Project
averaged GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for PAX7_NOBOX
Z-value: 1.07
Transcription factors associated with PAX7_NOBOX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
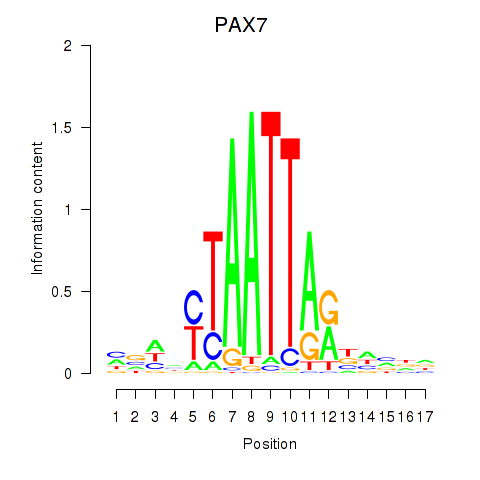
PAX7
|
ENSG00000009709.7 | paired box 7 |
|
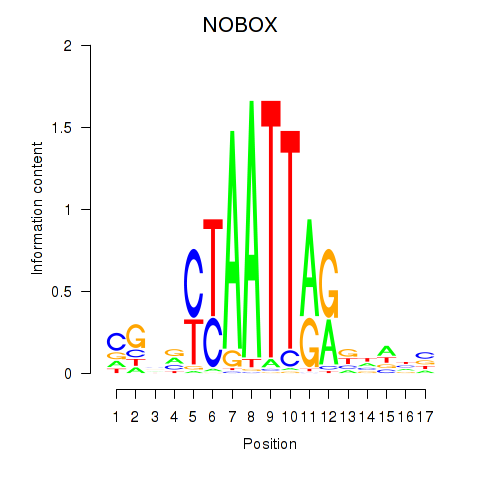
NOBOX
|
ENSG00000106410.10 | NOBOX oogenesis homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PAX7 | hg19_v2_chr1_+_18958008_18958023 | 0.54 | 4.0e-18 | Click! |
Activity profile of PAX7_NOBOX motif
Sorted Z-values of PAX7_NOBOX motif
Network of associatons between targets according to the STRING database.
First level regulatory network of PAX7_NOBOX
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.1 | 24.3 | GO:0019859 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 4.1 | 12.2 | GO:0046125 | thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 3.5 | 17.4 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 2.7 | 8.1 | GO:0071140 | resolution of recombination intermediates(GO:0071139) resolution of mitotic recombination intermediates(GO:0071140) |
| 2.5 | 10.0 | GO:0072166 | posterior mesonephric tubule development(GO:0072166) negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) |
| 2.3 | 16.2 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 2.3 | 6.9 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 2.0 | 8.1 | GO:0061743 | motor learning(GO:0061743) |
| 1.9 | 22.4 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 1.8 | 23.4 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 1.8 | 5.3 | GO:0052331 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 1.7 | 10.4 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 1.6 | 4.9 | GO:1900114 | positive regulation of histone H3-K9 trimethylation(GO:1900114) |
| 1.6 | 8.0 | GO:0060979 | vasculogenesis involved in coronary vascular morphogenesis(GO:0060979) |
| 1.5 | 4.5 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 1.4 | 4.1 | GO:2000048 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 1.3 | 3.8 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 1.0 | 24.1 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 1.0 | 6.0 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 1.0 | 1.0 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 1.0 | 3.9 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 1.0 | 2.9 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.9 | 12.9 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.9 | 2.7 | GO:1990168 | protein K33-linked deubiquitination(GO:1990168) |
| 0.9 | 3.5 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.9 | 2.6 | GO:0002589 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) regulation of T cell antigen processing and presentation(GO:0002625) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) response to iron ion starvation(GO:1990641) |
| 0.8 | 2.5 | GO:0010700 | negative regulation of norepinephrine secretion(GO:0010700) |
| 0.8 | 6.6 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.8 | 10.2 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.8 | 6.3 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.8 | 6.2 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.8 | 10.8 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.7 | 4.4 | GO:0052565 | response to defense-related nitric oxide production by other organism involved in symbiotic interaction(GO:0052551) response to defense-related host nitric oxide production(GO:0052565) |
| 0.7 | 3.6 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.7 | 5.0 | GO:0044800 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.7 | 11.3 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.7 | 2.1 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.7 | 2.0 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.7 | 2.7 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.6 | 4.5 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.6 | 6.2 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.6 | 1.8 | GO:0007231 | osmosensory signaling pathway(GO:0007231) |
| 0.6 | 4.3 | GO:0048241 | epinephrine transport(GO:0048241) |
| 0.6 | 2.4 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.6 | 3.5 | GO:0033133 | fructose 2,6-bisphosphate metabolic process(GO:0006003) positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.6 | 6.4 | GO:1902172 | keratinocyte apoptotic process(GO:0097283) regulation of keratinocyte apoptotic process(GO:1902172) |
| 0.6 | 2.3 | GO:0003095 | pressure natriuresis(GO:0003095) |
| 0.6 | 5.8 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.5 | 4.7 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.5 | 11.5 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.5 | 1.0 | GO:1990418 | response to insulin-like growth factor stimulus(GO:1990418) |
| 0.5 | 4.3 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
| 0.5 | 12.1 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.5 | 1.9 | GO:1903719 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.5 | 5.4 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.4 | 4.3 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.4 | 3.4 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.4 | 9.5 | GO:0044126 | regulation of growth of symbiont in host(GO:0044126) |
| 0.4 | 2.0 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.4 | 0.8 | GO:0042320 | regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) |
| 0.4 | 1.6 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.4 | 2.0 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.4 | 2.6 | GO:0046836 | glycolipid transport(GO:0046836) |
| 0.4 | 1.5 | GO:0051563 | smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) |
| 0.4 | 3.6 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.4 | 1.4 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.3 | 7.1 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.3 | 6.1 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.3 | 1.0 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.3 | 2.9 | GO:0032224 | positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.3 | 0.9 | GO:0060369 | natural killer cell cytokine production(GO:0002370) regulation of natural killer cell cytokine production(GO:0002727) positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.3 | 2.4 | GO:1902750 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) negative regulation of cell cycle G2/M phase transition(GO:1902750) |
| 0.3 | 1.8 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.3 | 1.2 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.3 | 1.2 | GO:0070092 | regulation of glucagon secretion(GO:0070092) |
| 0.3 | 2.7 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.3 | 2.3 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.3 | 1.1 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.3 | 3.1 | GO:0015074 | DNA integration(GO:0015074) |
| 0.3 | 1.1 | GO:0021784 | postganglionic parasympathetic fiber development(GO:0021784) |
| 0.3 | 1.6 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.3 | 3.8 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.3 | 3.7 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.3 | 0.8 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.3 | 13.8 | GO:0006458 | 'de novo' protein folding(GO:0006458) |
| 0.2 | 0.7 | GO:0042109 | negative regulation of tolerance induction(GO:0002644) lymphotoxin A production(GO:0032641) interleukin-4 biosynthetic process(GO:0042097) lymphotoxin A biosynthetic process(GO:0042109) regulation of interleukin-4 biosynthetic process(GO:0045402) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.2 | 1.5 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.2 | 3.8 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.2 | 0.9 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.2 | 1.1 | GO:0002826 | negative regulation of T-helper 1 type immune response(GO:0002826) |
| 0.2 | 4.9 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.2 | 38.5 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.2 | 3.9 | GO:0042246 | tissue regeneration(GO:0042246) |
| 0.2 | 0.9 | GO:0097527 | microglial cell activation involved in immune response(GO:0002282) type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) positive regulation of interferon-alpha biosynthetic process(GO:0045356) necroptotic signaling pathway(GO:0097527) |
| 0.2 | 2.1 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.2 | 1.3 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.2 | 3.7 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.2 | 1.0 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.2 | 1.2 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.2 | 3.8 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.2 | 2.9 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.2 | 1.0 | GO:0050955 | thermoception(GO:0050955) |
| 0.2 | 0.8 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.2 | 4.0 | GO:0030220 | platelet formation(GO:0030220) |
| 0.2 | 2.6 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.2 | 1.3 | GO:0006772 | thiamine metabolic process(GO:0006772) |
| 0.2 | 1.8 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.2 | 1.1 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.2 | 1.7 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.2 | 1.7 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.2 | 11.7 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.2 | 1.8 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.2 | 2.0 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.2 | 2.3 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.2 | 3.9 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.2 | 4.3 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.2 | 3.6 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.2 | 0.8 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 | 0.7 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.1 | 4.3 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.1 | 1.9 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.1 | 0.7 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.1 | 5.8 | GO:0019835 | cytolysis(GO:0019835) |
| 0.1 | 0.9 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.1 | 1.7 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 2.1 | GO:0010804 | negative regulation of tumor necrosis factor-mediated signaling pathway(GO:0010804) |
| 0.1 | 2.8 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.1 | 7.6 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.1 | 2.3 | GO:0060080 | inhibitory postsynaptic potential(GO:0060080) |
| 0.1 | 1.2 | GO:0007351 | tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.1 | 0.9 | GO:0021940 | positive regulation of cerebellar granule cell precursor proliferation(GO:0021940) |
| 0.1 | 2.2 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 1.2 | GO:0072178 | negative regulation of synapse assembly(GO:0051964) nephric duct morphogenesis(GO:0072178) |
| 0.1 | 0.3 | GO:0042746 | regulation of circadian sleep/wake cycle, wakefulness(GO:0010840) circadian sleep/wake cycle, wakefulness(GO:0042746) |
| 0.1 | 1.8 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.1 | 1.6 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.1 | 1.1 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.1 | 8.3 | GO:0042795 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.1 | 1.0 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.1 | 0.4 | GO:0036343 | negative regulation of extracellular matrix disassembly(GO:0010716) psychomotor behavior(GO:0036343) |
| 0.1 | 2.3 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.1 | 1.1 | GO:0098856 | intestinal cholesterol absorption(GO:0030299) intestinal lipid absorption(GO:0098856) |
| 0.1 | 0.8 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.1 | 2.9 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 1.9 | GO:0018146 | keratan sulfate biosynthetic process(GO:0018146) |
| 0.1 | 3.9 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 1.5 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.1 | 0.6 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) |
| 0.1 | 1.8 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.1 | 2.7 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 0.7 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.1 | 3.8 | GO:0009790 | embryo development(GO:0009790) |
| 0.1 | 3.8 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.1 | 26.9 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.1 | 1.0 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.1 | 1.7 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.1 | 8.3 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.1 | 0.7 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 1.5 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.1 | 1.0 | GO:0050974 | detection of mechanical stimulus involved in sensory perception(GO:0050974) |
| 0.1 | 2.1 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.1 | 1.7 | GO:0030728 | ovulation(GO:0030728) |
| 0.1 | 4.3 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.1 | 0.4 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.1 | 4.9 | GO:0000725 | double-strand break repair via homologous recombination(GO:0000724) recombinational repair(GO:0000725) |
| 0.1 | 1.6 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.1 | 1.3 | GO:0008211 | glucocorticoid metabolic process(GO:0008211) |
| 0.1 | 3.1 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.1 | 0.4 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.1 | 1.7 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 0.8 | GO:0006067 | ethanol metabolic process(GO:0006067) ethanol catabolic process(GO:0006068) |
| 0.1 | 0.3 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.1 | 1.5 | GO:0010762 | regulation of fibroblast migration(GO:0010762) |
| 0.0 | 1.7 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 1.8 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 0.1 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.0 | 0.6 | GO:0036315 | cellular response to sterol(GO:0036315) |
| 0.0 | 0.6 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.0 | 26.1 | GO:0045596 | negative regulation of cell differentiation(GO:0045596) |
| 0.0 | 1.2 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.4 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 6.3 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.0 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 6.0 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.1 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.4 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.9 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.4 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.9 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 1.8 | GO:0030218 | erythrocyte differentiation(GO:0030218) |
| 0.0 | 5.9 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.0 | 1.2 | GO:0031295 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.5 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 0.7 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.7 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.3 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.0 | 1.9 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.0 | 0.6 | GO:0006986 | response to unfolded protein(GO:0006986) |
| 0.0 | 2.5 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.5 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 2.0 | GO:0060271 | cilium morphogenesis(GO:0060271) |
| 0.0 | 0.1 | GO:0042355 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.4 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 1.7 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.4 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.0 | 0.2 | GO:0045332 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 10.4 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 1.7 | 10.3 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 1.6 | 22.4 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 1.4 | 8.1 | GO:0033061 | DNA recombinase mediator complex(GO:0033061) Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 1.1 | 3.4 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 1.1 | 3.3 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 1.1 | 4.3 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 1.0 | 4.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 1.0 | 2.9 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.8 | 6.6 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.8 | 7.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.7 | 2.1 | GO:0097679 | other organism cytoplasm(GO:0097679) |
| 0.6 | 2.6 | GO:1990357 | terminal web(GO:1990357) |
| 0.6 | 1.9 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.6 | 8.0 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.6 | 1.7 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.5 | 17.1 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.5 | 3.5 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.5 | 2.3 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.4 | 5.2 | GO:0043203 | axon hillock(GO:0043203) |
| 0.4 | 4.5 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.4 | 3.7 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.4 | 9.9 | GO:0032982 | myosin filament(GO:0032982) |
| 0.3 | 2.0 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.3 | 5.7 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.3 | 4.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.3 | 3.5 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.3 | 6.9 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.3 | 16.8 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.3 | 7.5 | GO:0005605 | basal lamina(GO:0005605) |
| 0.3 | 7.6 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.3 | 2.3 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.3 | 2.0 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.3 | 2.3 | GO:0005638 | lamin filament(GO:0005638) |
| 0.3 | 2.0 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.2 | 1.5 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.2 | 4.9 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.2 | 4.4 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.2 | 6.3 | GO:0030673 | axolemma(GO:0030673) |
| 0.2 | 1.8 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.2 | 3.0 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.2 | 2.3 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.2 | 2.0 | GO:0036038 | MKS complex(GO:0036038) |
| 0.2 | 5.4 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.1 | 1.0 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.1 | 1.8 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.1 | 1.0 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 0.9 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.1 | 1.6 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 1.5 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 1.5 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 4.9 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.1 | 0.8 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.1 | 1.8 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 2.1 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.1 | 19.6 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.1 | 1.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 0.6 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 4.8 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 1.8 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 8.6 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.1 | 3.9 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 2.7 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 1.7 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 2.3 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 2.7 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 2.9 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 0.9 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.1 | 10.8 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 18.7 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 23.5 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 0.6 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.3 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 1.3 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 4.4 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 29.2 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 3.8 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 50.2 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 1.3 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 1.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 6.8 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 3.9 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.7 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.5 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.9 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 1.0 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 4.3 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.0 | 2.9 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 1.0 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 0.7 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.2 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.7 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 1.4 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.3 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 12.2 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 4.1 | 16.2 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 3.4 | 10.2 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 2.4 | 14.3 | GO:0000285 | 1-phosphatidylinositol-3-phosphate 5-kinase activity(GO:0000285) |
| 2.4 | 23.5 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 2.0 | 10.0 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 1.9 | 5.6 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 1.8 | 5.4 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 1.5 | 4.5 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 1.4 | 10.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 1.3 | 10.7 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 1.1 | 4.3 | GO:0005277 | acetylcholine transmembrane transporter activity(GO:0005277) secondary active organic cation transmembrane transporter activity(GO:0008513) acetate ester transmembrane transporter activity(GO:1901375) |
| 1.0 | 8.1 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 1.0 | 24.3 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.9 | 2.7 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.9 | 12.7 | GO:0031432 | titin binding(GO:0031432) |
| 0.9 | 6.3 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.8 | 6.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.8 | 3.1 | GO:0004803 | transposase activity(GO:0004803) |
| 0.7 | 3.6 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.7 | 3.6 | GO:0004771 | sterol esterase activity(GO:0004771) methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.7 | 4.9 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.7 | 3.4 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.7 | 4.7 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.6 | 2.5 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.6 | 1.9 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.6 | 1.8 | GO:0030366 | molybdopterin synthase activity(GO:0030366) |
| 0.6 | 3.5 | GO:0004331 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.6 | 2.3 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.6 | 6.2 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.6 | 2.8 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.5 | 11.5 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.5 | 2.1 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.5 | 4.1 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.5 | 8.5 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.5 | 6.0 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.4 | 1.3 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.4 | 1.3 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.4 | 4.3 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.4 | 4.1 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.4 | 2.9 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.4 | 5.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.4 | 1.1 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.4 | 5.0 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.3 | 3.8 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.3 | 23.0 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.3 | 1.7 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.3 | 1.1 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.3 | 1.3 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.3 | 1.8 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.3 | 5.0 | GO:0043495 | protein anchor(GO:0043495) |
| 0.2 | 1.9 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.2 | 1.0 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.2 | 2.6 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.2 | 2.8 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.2 | 0.9 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.2 | 2.3 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.2 | 1.5 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.2 | 6.9 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.2 | 47.6 | GO:0003823 | antigen binding(GO:0003823) |
| 0.2 | 4.7 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.2 | 10.4 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.2 | 2.0 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.2 | 0.8 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.2 | 5.7 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 21.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 1.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.2 | 1.8 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.2 | 1.0 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.2 | 1.7 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.2 | 4.4 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.2 | 15.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.2 | 2.3 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.2 | 1.0 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.2 | 0.6 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.2 | 0.8 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.2 | 1.2 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.1 | 0.4 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.1 | 2.2 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.1 | 3.8 | GO:0071949 | FAD binding(GO:0071949) |
| 0.1 | 7.6 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 0.1 | GO:0016880 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 0.8 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) steroid sulfotransferase activity(GO:0050294) |
| 0.1 | 2.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 1.2 | GO:0016918 | retinal binding(GO:0016918) |
| 0.1 | 0.4 | GO:0008422 | beta-glucuronidase activity(GO:0004566) beta-glucosidase activity(GO:0008422) |
| 0.1 | 38.1 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 8.0 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.1 | 0.6 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.1 | 5.5 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 1.6 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.1 | 1.8 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 2.6 | GO:0032934 | cholesterol binding(GO:0015485) sterol binding(GO:0032934) |
| 0.1 | 4.3 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 2.0 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.1 | 1.6 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.1 | 0.2 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 4.1 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 2.9 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 0.3 | GO:0016499 | orexin receptor activity(GO:0016499) |
| 0.1 | 2.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 2.1 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.1 | 3.8 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.1 | 1.5 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 5.5 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 1.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 0.7 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.1 | 5.2 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.1 | 0.4 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.1 | 0.7 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.1 | 15.3 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.1 | 1.5 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.1 | 4.3 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 2.0 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 1.8 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 1.8 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 1.0 | GO:0019001 | guanyl nucleotide binding(GO:0019001) guanyl ribonucleotide binding(GO:0032561) |
| 0.1 | 8.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 5.2 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.0 | 1.5 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 4.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.4 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.5 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 5.4 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 1.1 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 11.3 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 1.1 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 2.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 12.8 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 0.8 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.4 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 1.0 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 0.3 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.6 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.0 | 0.5 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.7 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.8 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.0 | 0.4 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) lysophosphatidic acid acyltransferase activity(GO:0042171) lysophospholipid acyltransferase activity(GO:0071617) |
| 0.0 | 0.6 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.9 | GO:0005319 | lipid transporter activity(GO:0005319) |
| 0.0 | 0.1 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 3.5 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 2.4 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.7 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 0.9 | GO:0005178 | integrin binding(GO:0005178) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 34.2 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.3 | 25.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.3 | 11.8 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.2 | 27.5 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.2 | 7.6 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.2 | 10.6 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.2 | 9.1 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.2 | 3.6 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.2 | 7.3 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.1 | 1.2 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 2.8 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 17.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 3.4 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 19.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 5.0 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.1 | 21.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 2.2 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 2.4 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 1.5 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 3.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 3.4 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 2.6 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 1.2 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 1.8 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 5.0 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.1 | 4.1 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 0.7 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 4.1 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 0.8 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 1.8 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 3.2 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 3.4 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 4.1 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 1.6 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 2.0 | NABA CORE MATRISOME | Ensemble of genes encoding core extracellular matrix including ECM glycoproteins, collagens and proteoglycans |
| 0.0 | 1.1 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.9 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.9 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.1 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 2.0 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.7 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 1.0 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.4 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.6 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 5.0 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.9 | 10.4 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.8 | 22.4 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.7 | 19.2 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.6 | 7.4 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.6 | 10.7 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.5 | 11.5 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.3 | 3.8 | REACTOME OPSINS | Genes involved in Opsins |
| 0.3 | 5.5 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.3 | 26.4 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.3 | 2.3 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.2 | 3.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.2 | 3.8 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.2 | 3.6 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.2 | 7.2 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.2 | 6.4 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.2 | 4.7 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.2 | 12.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 2.7 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.2 | 6.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.2 | 28.5 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 7.1 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 2.5 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 2.1 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 5.0 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 3.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 3.0 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 5.4 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 5.4 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 5.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 3.8 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 0.9 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 4.1 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.1 | 4.0 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 6.3 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 0.8 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.1 | 1.7 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.1 | 1.5 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.1 | 3.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 4.0 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.1 | 2.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 1.9 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.1 | 1.9 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 0.9 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 0.7 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 1.5 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 1.9 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 1.2 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.1 | 1.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 2.2 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 2.3 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 0.7 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 2.0 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.1 | 2.1 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.1 | 1.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 1.8 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 2.3 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 1.8 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 1.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 1.1 | REACTOME CELL DEATH SIGNALLING VIA NRAGE NRIF AND NADE | Genes involved in Cell death signalling via NRAGE, NRIF and NADE |
| 0.0 | 1.1 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.0 | 4.9 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 2.0 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.0 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 3.0 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 1.1 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 1.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 1.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.8 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 1.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.7 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.0 | 0.6 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.6 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.8 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 6.7 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 0.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.9 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.4 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.0 | 0.4 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |