884 | 16.81 |
ENST00000302347.5
ENST00000517819.1 |
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chr15_-_79237433 | 16.46 |
ENST00000220166.5
|
CTSH
|
cathepsin H |
| chr3_-_121379739 | 15.75 |
ENST00000428394.2
ENST00000314583.3 |
HCLS1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr6_-_41909191 | 15.74 |
ENST00000512426.1
ENST00000372987.4 |
CCND3
|
cyclin D3 |
| chr22_+_22764088 | 15.69 |
ENST00000390299.2
|
IGLV1-40
|
immunoglobulin lambda variable 1-40 |
| chr14_-_106573756 | 15.57 |
ENST00000390601.2
|
IGHV3-11
|
immunoglobulin heavy variable 3-11 (gene/pseudogene) |
| chr16_-_33647696 | 15.30 |
ENST00000558425.1
ENST00000569103.2 |
RP11-812E19.9
|
Uncharacterized protein |
| chr1_-_207095324 | 15.17 |
ENST00000530505.1
ENST00000367091.3 ENST00000442471.2 |
FAIM3
|
Fas apoptotic inhibitory molecule 3 |
| chr4_+_15779901 | 14.87 |
ENST00000226279.3
|
CD38
|
CD38 molecule |
| chr1_+_22979676 | 14.69 |
ENST00000432749.2
ENST00000314933.6 |
C1QB
|
complement component 1, q subcomponent, B chain |
| chr9_-_97402413 | 14.66 |
ENST00000414122.1
|
FBP1
|
fructose-1,6-bisphosphatase 1 |
| chr16_+_33020496 | 14.37 |
ENST00000565407.2
|
IGHV3OR16-8
|
immunoglobulin heavy variable 3/OR16-8 (non-functional) |
| chr14_-_106668095 | 14.11 |
ENST00000390606.2
|
IGHV3-20
|
immunoglobulin heavy variable 3-20 |
| chr16_+_1578674 | 13.75 |
ENST00000253934.5
|
TMEM204
|
transmembrane protein 204 |
| chr11_+_5710919 | 13.65 |
ENST00000379965.3
ENST00000425490.1 |
TRIM22
|
tripartite motif containing 22 |
| chr14_-_106816253 | 13.64 |
ENST00000390615.2
|
IGHV3-33
|
immunoglobulin heavy variable 3-33 |
| chr19_+_42381173 | 13.29 |
ENST00000221972.3
|
CD79A
|
CD79a molecule, immunoglobulin-associated alpha |
| chr6_+_32605134 | 13.10 |
ENST00000343139.5
ENST00000395363.1 ENST00000496318.1 |
HLA-DQA1
|
major histocompatibility complex, class II, DQ alpha 1 |
| chr12_-_7245125 | 12.99 |
ENST00000542285.1
ENST00000540610.1 |
C1R
|
complement component 1, r subcomponent |
| chr6_+_29795595 | 12.78 |
ENST00000360323.6
ENST00000376818.3 ENST00000376815.3 |
HLA-G
|
major histocompatibility complex, class I, G |
| chr19_+_13261216 | 12.41 |
ENST00000587885.1
ENST00000292433.3 |
IER2
|
immediate early response 2 |
| chr19_-_2041159 | 12.36 |
ENST00000589441.1
|
MKNK2
|
MAP kinase interacting serine/threonine kinase 2 |
| chr2_+_114163945 | 12.23 |
ENST00000453673.3
|
IGKV1OR2-108
|
immunoglobulin kappa variable 1/OR2-108 (non-functional) |
| chr6_-_31324943 | 12.07 |
ENST00000412585.2
ENST00000434333.1 |
HLA-B
|
major histocompatibility complex, class I, B |
| chr16_+_32063311 | 12.06 |
ENST00000426099.1
|
AC142381.1
|
AC142381.1 |
| chr19_-_10450328 | 11.86 |
ENST00000160262.5
|
ICAM3
|
intercellular adhesion molecule 3 |
| chr19_-_10450287 | 11.82 |
ENST00000589261.1
ENST00000590569.1 ENST00000589580.1 ENST00000589249.1 |
ICAM3
|
intercellular adhesion molecule 3 |
| chr6_+_32811885 | 11.74 |
ENST00000458296.1
ENST00000413039.1 ENST00000429600.1 ENST00000412095.1 ENST00000415067.1 ENST00000395330.1 |
TAPSAR1
PSMB9
|
TAP1 and PSMB8 antisense RNA 1 proteasome (prosome, macropain) subunit, beta type, 9 |
| chr14_-_106733624 | 11.66 |
ENST00000390610.2
|
IGHV1-24
|
immunoglobulin heavy variable 1-24 |
| chr19_-_54876558 | 11.42 |
ENST00000391742.2
ENST00000434277.2 |
LAIR1
|
leukocyte-associated immunoglobulin-like receptor 1 |
| chr14_-_107283278 | 11.33 |
ENST00000390639.2
|
IGHV7-81
|
immunoglobulin heavy variable 7-81 (non-functional) |
| chr20_-_62711259 | 11.16 |
ENST00000332298.5
|
RGS19
|
regulator of G-protein signaling 19 |
| chr16_+_33006369 | 11.14 |
ENST00000425181.3
|
IGHV3OR16-10
|
immunoglobulin heavy variable 3/OR16-10 (non-functional) |
| chr12_+_51318513 | 10.91 |
ENST00000332160.4
|
METTL7A
|
methyltransferase like 7A |
| chr2_-_158300556 | 10.86 |
ENST00000264192.3
|
CYTIP
|
cytohesin 1 interacting protein |
| chr3_-_18480260 | 10.66 |
ENST00000454909.2
|
SATB1
|
SATB homeobox 1 |
| chr1_+_22979474 | 10.51 |
ENST00000509305.1
|
C1QB
|
complement component 1, q subcomponent, B chain |
| chr17_-_6983594 | 10.43 |
ENST00000571664.1
ENST00000254868.4 |
CLEC10A
|
C-type lectin domain family 10, member A |
| chr2_+_89952792 | 10.41 |
ENST00000390265.2
|
IGKV1D-33
|
immunoglobulin kappa variable 1D-33 |
| chr5_-_42811986 | 10.33 |
ENST00000511224.1
ENST00000507920.1 ENST00000510965.1 |
SEPP1
|
selenoprotein P, plasma, 1 |
| chr15_+_91073111 | 10.26 |
ENST00000420329.2
|
CRTC3
|
CREB regulated transcription coactivator 3 |
| chr6_+_31783291 | 9.93 |
ENST00000375651.5
ENST00000608703.1 ENST00000458062.2 |
HSPA1A
|
heat shock 70kDa protein 1A |
| chr6_-_109777128 | 9.89 |
ENST00000358807.3
ENST00000358577.3 |
MICAL1
|
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
| chr14_-_106478603 | 9.88 |
ENST00000390596.2
|
IGHV4-4
|
immunoglobulin heavy variable 4-4 |
| chr1_+_158969752 | 9.64 |
ENST00000566111.1
|
IFI16
|
interferon, gamma-inducible protein 16 |
| chr17_-_6983550 | 9.43 |
ENST00000576617.1
ENST00000416562.2 |
CLEC10A
|
C-type lectin domain family 10, member A |
| chr6_+_32821924 | 9.28 |
ENST00000374859.2
ENST00000453265.2 |
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr15_+_81475047 | 9.23 |
ENST00000559388.1
|
IL16
|
interleukin 16 |
| chr10_+_17270214 | 9.19 |
ENST00000544301.1
|
VIM
|
vimentin |
| chr1_-_207095212 | 9.10 |
ENST00000420007.2
|
FAIM3
|
Fas apoptotic inhibitory molecule 3 |
| chr2_+_233925064 | 9.07 |
ENST00000359570.5
ENST00000538935.1 |
INPP5D
|
inositol polyphosphate-5-phosphatase, 145kDa |
| chr12_-_51718436 | 9.03 |
ENST00000544402.1
|
BIN2
|
bridging integrator 2 |
| chr5_+_49961727 | 8.97 |
ENST00000505697.2
ENST00000503750.2 ENST00000514342.2 |
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr19_-_19774473 | 8.93 |
ENST00000357324.6
|
ATP13A1
|
ATPase type 13A1 |
| chr6_+_6588902 | 8.85 |
ENST00000230568.4
|
LY86
|
lymphocyte antigen 86 |
| chr2_-_89568263 | 8.75 |
ENST00000473726.1
|
IGKV1-33
|
immunoglobulin kappa variable 1-33 |
| chr9_+_139873264 | 8.71 |
ENST00000446677.1
|
PTGDS
|
prostaglandin D2 synthase 21kDa (brain) |
| chr11_+_63304273 | 8.70 |
ENST00000439013.2
ENST00000255688.3 |
RARRES3
|
retinoic acid receptor responder (tazarotene induced) 3 |
| chr20_+_46130619 | 8.70 |
ENST00000372004.3
|
NCOA3
|
nuclear receptor coactivator 3 |
| chr6_+_31795506 | 8.63 |
ENST00000375650.3
|
HSPA1B
|
heat shock 70kDa protein 1B |
| chr19_-_36643329 | 8.61 |
ENST00000589154.1
|
COX7A1
|
cytochrome c oxidase subunit VIIa polypeptide 1 (muscle) |
| chr1_+_44115814 | 8.56 |
ENST00000372396.3
|
KDM4A
|
lysine (K)-specific demethylase 4A |
| chr1_+_158900568 | 8.52 |
ENST00000458222.1
|
PYHIN1
|
pyrin and HIN domain family, member 1 |
| chrY_+_16168097 | 8.50 |
ENST00000250823.4
|
VCY1B
|
variable charge, Y-linked 1B |
| chr14_-_106471723 | 8.31 |
ENST00000390595.2
|
IGHV1-3
|
immunoglobulin heavy variable 1-3 |
| chr22_+_37309662 | 8.26 |
ENST00000403662.3
ENST00000262825.5 |
CSF2RB
|
colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) |
| chr2_+_90458201 | 8.05 |
ENST00000603238.1
|
CH17-132F21.1
|
Uncharacterized protein |
| chr2_+_27593389 | 8.02 |
ENST00000233575.2
ENST00000543024.1 ENST00000537606.1 |
SNX17
|
sorting nexin 17 |
| chr11_-_85376121 | 7.99 |
ENST00000527447.1
|
CREBZF
|
CREB/ATF bZIP transcription factor |
| chr19_-_5340730 | 7.85 |
ENST00000372412.4
ENST00000357368.4 ENST00000262963.6 ENST00000348075.2 ENST00000353284.2 |
PTPRS
|
protein tyrosine phosphatase, receptor type, S |
| chr6_-_99873145 | 7.69 |
ENST00000369239.5
ENST00000438806.1 |
PNISR
|
PNN-interacting serine/arginine-rich protein |
| chr19_-_40440533 | 7.65 |
ENST00000221347.6
|
FCGBP
|
Fc fragment of IgG binding protein |
| chr10_-_135090338 | 7.47 |
ENST00000415217.3
|
ADAM8
|
ADAM metallopeptidase domain 8 |
| chr17_-_19290117 | 7.44 |
ENST00000497081.2
|
MFAP4
|
microfibrillar-associated protein 4 |
| chr16_+_31366536 | 7.30 |
ENST00000562522.1
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit) |
| chr17_-_34313685 | 7.23 |
ENST00000435911.2
ENST00000586216.1 ENST00000394509.4 |
CCL14
|
chemokine (C-C motif) ligand 14 |
| chr20_+_35201993 | 7.22 |
ENST00000373872.4
|
TGIF2
|
TGFB-induced factor homeobox 2 |
| chr17_-_76123101 | 7.15 |
ENST00000392467.3
|
TMC6
|
transmembrane channel-like 6 |
| chr1_+_233086326 | 7.11 |
ENST00000366628.5
ENST00000366627.4 |
NTPCR
|
nucleoside-triphosphatase, cancer-related |
| chr17_+_7452442 | 7.10 |
ENST00000557233.1
|
TNFSF12
|
tumor necrosis factor (ligand) superfamily, member 12 |
| chr9_+_139874683 | 7.09 |
ENST00000444903.1
|
PTGDS
|
prostaglandin D2 synthase 21kDa (brain) |
| chr8_-_70745575 | 7.08 |
ENST00000524945.1
|
SLCO5A1
|
solute carrier organic anion transporter family, member 5A1 |
| chr17_-_3599327 | 7.03 |
ENST00000551178.1
ENST00000552276.1 ENST00000547178.1 |
P2RX5
|
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr6_-_133079022 | 6.91 |
ENST00000525289.1
ENST00000326499.6 |
VNN2
|
vanin 2 |
| chr22_-_39637135 | 6.82 |
ENST00000440375.1
|
PDGFB
|
platelet-derived growth factor beta polypeptide |
| chr18_-_59854203 | 6.80 |
ENST00000589339.1
ENST00000357637.5 ENST00000585458.1 ENST00000400334.3 ENST00000587134.1 ENST00000585923.1 ENST00000590765.1 ENST00000589720.1 ENST00000588571.1 ENST00000585344.1 |
PIGN
|
phosphatidylinositol glycan anchor biosynthesis, class N |
| chr6_-_39693111 | 6.74 |
ENST00000373215.3
ENST00000538893.1 ENST00000287152.7 ENST00000373216.3 |
KIF6
|
kinesin family member 6 |
| chr2_+_90259593 | 6.66 |
ENST00000471857.1
|
IGKV1D-8
|
immunoglobulin kappa variable 1D-8 |
| chr9_-_35619539 | 6.61 |
ENST00000396757.1
|
CD72
|
CD72 molecule |
| chr16_+_31366455 | 6.57 |
ENST00000268296.4
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit) |
| chrX_+_56259316 | 6.56 |
ENST00000468660.1
|
KLF8
|
Kruppel-like factor 8 |
| chr6_-_32811771 | 6.54 |
ENST00000395339.3
ENST00000374882.3 |
PSMB8
|
proteasome (prosome, macropain) subunit, beta type, 8 |
| chr17_-_76124711 | 6.53 |
ENST00000306591.7
ENST00000590602.1 |
TMC6
|
transmembrane channel-like 6 |
| chr8_+_38614807 | 6.41 |
ENST00000330691.6
ENST00000348567.4 |
TACC1
|
transforming, acidic coiled-coil containing protein 1 |
| chr1_-_207119738 | 6.41 |
ENST00000356495.4
|
PIGR
|
polymeric immunoglobulin receptor |
| chr3_-_151047327 | 6.36 |
ENST00000325602.5
|
P2RY13
|
purinergic receptor P2Y, G-protein coupled, 13 |
| chr19_-_54876414 | 6.35 |
ENST00000474878.1
ENST00000348231.4 |
LAIR1
|
leukocyte-associated immunoglobulin-like receptor 1 |
| chr22_+_23046750 | 6.33 |
ENST00000390307.2
|
IGLV3-22
|
immunoglobulin lambda variable 3-22 (gene/pseudogene) |
| chr15_-_55700522 | 6.30 |
ENST00000564092.1
ENST00000310958.6 |
CCPG1
|
cell cycle progression 1 |
| chr17_+_7462103 | 6.29 |
ENST00000396545.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr16_+_68119764 | 6.28 |
ENST00000570212.1
ENST00000562926.1 |
NFATC3
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3 |
| chr6_-_32821599 | 6.25 |
ENST00000354258.4
|
TAP1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr8_-_125740730 | 6.21 |
ENST00000354184.4
|
MTSS1
|
metastasis suppressor 1 |
| chr13_-_36920872 | 6.18 |
ENST00000451493.1
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome) |
| chrX_+_2609207 | 6.13 |
ENST00000381192.3
|
CD99
|
CD99 molecule |
| chr12_-_11036844 | 6.12 |
ENST00000428168.2
|
PRH1
|
proline-rich protein HaeIII subfamily 1 |
| chr1_-_153013588 | 6.12 |
ENST00000360379.3
|
SPRR2D
|
small proline-rich protein 2D |
| chr1_+_90460661 | 6.08 |
ENST00000340281.4
ENST00000361911.5 ENST00000370447.3 ENST00000455342.2 |
ZNF326
|
zinc finger protein 326 |
| chr8_-_125740514 | 6.05 |
ENST00000325064.5
ENST00000518547.1 |
MTSS1
|
metastasis suppressor 1 |
| chr5_-_39219705 | 6.03 |
ENST00000351578.6
|
FYB
|
FYN binding protein |
| chr17_+_7461849 | 5.88 |
ENST00000338784.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chrX_+_64887512 | 5.81 |
ENST00000360270.5
|
MSN
|
moesin |
| chr3_+_9834179 | 5.79 |
ENST00000498623.2
|
ARPC4
|
actin related protein 2/3 complex, subunit 4, 20kDa |
| chr11_+_121447469 | 5.67 |
ENST00000532694.1
ENST00000534286.1 |
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing |
| chr3_+_38179969 | 5.67 |
ENST00000396334.3
ENST00000417037.2 ENST00000424893.1 ENST00000495303.1 ENST00000443433.2 ENST00000421516.1 |
MYD88
|
myeloid differentiation primary response 88 |
| chr14_+_24641062 | 5.66 |
ENST00000311457.3
ENST00000557806.1 ENST00000559919.1 |
REC8
|
REC8 meiotic recombination protein |
| chr1_+_149230680 | 5.61 |
ENST00000443018.1
|
RP11-403I13.5
|
RP11-403I13.5 |
| chr1_+_24969755 | 5.60 |
ENST00000447431.2
ENST00000374389.4 |
SRRM1
|
serine/arginine repetitive matrix 1 |
| chr9_-_37034028 | 5.58 |
ENST00000520281.1
ENST00000446742.1 ENST00000522003.1 ENST00000523145.1 ENST00000414447.1 ENST00000377847.2 ENST00000377853.2 ENST00000377852.2 ENST00000523241.1 ENST00000520154.1 ENST00000358127.4 |
PAX5
|
paired box 5 |
| chr15_-_55700457 | 5.52 |
ENST00000442196.3
ENST00000563171.1 ENST00000425574.3 |
CCPG1
|
cell cycle progression 1 |
| chr17_-_73975444 | 5.49 |
ENST00000293217.5
ENST00000537812.1 |
ACOX1
|
acyl-CoA oxidase 1, palmitoyl |
| chr19_-_52255107 | 5.42 |
ENST00000595042.1
ENST00000304748.4 |
FPR1
|
formyl peptide receptor 1 |
| chr17_-_19290483 | 5.42 |
ENST00000395592.2
ENST00000299610.4 |
MFAP4
|
microfibrillar-associated protein 4 |
| chrX_+_2609317 | 5.38 |
ENST00000381187.3
ENST00000381184.1 |
CD99
|
CD99 molecule |
| chr6_-_49712147 | 5.29 |
ENST00000433368.2
ENST00000354620.4 |
CRISP3
|
cysteine-rich secretory protein 3 |
| chr15_+_99791567 | 5.28 |
ENST00000558879.1
ENST00000301981.3 ENST00000422500.2 ENST00000447360.2 ENST00000442993.2 |
LRRC28
|
leucine rich repeat containing 28 |
| chr10_+_75545329 | 5.20 |
ENST00000604729.1
ENST00000603114.1 |
ZSWIM8
|
zinc finger, SWIM-type containing 8 |
| chr10_-_135090360 | 5.20 |
ENST00000486609.1
ENST00000445355.3 ENST00000485491.2 |
ADAM8
|
ADAM metallopeptidase domain 8 |
| chr17_+_4843303 | 5.18 |
ENST00000571816.1
|
RNF167
|
ring finger protein 167 |
| chr3_-_58200398 | 5.14 |
ENST00000318316.3
ENST00000460422.1 ENST00000483681.1 |
DNASE1L3
|
deoxyribonuclease I-like 3 |
| chr1_-_182360918 | 5.13 |
ENST00000339526.4
|
GLUL
|
glutamate-ammonia ligase |
| chr17_+_4843413 | 5.11 |
ENST00000572430.1
ENST00000262482.6 |
RNF167
|
ring finger protein 167 |
| chrY_-_16098393 | 5.06 |
ENST00000250825.4
|
VCY
|
variable charge, Y-linked |
| chr17_+_7461781 | 5.04 |
ENST00000349228.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr15_+_74833518 | 5.03 |
ENST00000346246.5
|
ARID3B
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr2_-_175499294 | 5.01 |
ENST00000392547.2
|
WIPF1
|
WAS/WASL interacting protein family, member 1 |
| chr5_-_179499086 | 4.99 |
ENST00000261947.4
|
RNF130
|
ring finger protein 130 |
| chr7_-_40174201 | 4.95 |
ENST00000306984.6
|
MPLKIP
|
M-phase specific PLK1 interacting protein |
| chr4_+_7045042 | 4.93 |
ENST00000310074.7
ENST00000512388.1 |
TADA2B
|
transcriptional adaptor 2B |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR3C1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 19.1 | 191.3 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 10.4 | 31.1 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 8.8 | 43.9 | GO:0002901 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 8.7 | 602.0 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 6.6 | 46.0 | GO:0044800 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 5.5 | 16.5 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 5.3 | 21.0 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 4.6 | 18.6 | GO:0070434 | positive regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070426) positive regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070434) |
| 4.6 | 463.2 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 4.2 | 12.7 | GO:2000412 | positive regulation of tumor necrosis factor (ligand) superfamily member 11 production(GO:2000309) positive regulation of thymocyte migration(GO:2000412) |
| 3.7 | 14.7 | GO:0005986 | sucrose biosynthetic process(GO:0005986) |
| 3.1 | 15.6 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 3.0 | 9.1 | GO:0045659 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 2.8 | 8.3 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
| 2.6 | 7.8 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 2.5 | 25.2 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 2.5 | 24.9 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 2.4 | 7.2 | GO:0097327 | response to antineoplastic agent(GO:0097327) |
| 2.3 | 6.9 | GO:0090283 | regulation of protein glycosylation in Golgi(GO:0090283) |
| 2.3 | 9.2 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 2.1 | 12.4 | GO:0030035 | microspike assembly(GO:0030035) |
| 2.1 | 24.7 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 2.0 | 7.9 | GO:0032058 | positive regulation of translational initiation in response to stress(GO:0032058) |
| 1.9 | 5.8 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 1.9 | 5.7 | GO:1902955 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) regulation of neurofibrillary tangle assembly(GO:1902996) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 1.9 | 5.7 | GO:0051697 | protein delipidation(GO:0051697) |
| 1.9 | 33.6 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 1.7 | 6.8 | GO:0090677 | metanephric glomerular mesangial cell development(GO:0072255) reversible differentiation(GO:0090677) cell dedifferentiation involved in phenotypic switching(GO:0090678) positive regulation of phenotypic switching(GO:1900241) regulation of vascular smooth muscle cell dedifferentiation(GO:1905174) positive regulation of vascular smooth muscle cell dedifferentiation(GO:1905176) vascular smooth muscle cell dedifferentiation(GO:1990936) |
| 1.6 | 6.3 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 1.5 | 6.2 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 1.5 | 6.1 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 1.5 | 8.9 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 1.5 | 5.8 | GO:1902965 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 1.4 | 5.7 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 1.4 | 18.1 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 1.4 | 5.6 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 1.4 | 19.3 | GO:0045779 | negative regulation of bone resorption(GO:0045779) |
| 1.3 | 6.5 | GO:2000845 | testosterone secretion(GO:0035936) regulation of testosterone secretion(GO:2000843) positive regulation of testosterone secretion(GO:2000845) |
| 1.3 | 6.4 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 1.3 | 8.9 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 1.3 | 3.8 | GO:0031064 | negative regulation of histone deacetylation(GO:0031064) |
| 1.2 | 8.7 | GO:0035624 | receptor transactivation(GO:0035624) |
| 1.2 | 18.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 1.2 | 19.4 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 1.2 | 134.9 | GO:0031295 | T cell costimulation(GO:0031295) |
| 1.2 | 3.6 | GO:0070272 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 1.2 | 177.5 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 1.2 | 3.5 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 1.1 | 4.5 | GO:1990668 | vesicle fusion with endoplasmic reticulum-Golgi intermediate compartment (ERGIC) membrane(GO:1990668) |
| 1.1 | 21.4 | GO:2001204 | regulation of osteoclast development(GO:2001204) |
| 1.1 | 15.8 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 1.1 | 4.4 | GO:1903238 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) positive regulation of leukocyte tethering or rolling(GO:1903238) |
| 1.1 | 12.9 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 1.1 | 8.6 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 1.1 | 19.3 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 1.1 | 2.1 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 1.0 | 4.2 | GO:0035900 | response to isolation stress(GO:0035900) |
| 1.0 | 9.2 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 1.0 | 3.9 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 1.0 | 4.8 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.9 | 2.8 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 0.9 | 2.8 | GO:0070253 | somatostatin secretion(GO:0070253) |
| 0.9 | 7.1 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.8 | 2.5 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.8 | 2.5 | GO:0019477 | L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.8 | 1.7 | GO:0007623 | circadian rhythm(GO:0007623) |
| 0.8 | 8.1 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.8 | 4.0 | GO:1904098 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.8 | 2.4 | GO:0051595 | response to methylglyoxal(GO:0051595) |
| 0.8 | 7.9 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.8 | 4.6 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.8 | 10.7 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.8 | 3.8 | GO:0034351 | regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.7 | 4.5 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.7 | 2.2 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.7 | 2.9 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.7 | 228.2 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.7 | 7.0 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.7 | 2.1 | GO:0042320 | regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) |
| 0.7 | 4.8 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.7 | 3.4 | GO:0015744 | succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) |
| 0.7 | 2.6 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.6 | 1.9 | GO:0043012 | regulation of fusion of sperm to egg plasma membrane(GO:0043012) |
| 0.6 | 1.9 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.6 | 2.5 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.6 | 4.4 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.6 | 1.9 | GO:0010520 | meiotic gene conversion(GO:0006311) regulation of reciprocal meiotic recombination(GO:0010520) positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.6 | 1.9 | GO:0043105 | regulation of GTP cyclohydrolase I activity(GO:0043095) negative regulation of GTP cyclohydrolase I activity(GO:0043105) |
| 0.6 | 1.2 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) positive regulation of chondrocyte proliferation(GO:1902732) |
| 0.6 | 2.9 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.6 | 12.4 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.6 | 8.6 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.6 | 1.7 | GO:0061055 | myotome development(GO:0061055) |
| 0.6 | 2.2 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.5 | 5.4 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.5 | 1.6 | GO:0042418 | epinephrine metabolic process(GO:0042414) epinephrine biosynthetic process(GO:0042418) |
| 0.5 | 1.6 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.5 | 4.2 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.5 | 6.8 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.5 | 2.1 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.5 | 2.0 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.5 | 13.4 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.5 | 12.9 | GO:0030947 | regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030947) |
| 0.5 | 2.0 | GO:0045064 | T-helper 2 cell differentiation(GO:0045064) regulation of T-helper 2 cell differentiation(GO:0045628) positive regulation of T-helper 2 cell differentiation(GO:0045630) Sertoli cell fate commitment(GO:0060010) |
| 0.5 | 8.3 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.5 | 1.4 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.5 | 9.5 | GO:0050855 | regulation of B cell receptor signaling pathway(GO:0050855) |
| 0.4 | 27.6 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.4 | 18.5 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.4 | 1.3 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.4 | 3.0 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.4 | 9.0 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.4 | 5.1 | GO:0010623 | programmed cell death involved in cell development(GO:0010623) |
| 0.4 | 3.4 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.4 | 2.9 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.4 | 3.7 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.4 | 2.9 | GO:0046087 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.4 | 2.4 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.4 | 3.6 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.4 | 4.4 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.4 | 1.6 | GO:0046725 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of nephron tubule epithelial cell differentiation(GO:0072183) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) negative regulation of epithelial cell differentiation involved in kidney development(GO:2000697) |
| 0.4 | 1.2 | GO:0021558 | trochlear nerve development(GO:0021558) |
| 0.4 | 1.2 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.4 | 2.7 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.4 | 9.8 | GO:0036152 | phosphatidylethanolamine acyl-chain remodeling(GO:0036152) |
| 0.4 | 4.8 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.4 | 4.1 | GO:0032364 | oxygen homeostasis(GO:0032364) |
| 0.4 | 1.5 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.4 | 1.5 | GO:0046015 | regulation of transcription by glucose(GO:0046015) |
| 0.4 | 3.3 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.4 | 10.3 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.4 | 1.1 | GO:1901420 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) negative regulation of response to alcohol(GO:1901420) |
| 0.4 | 5.0 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 0.4 | 1.4 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.3 | 6.3 | GO:1902894 | negative regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902894) |
| 0.3 | 5.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.3 | 5.8 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.3 | 1.0 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.3 | 4.7 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.3 | 1.0 | GO:0072560 | glandular epithelial cell maturation(GO:0002071) type B pancreatic cell maturation(GO:0072560) |
| 0.3 | 0.3 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.3 | 2.5 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.3 | 0.6 | GO:0038091 | VEGF-activated platelet-derived growth factor receptor signaling pathway(GO:0038086) positive regulation of cell proliferation by VEGF-activated platelet derived growth factor receptor signaling pathway(GO:0038091) |
| 0.3 | 1.6 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.3 | 1.2 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.3 | 1.5 | GO:0060214 | stem cell fate commitment(GO:0048865) endocardium formation(GO:0060214) |
| 0.3 | 2.5 | GO:0002441 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.3 | 2.1 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.3 | 1.8 | GO:0052203 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.3 | 2.4 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.3 | 9.5 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.3 | 1.2 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.3 | 2.3 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.3 | 2.8 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.3 | 0.6 | GO:0071332 | cellular response to fructose stimulus(GO:0071332) |
| 0.3 | 0.3 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.3 | 2.4 | GO:0032695 | negative regulation of interleukin-12 production(GO:0032695) |
| 0.3 | 4.1 | GO:0034656 | nucleobase-containing small molecule catabolic process(GO:0034656) |
| 0.3 | 5.4 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.3 | 3.8 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.3 | 1.1 | GO:0016553 | base conversion or substitution editing(GO:0016553) cytidine to uridine editing(GO:0016554) |
| 0.3 | 2.7 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.3 | 6.9 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.3 | 15.6 | GO:0046626 | regulation of insulin receptor signaling pathway(GO:0046626) |
| 0.3 | 9.7 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.3 | 8.4 | GO:0042403 | thyroid hormone metabolic process(GO:0042403) |
| 0.3 | 5.7 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.3 | 1.0 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.3 | 11.3 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.2 | 14.6 | GO:0034113 | heterotypic cell-cell adhesion(GO:0034113) |
| 0.2 | 6.6 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.2 | 2.3 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.2 | 1.6 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.2 | 1.3 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.2 | 1.8 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.2 | 5.6 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.2 | 1.1 | GO:1990253 | cellular response to leucine(GO:0071233) cellular response to leucine starvation(GO:1990253) |
| 0.2 | 43.6 | GO:0006909 | phagocytosis(GO:0006909) |
| 0.2 | 0.9 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.2 | 0.4 | GO:0010641 | positive regulation of platelet-derived growth factor receptor signaling pathway(GO:0010641) |
| 0.2 | 1.4 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.2 | 2.0 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.2 | 8.6 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.2 | 1.2 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.2 | 1.5 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.2 | 3.2 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.2 | 0.6 | GO:0036343 | psychomotor behavior(GO:0036343) motor behavior(GO:0061744) |
| 0.2 | 2.2 | GO:0045577 | regulation of B cell differentiation(GO:0045577) |
| 0.2 | 0.4 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.2 | 7.4 | GO:0030204 | chondroitin sulfate metabolic process(GO:0030204) |
| 0.2 | 1.6 | GO:0030916 | otic vesicle formation(GO:0030916) pronephros development(GO:0048793) |
| 0.2 | 0.9 | GO:0060573 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.2 | 0.8 | GO:0048318 | axial mesoderm development(GO:0048318) |
| 0.2 | 1.2 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.2 | 0.3 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.2 | 1.1 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.1 | 2.1 | GO:0045124 | regulation of bone resorption(GO:0045124) |
| 0.1 | 1.9 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.1 | 2.1 | GO:0006693 | prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.1 | 1.9 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 3.8 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.1 | 6.1 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.1 | 2.1 | GO:0051972 | regulation of telomerase activity(GO:0051972) |
| 0.1 | 1.3 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.1 | 0.3 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.1 | 0.1 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
| 0.1 | 0.8 | GO:0090043 | tubulin deacetylation(GO:0090042) regulation of tubulin deacetylation(GO:0090043) |
| 0.1 | 1.9 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.1 | 1.8 | GO:0006705 | mineralocorticoid biosynthetic process(GO:0006705) mineralocorticoid metabolic process(GO:0008212) |
| 0.1 | 0.8 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.1 | 1.2 | GO:0050942 | positive regulation of pigment cell differentiation(GO:0050942) |
| 0.1 | 7.1 | GO:0001938 | positive regulation of endothelial cell proliferation(GO:0001938) |
| 0.1 | 1.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.1 | 0.5 | GO:0046125 | thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.1 | 5.4 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.1 | 0.6 | GO:0097396 | response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 0.1 | 1.7 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.1 | 2.0 | GO:0006023 | aminoglycan biosynthetic process(GO:0006023) glycosaminoglycan biosynthetic process(GO:0006024) |
| 0.1 | 0.3 | GO:2000400 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.1 | 2.0 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.1 | 1.6 | GO:1901750 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.1 | 1.2 | GO:0043116 | negative regulation of vascular permeability(GO:0043116) negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.1 | 2.2 | GO:0035307 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.1 | 13.3 | GO:0050807 | regulation of synapse organization(GO:0050807) |
| 0.1 | 0.5 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.1 | 6.5 | GO:0071357 | type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.1 | 3.0 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 1.1 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) |
| 0.1 | 1.1 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.1 | 0.6 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.1 | 2.8 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.1 | 3.3 | GO:0060976 | coronary vasculature development(GO:0060976) |
| 0.1 | 1.0 | GO:0045008 | depyrimidination(GO:0045008) |
| 0.1 | 0.9 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 1.4 | GO:0051444 | negative regulation of ubiquitin-protein transferase activity(GO:0051444) |
| 0.1 | 4.2 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.1 | 0.1 | GO:0032472 | Golgi calcium ion transport(GO:0032472) |
| 0.1 | 0.5 | GO:0042364 | water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.1 | 0.4 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 0.5 | GO:0002027 | regulation of heart rate(GO:0002027) |
| 0.1 | 1.9 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 1.1 | GO:0030878 | thyroid gland development(GO:0030878) |
| 0.1 | 0.2 | GO:0032773 | regulation of monophenol monooxygenase activity(GO:0032771) positive regulation of monophenol monooxygenase activity(GO:0032773) negative regulation of transforming growth factor beta2 production(GO:0032912) negative regulation of catagen(GO:0051796) regulation of hair cycle by canonical Wnt signaling pathway(GO:0060901) positive regulation of melanosome transport(GO:1902910) |
| 0.1 | 2.4 | GO:0018146 | keratan sulfate biosynthetic process(GO:0018146) |
| 0.1 | 1.0 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 4.5 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.1 | 0.5 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 2.9 | GO:0034724 | DNA replication-independent nucleosome assembly(GO:0006336) DNA replication-independent nucleosome organization(GO:0034724) |
| 0.1 | 0.2 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.1 | 3.9 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.1 | 4.7 | GO:0006672 | ceramide metabolic process(GO:0006672) |
| 0.1 | 1.0 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.1 | 0.2 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.1 | 0.5 | GO:0038042 | dimeric G-protein coupled receptor signaling pathway(GO:0038042) regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.1 | 1.6 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.1 | 0.3 | GO:0046618 | drug export(GO:0046618) |
| 0.1 | 0.1 | GO:0002438 | acute inflammatory response to antigenic stimulus(GO:0002438) |
| 0.1 | 1.9 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.1 | 1.0 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.1 | 1.0 | GO:0043330 | response to exogenous dsRNA(GO:0043330) |
| 0.1 | 6.4 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.1 | 0.4 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.1 | 1.8 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 0.8 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.2 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.0 | 0.4 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 1.7 | GO:0019915 | lipid storage(GO:0019915) |
| 0.0 | 2.0 | GO:0045814 | negative regulation of gene expression, epigenetic(GO:0045814) |
| 0.0 | 4.2 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 0.5 | GO:0043114 | regulation of vascular permeability(GO:0043114) |
| 0.0 | 0.9 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 1.8 | GO:0048662 | negative regulation of smooth muscle cell proliferation(GO:0048662) |
| 0.0 | 2.1 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.0 | 0.2 | GO:0002335 | mature B cell differentiation involved in immune response(GO:0002313) marginal zone B cell differentiation(GO:0002315) mature B cell differentiation(GO:0002335) |
| 0.0 | 1.1 | GO:0051017 | actin filament bundle assembly(GO:0051017) actin filament bundle organization(GO:0061572) |
| 0.0 | 1.5 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 0.2 | GO:0015824 | proline transport(GO:0015824) |
| 0.0 | 1.4 | GO:0046037 | GMP metabolic process(GO:0046037) |
| 0.0 | 0.2 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.2 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.0 | 1.2 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.5 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.0 | 0.4 | GO:0046135 | pyrimidine nucleoside catabolic process(GO:0046135) |
| 0.0 | 0.2 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.0 | 0.9 | GO:0007420 | brain development(GO:0007420) |
| 0.0 | 0.6 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.2 | GO:0006811 | ion transport(GO:0006811) |
| 0.0 | 0.2 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.5 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.4 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.3 | GO:0042267 | natural killer cell mediated cytotoxicity(GO:0042267) |
| 0.0 | 0.1 | GO:2000117 | negative regulation of cysteine-type endopeptidase activity(GO:2000117) |
| 0.0 | 0.2 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.0 | 2.3 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.1 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.0 | 1.3 | GO:0071427 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 0.1 | GO:0030238 | male sex determination(GO:0030238) |
| 0.0 | 0.4 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.6 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.3 | GO:0002062 | chondrocyte differentiation(GO:0002062) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 63.8 | 191.3 | GO:0071746 | IgA immunoglobulin complex(GO:0071745) IgA immunoglobulin complex, circulating(GO:0071746) monomeric IgA immunoglobulin complex(GO:0071748) polymeric IgA immunoglobulin complex(GO:0071749) secretory IgA immunoglobulin complex(GO:0071751) |
| 12.8 | 601.9 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 8.4 | 177.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 6.7 | 33.6 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 5.0 | 25.2 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 4.6 | 18.3 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 4.2 | 12.7 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 3.4 | 27.6 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 2.3 | 24.9 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 2.1 | 16.5 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 2.0 | 6.1 | GO:0044609 | DBIRD complex(GO:0044609) |
| 2.0 | 360.4 | GO:0072562 | blood microparticle(GO:0072562) |
| 1.9 | 13.3 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 1.9 | 5.7 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 1.8 | 10.8 | GO:0042825 | TAP complex(GO:0042825) |
| 1.7 | 6.9 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 1.4 | 29.3 | GO:0042588 | zymogen granule(GO:0042588) |
| 1.3 | 5.3 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 1.3 | 12.9 | GO:0071953 | elastic fiber(GO:0071953) |
| 1.1 | 4.4 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 1.1 | 9.7 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.9 | 8.5 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.7 | 2.9 | GO:0005713 | recombination nodule(GO:0005713) |
| 0.7 | 4.8 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.7 | 6.7 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.7 | 3.4 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.7 | 2.7 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.6 | 11.6 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.6 | 5.7 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.6 | 4.4 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.5 | 4.4 | GO:0031672 | A band(GO:0031672) |
| 0.5 | 3.3 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.5 | 7.8 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.5 | 9.0 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.4 | 1.7 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.4 | 13.9 | GO:0008305 | integrin complex(GO:0008305) |
| 0.4 | 1.1 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.4 | 18.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.3 | 2.4 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.3 | 4.8 | GO:0036038 | MKS complex(GO:0036038) |
| 0.3 | 5.3 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.3 | 4.0 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.3 | 1.3 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.3 | 3.8 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.3 | 0.9 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.3 | 24.3 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.3 | 31.3 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.3 | 2.4 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.3 | 13.0 | GO:0016235 | aggresome(GO:0016235) |
| 0.2 | 1.5 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.2 | 2.9 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.2 | 1.4 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.2 | 2.1 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.2 | 3.9 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.2 | 3.2 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.2 | 4.7 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.2 | 13.3 | GO:0015030 | Cajal body(GO:0015030) |
| 0.2 | 10.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.2 | 0.2 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.2 | 1.1 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.2 | 2.1 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.2 | 4.3 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.2 | 4.2 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.2 | 0.8 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.2 | 3.6 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.2 | 8.9 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.2 | 4.8 | GO:0090568 | nuclear transcriptional repressor complex(GO:0090568) |
| 0.1 | 1.9 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.1 | 211.2 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 31.7 | GO:0005770 | late endosome(GO:0005770) |
| 0.1 | 5.6 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 13.0 | GO:0005769 | early endosome(GO:0005769) |
| 0.1 | 0.3 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.8 | GO:0033063 | DNA recombinase mediator complex(GO:0033061) Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.1 | 16.8 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 1.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 1.8 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.1 | 0.5 | GO:0038038 | G-protein coupled receptor homodimeric complex(GO:0038038) |
| 0.1 | 5.7 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 6.0 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 6.3 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 1.5 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 2.8 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.1 | 1.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 2.4 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.1 | 1.1 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.1 | 2.0 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 10.2 | GO:0001726 | ruffle(GO:0001726) |
| 0.1 | 3.8 | GO:0030120 | vesicle coat(GO:0030120) |
| 0.1 | 4.2 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 0.5 | GO:0097361 | CIA complex(GO:0097361) |
| 0.1 | 1.1 | GO:0043235 | receptor complex(GO:0043235) |
| 0.1 | 6.5 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 0.2 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.1 | 6.6 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.1 | 2.3 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 1.5 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 4.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 9.0 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 0.7 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 1.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 1.0 | GO:0000782 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.1 | 1.1 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 3.8 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 5.3 | GO:0031410 | cytoplasmic vesicle(GO:0031410) intracellular vesicle(GO:0097708) |
| 0.1 | 0.7 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.1 | 1.8 | GO:0005657 | replication fork(GO:0005657) |
| 0.1 | 6.5 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 2.1 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.4 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 1.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 3.7 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 1.7 | GO:0031228 | intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 2.0 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 3.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 1.0 | GO:0005856 | cytoskeleton(GO:0005856) |
| 0.0 | 1.8 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 3.6 | GO:0098802 | plasma membrane receptor complex(GO:0098802) |
| 0.0 | 0.5 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.2 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 1.5 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 3.5 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 1.1 | GO:1902555 | endoribonuclease complex(GO:1902555) |
| 0.0 | 2.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.0 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 16.0 | GO:0005912 | adherens junction(GO:0005912) |
| 0.0 | 2.4 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.7 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.0 | 0.3 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 1.1 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 1.9 | GO:0101002 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.0 | 0.5 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.9 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.5 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.1 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.0 | 0.7 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.3 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.4 | 793.2 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 8.8 | 43.9 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 6.9 | 20.6 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 6.2 | 81.2 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 5.0 | 14.9 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) NAD(P)+ nucleosidase activity(GO:0050135) |
| 4.8 | 33.6 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 4.8 | 830.0 | GO:0003823 | antigen binding(GO:0003823) |
| 4.2 | 46.0 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 3.9 | 15.8 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 3.7 | 14.7 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 3.1 | 18.6 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 3.0 | 9.1 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 2.8 | 19.3 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 2.8 | 8.3 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 2.4 | 7.1 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 2.3 | 9.2 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 2.3 | 6.8 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 2.1 | 21.0 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 2.1 | 16.5 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 1.9 | 5.6 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 1.5 | 6.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 1.5 | 9.2 | GO:1990254 | keratin filament binding(GO:1990254) |
| 1.5 | 4.5 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 1.4 | 7.2 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 1.4 | 18.3 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 1.4 | 4.2 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 1.4 | 5.4 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 1.4 | 4.1 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 1.1 | 8.6 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 1.1 | 6.4 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 1.1 | 4.3 | GO:0017040 | ceramidase activity(GO:0017040) |
| 1.1 | 4.2 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 1.0 | 7.8 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 1.0 | 2.9 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 1.0 | 8.6 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.9 | 3.8 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.9 | 4.5 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.9 | 3.5 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.9 | 3.5 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.8 | 5.0 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.8 | 15.1 | GO:0010857 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) calcium-dependent protein kinase activity(GO:0010857) |
| 0.8 | 7.9 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.8 | 3.8 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.7 | 6.0 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.7 | 10.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.7 | 6.8 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.7 | 2.7 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.7 | 3.4 | GO:0015141 | succinate transmembrane transporter activity(GO:0015141) |
| 0.7 | 4.0 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.7 | 3.9 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.7 | 2.6 | GO:0016160 | amylase activity(GO:0016160) maltose alpha-glucosidase activity(GO:0032450) |
| 0.6 | 1.9 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.6 | 4.5 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.6 | 4.5 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.6 | 7.0 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.6 | 1.9 | GO:0044549 | GTP cyclohydrolase binding(GO:0044549) |
| 0.6 | 29.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.6 | 3.5 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.6 | 3.3 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.5 | 17.0 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.5 | 1.6 | GO:0004603 | phenylethanolamine N-methyltransferase activity(GO:0004603) |
| 0.5 | 1.6 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.5 | 15.7 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.5 | 1.6 | GO:0031859 | platelet activating factor receptor binding(GO:0031859) |
| 0.5 | 3.5 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.5 | 7.0 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.5 | 8.3 | GO:0004407 | histone deacetylase activity(GO:0004407) protein deacetylase activity(GO:0033558) |
| 0.5 | 2.4 | GO:0070728 | leucine binding(GO:0070728) |
| 0.4 | 9.2 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.4 | 1.7 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.4 | 2.4 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.4 | 1.9 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.4 | 1.1 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.4 | 3.4 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.4 | 2.9 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.4 | 1.4 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.3 | 1.7 | GO:0047374 | methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.3 | 1.0 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) procollagen galactosyltransferase activity(GO:0050211) |
| 0.3 | 4.4 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.3 | 31.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.3 | 1.0 | GO:0008534 | oxidized purine nucleobase lesion DNA N-glycosylase activity(GO:0008534) |
| 0.3 | 1.9 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.3 | 6.4 | GO:0016502 | G-protein coupled nucleotide receptor activity(GO:0001608) purinergic nucleotide receptor activity(GO:0001614) nucleotide receptor activity(GO:0016502) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.3 | 4.4 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.3 | 7.5 | GO:0071949 | FAD binding(GO:0071949) |
| 0.3 | 8.9 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.3 | 4.3 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.3 | 2.3 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.3 | 1.4 | GO:0004368 | glycerol-3-phosphate dehydrogenase activity(GO:0004368) oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.3 | 5.2 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.3 | 11.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.3 | 2.8 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.3 | 2.0 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.3 | 9.8 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.3 | 15.1 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.2 | 6.7 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.2 | 3.4 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.2 | 2.5 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.2 | 9.2 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.2 | 1.2 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.2 | 0.7 | GO:0050473 | hepoxilin-epoxide hydrolase activity(GO:0047977) arachidonate 15-lipoxygenase activity(GO:0050473) |
| 0.2 | 4.8 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.2 | 1.7 | GO:0043559 | insulin binding(GO:0043559) |
| 0.2 | 1.2 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.2 | 8.6 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.2 | 1.6 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.2 | 2.9 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.2 | 7.4 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.2 | 1.1 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.2 | 2.8 | GO:0031005 | filamin binding(GO:0031005) |
| 0.2 | 0.9 | GO:0034618 | arginine binding(GO:0034618) |
| 0.2 | 3.8 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.2 | 2.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.2 | 12.6 | GO:0043621 | protein self-association(GO:0043621) |
| 0.2 | 0.6 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.2 | 1.9 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.2 | 6.0 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.2 | 32.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.2 | 3.6 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.2 | 1.2 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.2 | 14.0 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.2 | 4.7 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.2 | 0.6 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.2 | 3.0 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.2 | 3.3 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.2 | 4.1 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.2 | 1.7 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.2 | 1.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.2 | 2.9 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.2 | 4.9 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 1.0 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.2 | 4.4 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.2 | 6.8 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.2 | 41.6 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.2 | 3.3 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.2 | 3.8 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.2 | 0.5 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.2 | 2.1 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.2 | 0.6 | GO:0001855 | complement component C4b binding(GO:0001855) |
| 0.2 | 0.8 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.2 | 0.5 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.1 | 5.2 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 2.0 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 1.9 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 15.8 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 1.0 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.1 | 36.0 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 2.1 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.1 | 1.6 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 1.5 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.1 | 0.9 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 1.6 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.1 | 17.5 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.1 | 5.0 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 2.6 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 3.9 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 2.5 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.1 | 2.9 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 6.9 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.1 | 0.8 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 2.3 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 0.2 | GO:0089720 | caspase binding(GO:0089720) |
| 0.1 | 31.4 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.1 | 2.4 | GO:0001637 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) |
| 0.1 | 18.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 4.3 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.1 | 0.6 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 1.2 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 3.4 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 1.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 1.2 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.1 | 1.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 1.6 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.1 | 0.3 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.1 | 3.2 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.1 | 2.1 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 0.5 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.1 | 6.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 5.7 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 0.5 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.1 | 0.6 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 1.4 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 0.3 | GO:0015307 | drug:proton antiporter activity(GO:0015307) |
| 0.1 | 0.2 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.5 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 1.7 | GO:0015144 | carbohydrate transmembrane transporter activity(GO:0015144) carbohydrate transporter activity(GO:1901476) |
| 0.0 | 0.3 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 2.7 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.2 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 1.2 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.3 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.3 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 1.6 | GO:0004520 | endodeoxyribonuclease activity(GO:0004520) |
| 0.0 | 0.2 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 1.1 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 2.6 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.0 | 0.4 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.1 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 1.0 | GO:0005548 | phospholipid transporter activity(GO:0005548) |
| 0.0 | 1.0 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.9 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.0 | 1.1 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.3 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.8 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 2.4 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.1 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 109.0 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 1.2 | 56.1 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.6 | 37.0 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.6 | 31.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.5 | 83.9 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.5 | 9.8 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.5 | 43.5 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.5 | 2.5 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.5 | 9.9 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.5 | 4.3 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.4 | 4.4 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.4 | 15.7 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.4 | 24.9 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.4 | 3.8 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.4 | 12.7 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.3 | 6.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.3 | 16.9 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.3 | 13.9 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.2 | 7.2 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.2 | 3.2 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.2 | 17.9 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.2 | 5.7 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.2 | 53.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.2 | 6.3 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.2 | 6.5 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.2 | 6.6 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.2 | 1.6 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.2 | 5.0 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.2 | 3.4 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.2 | 3.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 2.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 2.2 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.1 | 6.5 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 2.3 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.1 | 7.9 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 2.2 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.1 | 1.9 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.1 | 5.4 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 0.9 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 0.6 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 7.3 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 1.2 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.1 | 1.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 11.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 1.5 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 2.0 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 1.4 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 1.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 2.6 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 1.7 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 1.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.3 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.8 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 1.5 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.3 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 6.1 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.2 | 46.0 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 6.3 | 137.5 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 2.5 | 24.9 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 1.8 | 20.0 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 1.4 | 116.3 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 1.2 | 23.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 1.0 | 5.8 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.9 | 4.4 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.6 | 9.1 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.5 | 12.6 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.4 | 22.2 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.4 | 3.4 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.4 | 3.1 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.4 | 10.6 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.4 | 9.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.3 | 6.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.3 | 4.0 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.3 | 4.8 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.3 | 4.7 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.3 | 5.0 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.3 | 6.6 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.3 | 12.8 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.3 | 9.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.3 | 1.1 | REACTOME MRNA PROCESSING | Genes involved in mRNA Processing |
| 0.3 | 8.3 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.2 | 2.6 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.2 | 5.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.2 | 4.4 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.2 | 28.2 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.2 | 7.5 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.2 | 2.7 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.2 | 17.2 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.2 | 2.7 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.2 | 13.6 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.2 | 4.5 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.2 | 12.5 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.2 | 5.9 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.2 | 4.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.2 | 5.9 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.2 | 7.1 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.2 | 2.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.2 | 4.0 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 4.5 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 3.9 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.1 | 5.8 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.1 | 1.5 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.1 | 1.1 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.1 | 3.8 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 3.3 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 8.0 | REACTOME TRANS GOLGI NETWORK VESICLE BUDDING | Genes involved in trans-Golgi Network Vesicle Budding |
| 0.1 | 4.4 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 5.7 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 3.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 5.6 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.1 | 1.1 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.1 | 1.6 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 5.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 3.6 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.1 | 1.7 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 2.6 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 0.6 | REACTOME BILE ACID AND BILE SALT METABOLISM | Genes involved in Bile acid and bile salt metabolism |
| 0.1 | 1.5 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 4.1 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.1 | 0.2 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.1 | 2.6 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 2.4 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.1 | 11.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 10.4 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.1 | 9.7 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 2.8 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 1.2 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 1.0 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 1.9 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 2.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.1 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 4.6 | REACTOME TOLL RECEPTOR CASCADES | Genes involved in Toll Receptor Cascades |
| 0.0 | 7.8 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.0 | 1.7 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 1.3 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.5 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.8 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 4.1 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 0.7 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.4 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 1.0 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.1 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 1.2 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.3 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |