Project
averaged GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for PGR
Z-value: 0.79
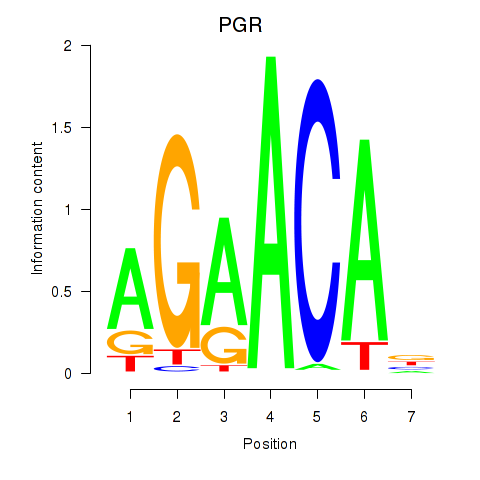
Transcription factors associated with PGR
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PGR
|
ENSG00000082175.10 | progesterone receptor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PGR | hg19_v2_chr11_-_100999775_100999801, hg19_v2_chr11_-_101000445_101000465 | 0.12 | 7.8e-02 | Click! |
Activity profile of PGR motif
Sorted Z-values of PGR motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_88450612 | 19.03 |
ENST00000418378.1
ENST00000282470.6 |
SPARCL1
|
SPARC-like 1 (hevin) |
| chr3_-_114343768 | 9.39 |
ENST00000393785.2
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chrX_+_38420623 | 7.70 |
ENST00000378482.2
|
TSPAN7
|
tetraspanin 7 |
| chr3_-_114343039 | 6.91 |
ENST00000481632.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr3_-_48057890 | 6.78 |
ENST00000434267.1
|
MAP4
|
microtubule-associated protein 4 |
| chr8_-_22089845 | 6.78 |
ENST00000454243.2
|
PHYHIP
|
phytanoyl-CoA 2-hydroxylase interacting protein |
| chr15_+_31619013 | 6.44 |
ENST00000307145.3
|
KLF13
|
Kruppel-like factor 13 |
| chr7_+_30960915 | 6.40 |
ENST00000441328.2
ENST00000409899.1 ENST00000409611.1 |
AQP1
|
aquaporin 1 (Colton blood group) |
| chr3_-_58563094 | 6.35 |
ENST00000464064.1
|
FAM107A
|
family with sequence similarity 107, member A |
| chr6_-_154831779 | 6.32 |
ENST00000607772.1
|
CNKSR3
|
CNKSR family member 3 |
| chr18_+_29171689 | 5.98 |
ENST00000237014.3
|
TTR
|
transthyretin |
| chr12_+_79258547 | 5.97 |
ENST00000457153.2
|
SYT1
|
synaptotagmin I |
| chr7_-_37026108 | 5.80 |
ENST00000396045.3
|
ELMO1
|
engulfment and cell motility 1 |
| chr11_-_66725837 | 5.50 |
ENST00000393958.2
ENST00000393960.1 ENST00000524491.1 ENST00000355677.3 |
PC
|
pyruvate carboxylase |
| chr6_-_32908765 | 5.39 |
ENST00000416244.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr3_-_114035026 | 5.16 |
ENST00000570269.1
|
RP11-553L6.5
|
RP11-553L6.5 |
| chr19_-_36247910 | 5.07 |
ENST00000587965.1
ENST00000004982.3 |
HSPB6
|
heat shock protein, alpha-crystallin-related, B6 |
| chr5_-_160279207 | 5.01 |
ENST00000327245.5
|
ATP10B
|
ATPase, class V, type 10B |
| chr17_-_26903900 | 4.92 |
ENST00000395319.3
ENST00000581807.1 ENST00000584086.1 ENST00000395321.2 |
ALDOC
|
aldolase C, fructose-bisphosphate |
| chr7_-_129845313 | 4.58 |
ENST00000397622.2
|
TMEM209
|
transmembrane protein 209 |
| chr8_+_133931648 | 4.57 |
ENST00000519178.1
ENST00000542445.1 |
TG
|
thyroglobulin |
| chr10_-_28571015 | 4.39 |
ENST00000375719.3
ENST00000375732.1 |
MPP7
|
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr7_-_129845188 | 4.27 |
ENST00000462753.1
ENST00000471077.1 ENST00000473456.1 ENST00000336804.8 |
TMEM209
|
transmembrane protein 209 |
| chr19_-_40440533 | 4.12 |
ENST00000221347.6
|
FCGBP
|
Fc fragment of IgG binding protein |
| chr19_-_14952689 | 3.88 |
ENST00000248058.1
|
OR7A10
|
olfactory receptor, family 7, subfamily A, member 10 |
| chr13_+_32838801 | 3.86 |
ENST00000542859.1
|
FRY
|
furry homolog (Drosophila) |
| chr9_+_87285539 | 3.74 |
ENST00000359847.3
|
NTRK2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr6_-_136788001 | 3.73 |
ENST00000544465.1
|
MAP7
|
microtubule-associated protein 7 |
| chr19_+_30863271 | 3.64 |
ENST00000355537.3
|
ZNF536
|
zinc finger protein 536 |
| chr15_+_31658349 | 3.60 |
ENST00000558844.1
|
KLF13
|
Kruppel-like factor 13 |
| chr12_-_11422630 | 3.58 |
ENST00000381842.3
ENST00000538488.1 |
PRB3
|
proline-rich protein BstNI subfamily 3 |
| chr19_+_19431490 | 3.56 |
ENST00000392313.6
ENST00000262815.8 ENST00000609122.1 |
MAU2
|
MAU2 sister chromatid cohesion factor |
| chr4_-_87028478 | 3.55 |
ENST00000515400.1
ENST00000395157.3 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chrX_+_46940254 | 3.43 |
ENST00000336169.3
|
RGN
|
regucalcin |
| chr11_+_64851729 | 3.42 |
ENST00000526791.1
ENST00000526945.1 |
ZFPL1
|
zinc finger protein-like 1 |
| chr2_+_99797542 | 3.39 |
ENST00000338148.3
ENST00000512183.2 |
MRPL30
C2orf15
|
mitochondrial ribosomal protein L30 chromosome 2 open reading frame 15 |
| chrX_-_78622805 | 3.39 |
ENST00000373298.2
|
ITM2A
|
integral membrane protein 2A |
| chr15_-_42783303 | 3.36 |
ENST00000565380.1
ENST00000564754.1 |
ZNF106
|
zinc finger protein 106 |
| chr3_+_111260954 | 3.30 |
ENST00000283285.5
|
CD96
|
CD96 molecule |
| chr3_+_187871060 | 3.26 |
ENST00000448637.1
|
LPP
|
LIM domain containing preferred translocation partner in lipoma |
| chr1_+_155829286 | 3.24 |
ENST00000368324.4
|
SYT11
|
synaptotagmin XI |
| chr13_+_102104952 | 3.23 |
ENST00000376180.3
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr3_+_111260856 | 3.18 |
ENST00000352690.4
|
CD96
|
CD96 molecule |
| chr2_+_60983361 | 3.17 |
ENST00000238714.3
|
PAPOLG
|
poly(A) polymerase gamma |
| chrX_+_23352133 | 3.08 |
ENST00000379361.4
|
PTCHD1
|
patched domain containing 1 |
| chr14_+_24641062 | 3.06 |
ENST00000311457.3
ENST00000557806.1 ENST00000559919.1 |
REC8
|
REC8 meiotic recombination protein |
| chr5_+_67584174 | 3.00 |
ENST00000320694.8
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr8_+_10530155 | 3.00 |
ENST00000521818.1
|
C8orf74
|
chromosome 8 open reading frame 74 |
| chr22_+_41253080 | 2.95 |
ENST00000541156.1
ENST00000414396.1 ENST00000357137.4 |
XPNPEP3
|
X-prolyl aminopeptidase (aminopeptidase P) 3, putative |
| chr2_-_175711133 | 2.95 |
ENST00000409597.1
ENST00000413882.1 |
CHN1
|
chimerin 1 |
| chr8_-_26371608 | 2.94 |
ENST00000522362.2
|
PNMA2
|
paraneoplastic Ma antigen 2 |
| chr19_+_10531150 | 2.92 |
ENST00000352831.6
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific |
| chr12_+_125478241 | 2.91 |
ENST00000341446.8
|
BRI3BP
|
BRI3 binding protein |
| chr20_+_23016057 | 2.89 |
ENST00000255008.3
|
SSTR4
|
somatostatin receptor 4 |
| chr6_+_27107053 | 2.87 |
ENST00000354348.2
|
HIST1H4I
|
histone cluster 1, H4i |
| chr18_+_580367 | 2.87 |
ENST00000327228.3
|
CETN1
|
centrin, EF-hand protein, 1 |
| chr19_-_19144243 | 2.84 |
ENST00000594445.1
ENST00000452918.2 ENST00000600377.1 ENST00000337018.6 |
SUGP2
|
SURP and G patch domain containing 2 |
| chr7_+_73442422 | 2.84 |
ENST00000358929.4
ENST00000431562.1 ENST00000320492.7 ENST00000438906.1 |
ELN
|
elastin |
| chrX_-_130037198 | 2.78 |
ENST00000370935.1
ENST00000338144.3 ENST00000394363.1 |
ENOX2
|
ecto-NOX disulfide-thiol exchanger 2 |
| chr6_+_72926145 | 2.78 |
ENST00000425662.2
ENST00000453976.2 |
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr15_-_88799661 | 2.73 |
ENST00000360948.2
ENST00000357724.2 ENST00000355254.2 ENST00000317501.3 |
NTRK3
|
neurotrophic tyrosine kinase, receptor, type 3 |
| chr1_+_150229554 | 2.72 |
ENST00000369111.4
|
CA14
|
carbonic anhydrase XIV |
| chr5_+_102201687 | 2.70 |
ENST00000304400.7
|
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr4_+_128802016 | 2.60 |
ENST00000270861.5
ENST00000515069.1 ENST00000513090.1 ENST00000507249.1 |
PLK4
|
polo-like kinase 4 |
| chr1_+_111770232 | 2.60 |
ENST00000369744.2
|
CHI3L2
|
chitinase 3-like 2 |
| chr13_+_49822041 | 2.60 |
ENST00000538056.1
ENST00000251108.6 ENST00000444959.1 ENST00000429346.1 |
CDADC1
|
cytidine and dCMP deaminase domain containing 1 |
| chr1_+_111770278 | 2.55 |
ENST00000369748.4
|
CHI3L2
|
chitinase 3-like 2 |
| chr5_-_146435694 | 2.54 |
ENST00000356826.3
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta |
| chr2_+_234602305 | 2.51 |
ENST00000406651.1
|
UGT1A6
|
UDP glucuronosyltransferase 1 family, polypeptide A6 |
| chr12_-_71551652 | 2.51 |
ENST00000546561.1
|
TSPAN8
|
tetraspanin 8 |
| chr9_-_113761720 | 2.46 |
ENST00000541779.1
ENST00000374430.2 |
LPAR1
|
lysophosphatidic acid receptor 1 |
| chr1_-_201390846 | 2.45 |
ENST00000367312.1
ENST00000555340.2 ENST00000361379.4 |
TNNI1
|
troponin I type 1 (skeletal, slow) |
| chr2_+_173686303 | 2.45 |
ENST00000397087.3
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr14_-_106692191 | 2.45 |
ENST00000390607.2
|
IGHV3-21
|
immunoglobulin heavy variable 3-21 |
| chr15_-_88799384 | 2.43 |
ENST00000540489.2
ENST00000557856.1 ENST00000558676.1 |
NTRK3
|
neurotrophic tyrosine kinase, receptor, type 3 |
| chr12_-_11422739 | 2.41 |
ENST00000279573.7
|
PRB3
|
proline-rich protein BstNI subfamily 3 |
| chr4_-_71532339 | 2.41 |
ENST00000254801.4
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr2_-_179672142 | 2.38 |
ENST00000342992.6
ENST00000360870.5 ENST00000460472.2 ENST00000589042.1 ENST00000591111.1 ENST00000342175.6 ENST00000359218.5 |
TTN
|
titin |
| chr5_-_131347501 | 2.37 |
ENST00000543479.1
|
ACSL6
|
acyl-CoA synthetase long-chain family member 6 |
| chr3_+_137906154 | 2.36 |
ENST00000466749.1
ENST00000358441.2 ENST00000489213.1 |
ARMC8
|
armadillo repeat containing 8 |
| chr8_-_87755878 | 2.35 |
ENST00000320005.5
|
CNGB3
|
cyclic nucleotide gated channel beta 3 |
| chrX_+_57618269 | 2.33 |
ENST00000374888.1
|
ZXDB
|
zinc finger, X-linked, duplicated B |
| chr16_-_3306587 | 2.33 |
ENST00000541159.1
ENST00000536379.1 ENST00000219596.1 ENST00000339854.4 |
MEFV
|
Mediterranean fever |
| chr5_-_115872142 | 2.27 |
ENST00000510263.1
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr5_-_146435501 | 2.27 |
ENST00000336640.6
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta |
| chr5_+_140261703 | 2.26 |
ENST00000409494.1
ENST00000289272.2 |
PCDHA13
|
protocadherin alpha 13 |
| chr6_-_46922659 | 2.20 |
ENST00000265417.7
|
GPR116
|
G protein-coupled receptor 116 |
| chr7_+_128116783 | 2.19 |
ENST00000262432.8
ENST00000480046.1 |
METTL2B
|
methyltransferase like 2B |
| chr3_+_46412345 | 2.17 |
ENST00000292303.4
|
CCR5
|
chemokine (C-C motif) receptor 5 (gene/pseudogene) |
| chr12_-_49110613 | 2.16 |
ENST00000261900.3
|
CCNT1
|
cyclin T1 |
| chr5_-_146435572 | 2.16 |
ENST00000394414.1
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta |
| chr3_-_149470229 | 2.15 |
ENST00000473414.1
|
COMMD2
|
COMM domain containing 2 |
| chr19_+_41119323 | 2.11 |
ENST00000599724.1
ENST00000597071.1 ENST00000243562.9 |
LTBP4
|
latent transforming growth factor beta binding protein 4 |
| chr17_-_15168624 | 2.10 |
ENST00000312280.3
ENST00000494511.1 ENST00000580584.1 |
PMP22
|
peripheral myelin protein 22 |
| chr3_-_49722523 | 2.10 |
ENST00000448220.1
|
MST1
|
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr15_-_58306295 | 2.09 |
ENST00000559517.1
|
ALDH1A2
|
aldehyde dehydrogenase 1 family, member A2 |
| chr11_+_123396528 | 2.06 |
ENST00000322282.7
ENST00000529750.1 |
GRAMD1B
|
GRAM domain containing 1B |
| chr6_+_39760129 | 2.06 |
ENST00000274867.4
|
DAAM2
|
dishevelled associated activator of morphogenesis 2 |
| chr7_-_135433460 | 2.05 |
ENST00000415751.1
|
FAM180A
|
family with sequence similarity 180, member A |
| chr1_+_192778161 | 2.04 |
ENST00000235382.5
|
RGS2
|
regulator of G-protein signaling 2, 24kDa |
| chr2_-_73869508 | 2.04 |
ENST00000272425.3
|
NAT8
|
N-acetyltransferase 8 (GCN5-related, putative) |
| chr22_+_40297105 | 2.04 |
ENST00000540310.1
|
GRAP2
|
GRB2-related adaptor protein 2 |
| chr6_-_32908792 | 2.03 |
ENST00000418107.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chrX_-_102348017 | 1.99 |
ENST00000425644.1
ENST00000395065.3 ENST00000425463.2 |
NXF3
|
nuclear RNA export factor 3 |
| chr22_-_31688431 | 1.97 |
ENST00000402249.3
ENST00000443175.1 ENST00000215912.5 ENST00000441972.1 |
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr9_-_37034028 | 1.94 |
ENST00000520281.1
ENST00000446742.1 ENST00000522003.1 ENST00000523145.1 ENST00000414447.1 ENST00000377847.2 ENST00000377853.2 ENST00000377852.2 ENST00000523241.1 ENST00000520154.1 ENST00000358127.4 |
PAX5
|
paired box 5 |
| chr2_-_89266286 | 1.93 |
ENST00000464162.1
|
IGKV1-6
|
immunoglobulin kappa variable 1-6 |
| chr19_+_35773242 | 1.89 |
ENST00000222304.3
|
HAMP
|
hepcidin antimicrobial peptide |
| chr19_+_41768561 | 1.87 |
ENST00000599719.1
ENST00000601309.1 |
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr11_+_64851666 | 1.86 |
ENST00000525509.1
ENST00000294258.3 ENST00000526334.1 |
ZFPL1
|
zinc finger protein-like 1 |
| chr7_+_73442457 | 1.86 |
ENST00000438880.1
ENST00000414324.1 ENST00000380562.4 |
ELN
|
elastin |
| chr19_+_44488330 | 1.85 |
ENST00000591532.1
ENST00000407951.2 ENST00000270014.2 ENST00000590615.1 ENST00000586454.1 |
ZNF155
|
zinc finger protein 155 |
| chrX_-_49965663 | 1.85 |
ENST00000376056.2
ENST00000376058.2 ENST00000358526.2 |
AKAP4
|
A kinase (PRKA) anchor protein 4 |
| chr1_-_32210275 | 1.84 |
ENST00000440175.2
|
BAI2
|
brain-specific angiogenesis inhibitor 2 |
| chr5_+_156607829 | 1.79 |
ENST00000422843.3
|
ITK
|
IL2-inducible T-cell kinase |
| chr17_-_46608272 | 1.74 |
ENST00000577092.1
ENST00000239174.6 |
HOXB1
|
homeobox B1 |
| chr21_+_34398153 | 1.74 |
ENST00000382357.3
ENST00000430860.1 ENST00000333337.3 |
OLIG2
|
oligodendrocyte lineage transcription factor 2 |
| chr19_-_6333614 | 1.73 |
ENST00000301452.4
|
ACER1
|
alkaline ceramidase 1 |
| chr15_-_45406385 | 1.72 |
ENST00000389039.6
|
DUOX2
|
dual oxidase 2 |
| chr1_+_43803475 | 1.72 |
ENST00000372470.3
ENST00000413998.2 |
MPL
|
myeloproliferative leukemia virus oncogene |
| chr16_+_85942594 | 1.72 |
ENST00000566369.1
|
IRF8
|
interferon regulatory factor 8 |
| chr6_+_108487245 | 1.71 |
ENST00000368986.4
|
NR2E1
|
nuclear receptor subfamily 2, group E, member 1 |
| chr17_-_3301704 | 1.71 |
ENST00000322608.2
|
OR1E1
|
olfactory receptor, family 1, subfamily E, member 1 |
| chr5_-_131347583 | 1.70 |
ENST00000379255.1
ENST00000430403.1 ENST00000544770.1 ENST00000379246.1 ENST00000414078.1 ENST00000441995.1 |
ACSL6
|
acyl-CoA synthetase long-chain family member 6 |
| chr9_+_82188077 | 1.69 |
ENST00000425506.1
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr22_-_30162924 | 1.68 |
ENST00000344318.3
ENST00000397781.3 |
ZMAT5
|
zinc finger, matrin-type 5 |
| chrX_+_135570046 | 1.67 |
ENST00000370648.3
|
BRS3
|
bombesin-like receptor 3 |
| chr2_-_219537134 | 1.65 |
ENST00000295704.2
|
RNF25
|
ring finger protein 25 |
| chrX_-_112084043 | 1.63 |
ENST00000304758.1
|
AMOT
|
angiomotin |
| chr7_-_5821314 | 1.63 |
ENST00000425013.2
ENST00000389902.3 |
RNF216
|
ring finger protein 216 |
| chr22_+_40297079 | 1.61 |
ENST00000344138.4
ENST00000543252.1 |
GRAP2
|
GRB2-related adaptor protein 2 |
| chr2_-_208994548 | 1.60 |
ENST00000282141.3
|
CRYGC
|
crystallin, gamma C |
| chr1_+_209859510 | 1.59 |
ENST00000367028.2
ENST00000261465.1 |
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1 |
| chr2_+_48796120 | 1.56 |
ENST00000394754.1
|
STON1-GTF2A1L
|
STON1-GTF2A1L readthrough |
| chr18_+_43304092 | 1.54 |
ENST00000321925.4
ENST00000587601.1 |
SLC14A1
|
solute carrier family 14 (urea transporter), member 1 (Kidd blood group) |
| chr7_+_73442487 | 1.52 |
ENST00000380575.4
ENST00000380584.4 ENST00000458204.1 ENST00000357036.5 ENST00000417091.1 ENST00000429192.1 ENST00000442310.1 ENST00000380553.4 ENST00000380576.5 ENST00000428787.1 ENST00000320399.6 |
ELN
|
elastin |
| chr14_-_107219365 | 1.51 |
ENST00000424969.2
|
IGHV3-74
|
immunoglobulin heavy variable 3-74 |
| chr22_-_31688381 | 1.51 |
ENST00000487265.2
|
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr16_-_20338748 | 1.42 |
ENST00000575582.1
ENST00000341642.5 ENST00000381362.4 ENST00000572347.1 ENST00000572478.1 ENST00000302555.5 |
GP2
|
glycoprotein 2 (zymogen granule membrane) |
| chr3_+_52454971 | 1.41 |
ENST00000465863.1
|
PHF7
|
PHD finger protein 7 |
| chr7_+_73442102 | 1.39 |
ENST00000445912.1
ENST00000252034.7 |
ELN
|
elastin |
| chr1_-_233431458 | 1.38 |
ENST00000258229.9
ENST00000430153.1 |
PCNXL2
|
pecanex-like 2 (Drosophila) |
| chr5_+_140797296 | 1.37 |
ENST00000398594.2
|
PCDHGB7
|
protocadherin gamma subfamily B, 7 |
| chr20_+_30063067 | 1.37 |
ENST00000201979.2
|
REM1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr17_-_3337135 | 1.36 |
ENST00000248384.1
|
OR1E2
|
olfactory receptor, family 1, subfamily E, member 2 |
| chr15_-_43877062 | 1.35 |
ENST00000381885.1
ENST00000396923.3 |
PPIP5K1
|
diphosphoinositol pentakisphosphate kinase 1 |
| chr10_-_104178857 | 1.35 |
ENST00000020673.5
|
PSD
|
pleckstrin and Sec7 domain containing |
| chr6_-_53013620 | 1.33 |
ENST00000259803.7
|
GCM1
|
glial cells missing homolog 1 (Drosophila) |
| chr19_+_16771936 | 1.32 |
ENST00000187762.2
ENST00000599479.1 |
TMEM38A
|
transmembrane protein 38A |
| chr9_+_131703757 | 1.32 |
ENST00000482796.1
|
RP11-101E3.5
|
RP11-101E3.5 |
| chr20_+_52105495 | 1.31 |
ENST00000439873.2
|
AL354993.1
|
Cell growth-inhibiting protein 7; HCG1784586; Uncharacterized protein |
| chrX_-_100641155 | 1.29 |
ENST00000372880.1
ENST00000308731.7 |
BTK
|
Bruton agammaglobulinemia tyrosine kinase |
| chr3_-_52001448 | 1.29 |
ENST00000461554.1
ENST00000395013.3 ENST00000428823.2 ENST00000483411.1 ENST00000461544.1 ENST00000355852.2 |
PCBP4
|
poly(rC) binding protein 4 |
| chr10_-_72142345 | 1.25 |
ENST00000373224.1
ENST00000446961.1 ENST00000358141.2 ENST00000357631.2 |
LRRC20
|
leucine rich repeat containing 20 |
| chr5_-_148442584 | 1.25 |
ENST00000394358.2
ENST00000512049.1 |
SH3TC2
|
SH3 domain and tetratricopeptide repeats 2 |
| chr18_+_6729725 | 1.24 |
ENST00000400091.2
ENST00000583410.1 ENST00000584387.1 |
ARHGAP28
|
Rho GTPase activating protein 28 |
| chr2_+_73144604 | 1.24 |
ENST00000258106.6
|
EMX1
|
empty spiracles homeobox 1 |
| chr17_-_26694979 | 1.24 |
ENST00000438614.1
|
VTN
|
vitronectin |
| chr1_+_155146318 | 1.23 |
ENST00000368385.4
ENST00000545012.1 ENST00000392451.2 ENST00000368383.3 ENST00000368382.1 ENST00000334634.4 |
TRIM46
|
tripartite motif containing 46 |
| chr17_-_26879567 | 1.23 |
ENST00000581945.1
ENST00000444148.1 ENST00000301032.4 ENST00000335765.4 |
UNC119
|
unc-119 homolog (C. elegans) |
| chr18_-_44336998 | 1.21 |
ENST00000315087.7
|
ST8SIA5
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 5 |
| chr3_+_29323043 | 1.21 |
ENST00000452462.1
ENST00000456853.1 |
RBMS3
|
RNA binding motif, single stranded interacting protein 3 |
| chrX_-_45060135 | 1.21 |
ENST00000398000.2
ENST00000377934.4 |
CXorf36
|
chromosome X open reading frame 36 |
| chr4_-_90759440 | 1.19 |
ENST00000336904.3
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr3_+_137906353 | 1.18 |
ENST00000461822.1
ENST00000485396.1 ENST00000471453.1 ENST00000470821.1 ENST00000471709.1 ENST00000538260.1 ENST00000393058.3 ENST00000463485.1 |
ARMC8
|
armadillo repeat containing 8 |
| chr1_+_197237352 | 1.15 |
ENST00000538660.1
ENST00000367400.3 ENST00000367399.2 |
CRB1
|
crumbs homolog 1 (Drosophila) |
| chr17_-_46691990 | 1.13 |
ENST00000576562.1
|
HOXB8
|
homeobox B8 |
| chr17_-_15522826 | 1.12 |
ENST00000395906.3
|
CDRT1
|
CMT1A duplicated region transcript 1 |
| chrX_+_69672136 | 1.12 |
ENST00000374355.3
|
DLG3
|
discs, large homolog 3 (Drosophila) |
| chr2_-_183387430 | 1.09 |
ENST00000410103.1
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr2_+_234601512 | 1.09 |
ENST00000305139.6
|
UGT1A6
|
UDP glucuronosyltransferase 1 family, polypeptide A6 |
| chr2_+_27237615 | 1.08 |
ENST00000458529.1
ENST00000402218.1 |
MAPRE3
|
microtubule-associated protein, RP/EB family, member 3 |
| chr12_-_88974236 | 1.06 |
ENST00000228280.5
ENST00000552044.1 ENST00000357116.4 |
KITLG
|
KIT ligand |
| chr5_+_66124590 | 1.05 |
ENST00000490016.2
ENST00000403666.1 ENST00000450827.1 |
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr17_-_4464081 | 1.05 |
ENST00000574154.1
|
GGT6
|
gamma-glutamyltransferase 6 |
| chr5_-_131347306 | 1.04 |
ENST00000296869.4
ENST00000379249.3 ENST00000379272.2 ENST00000379264.2 |
ACSL6
|
acyl-CoA synthetase long-chain family member 6 |
| chr18_+_6729698 | 1.04 |
ENST00000383472.4
|
ARHGAP28
|
Rho GTPase activating protein 28 |
| chr17_+_25958174 | 1.03 |
ENST00000313648.6
ENST00000577392.1 ENST00000584661.1 ENST00000413914.2 |
LGALS9
|
lectin, galactoside-binding, soluble, 9 |
| chr14_-_23877474 | 1.03 |
ENST00000405093.3
|
MYH6
|
myosin, heavy chain 6, cardiac muscle, alpha |
| chr3_+_149530836 | 1.01 |
ENST00000466478.1
ENST00000491086.1 ENST00000467977.1 |
RNF13
|
ring finger protein 13 |
| chrX_+_15525426 | 0.99 |
ENST00000342014.6
|
BMX
|
BMX non-receptor tyrosine kinase |
| chr5_+_133861339 | 0.99 |
ENST00000282605.4
ENST00000361895.2 ENST00000402835.1 |
PHF15
|
jade family PHD finger 2 |
| chrX_-_101718085 | 0.98 |
ENST00000372750.1
ENST00000372752.1 |
NXF2B
|
nuclear RNA export factor 2B |
| chrX_+_16964985 | 0.96 |
ENST00000303843.7
|
REPS2
|
RALBP1 associated Eps domain containing 2 |
| chr19_-_51529849 | 0.95 |
ENST00000600362.1
ENST00000453757.3 ENST00000601671.1 |
KLK11
|
kallikrein-related peptidase 11 |
| chr19_-_54726850 | 0.95 |
ENST00000245620.9
ENST00000346401.6 ENST00000424807.1 ENST00000445347.1 |
LILRB3
|
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3 |
| chr2_-_183387283 | 0.93 |
ENST00000435564.1
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr2_+_64681219 | 0.92 |
ENST00000238875.5
|
LGALSL
|
lectin, galactoside-binding-like |
| chr17_+_34136459 | 0.92 |
ENST00000588240.1
ENST00000590273.1 ENST00000588441.1 ENST00000587272.1 ENST00000592237.1 ENST00000311979.3 |
TAF15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa |
| chr7_+_105603657 | 0.90 |
ENST00000542731.1
ENST00000343407.5 |
CDHR3
|
cadherin-related family member 3 |
| chr8_+_23104130 | 0.90 |
ENST00000313219.7
ENST00000519984.1 |
CHMP7
|
charged multivesicular body protein 7 |
| chr1_+_16010779 | 0.90 |
ENST00000375799.3
ENST00000375793.2 |
PLEKHM2
|
pleckstrin homology domain containing, family M (with RUN domain) member 2 |
| chr4_+_177241094 | 0.90 |
ENST00000503362.1
|
SPCS3
|
signal peptidase complex subunit 3 homolog (S. cerevisiae) |
| chr1_-_145039949 | 0.87 |
ENST00000313382.9
|
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr14_-_23876801 | 0.86 |
ENST00000356287.3
|
MYH6
|
myosin, heavy chain 6, cardiac muscle, alpha |
| chr13_-_99667960 | 0.83 |
ENST00000448493.2
|
DOCK9
|
dedicator of cytokinesis 9 |
| chr2_+_27193480 | 0.81 |
ENST00000233121.2
ENST00000405074.3 |
MAPRE3
|
microtubule-associated protein, RP/EB family, member 3 |
| chr1_+_158815588 | 0.81 |
ENST00000438394.1
|
MNDA
|
myeloid cell nuclear differentiation antigen |
| chr21_-_34186006 | 0.81 |
ENST00000490358.1
|
C21orf62
|
chromosome 21 open reading frame 62 |
| chr7_+_143657027 | 0.81 |
ENST00000392899.1
|
OR2F1
|
olfactory receptor, family 2, subfamily F, member 1 (gene/pseudogene) |
Network of associatons between targets according to the STRING database.
First level regulatory network of PGR
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 3.6 | GO:0034088 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 2.8 | 8.4 | GO:2001190 | positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 2.2 | 6.5 | GO:0002370 | natural killer cell cytokine production(GO:0002370) regulation of natural killer cell cytokine production(GO:0002727) |
| 2.1 | 6.4 | GO:0072019 | carbon dioxide transmembrane transport(GO:0035378) proximal convoluted tubule development(GO:0072019) metanephric proximal convoluted tubule development(GO:0072229) |
| 2.0 | 6.0 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 1.8 | 5.5 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 1.7 | 5.2 | GO:0048687 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 1.3 | 5.1 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 1.2 | 3.5 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 1.1 | 3.4 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) negative regulation of DNA catabolic process(GO:1903625) regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 1.1 | 6.8 | GO:0051012 | microtubule sliding(GO:0051012) |
| 1.0 | 10.0 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.9 | 3.6 | GO:0031179 | peptide amidation(GO:0001519) protein amidation(GO:0018032) peptide modification(GO:0031179) |
| 0.9 | 1.7 | GO:0021571 | rhombomere 5 development(GO:0021571) |
| 0.9 | 1.7 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.8 | 3.4 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.8 | 2.4 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.8 | 2.3 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 0.8 | 2.3 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.8 | 2.3 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.7 | 3.7 | GO:0099538 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 0.6 | 1.9 | GO:1990641 | cellular response to bile acid(GO:1903413) negative regulation of intestinal absorption(GO:1904479) response to iron ion starvation(GO:1990641) |
| 0.6 | 1.9 | GO:1903033 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.6 | 4.4 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.6 | 2.5 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.6 | 1.7 | GO:0021780 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.6 | 2.9 | GO:0038169 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.6 | 1.7 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.5 | 1.1 | GO:0033025 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) |
| 0.5 | 2.6 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 0.5 | 4.6 | GO:0015705 | iodide transport(GO:0015705) |
| 0.5 | 3.9 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.5 | 5.2 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.5 | 6.0 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.5 | 1.4 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.4 | 1.3 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.4 | 3.5 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.4 | 1.7 | GO:0042335 | cuticle development(GO:0042335) |
| 0.4 | 1.3 | GO:0001812 | positive regulation of type I hypersensitivity(GO:0001812) |
| 0.4 | 0.4 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 0.3 | 1.2 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.3 | 2.2 | GO:0019064 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.3 | 1.5 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.3 | 16.3 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.3 | 2.0 | GO:0010519 | negative regulation of phospholipase activity(GO:0010519) regulation of adrenergic receptor signaling pathway(GO:0071877) |
| 0.3 | 2.8 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.3 | 3.6 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.3 | 4.9 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.3 | 3.6 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.2 | 1.2 | GO:0051622 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) regulation of peroxidase activity(GO:2000468) positive regulation of peroxidase activity(GO:2000470) |
| 0.2 | 1.9 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.2 | 2.1 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.2 | 3.2 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.2 | 3.1 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.2 | 2.2 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.2 | 5.1 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.2 | 1.8 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.2 | 3.1 | GO:0021794 | thalamus development(GO:0021794) |
| 0.2 | 0.6 | GO:0042418 | epinephrine biosynthetic process(GO:0042418) |
| 0.2 | 3.0 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) |
| 0.2 | 0.7 | GO:2000969 | positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.2 | 4.3 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.2 | 3.2 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.2 | 0.6 | GO:0071812 | regulation of fever generation by regulation of prostaglandin secretion(GO:0071810) positive regulation of fever generation by positive regulation of prostaglandin secretion(GO:0071812) positive regulation of ERK1 and ERK2 cascade via TNFSF11-mediated signaling(GO:0071848) regulation of fever generation by prostaglandin secretion(GO:0100009) |
| 0.2 | 1.1 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.2 | 0.9 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.2 | 0.7 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.2 | 0.2 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.2 | 0.6 | GO:0070253 | somatostatin secretion(GO:0070253) |
| 0.2 | 3.4 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.2 | 2.8 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.2 | 7.6 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.2 | 0.6 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.1 | 0.7 | GO:0007521 | muscle cell fate determination(GO:0007521) |
| 0.1 | 4.0 | GO:0045332 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.1 | 5.1 | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.1 | 5.8 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.1 | 2.1 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.1 | 1.9 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.1 | 1.2 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.1 | 1.2 | GO:0099612 | protein localization to axon(GO:0099612) |
| 0.1 | 4.0 | GO:0043949 | regulation of cAMP-mediated signaling(GO:0043949) |
| 0.1 | 6.9 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.1 | 1.7 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) sphingoid biosynthetic process(GO:0046520) |
| 0.1 | 2.4 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 0.6 | GO:0035624 | receptor transactivation(GO:0035624) |
| 0.1 | 0.4 | GO:0032223 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.1 | 1.6 | GO:0042074 | cell migration involved in gastrulation(GO:0042074) |
| 0.1 | 1.7 | GO:0044130 | negative regulation of growth of symbiont in host(GO:0044130) |
| 0.1 | 1.3 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.1 | 1.8 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 1.6 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.1 | 0.3 | GO:0036071 | N-glycan fucosylation(GO:0036071) |
| 0.1 | 7.6 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.1 | 0.7 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.1 | 3.0 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.1 | 6.3 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.1 | 2.6 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.1 | 0.7 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.1 | 1.4 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 0.7 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.1 | 5.2 | GO:0001558 | regulation of cell growth(GO:0001558) |
| 0.1 | 2.9 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.1 | 2.1 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.1 | 1.1 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.1 | 1.6 | GO:0032608 | interferon-beta production(GO:0032608) regulation of interferon-beta production(GO:0032648) |
| 0.1 | 0.3 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.1 | 0.4 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.1 | 0.6 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.1 | 0.8 | GO:0015868 | purine nucleotide transport(GO:0015865) purine ribonucleotide transport(GO:0015868) |
| 0.1 | 0.9 | GO:1904896 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.1 | 1.6 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.1 | 0.5 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.1 | 1.9 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.1 | 1.4 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 4.0 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 1.7 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.1 | 1.6 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.1 | 2.7 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.5 | GO:0032930 | positive regulation of superoxide anion generation(GO:0032930) |
| 0.0 | 0.2 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.1 | GO:0002582 | positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen(GO:0002585) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 0.0 | 1.2 | GO:0097503 | sialylation(GO:0097503) |
| 0.0 | 2.0 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.0 | 2.5 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 0.9 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.0 | 0.7 | GO:0006941 | striated muscle contraction(GO:0006941) |
| 0.0 | 0.8 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 0.0 | 0.9 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 3.9 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.8 | GO:0045725 | positive regulation of glycogen biosynthetic process(GO:0045725) |
| 0.0 | 0.8 | GO:0042136 | neurotransmitter biosynthetic process(GO:0042136) |
| 0.0 | 0.3 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.0 | 1.4 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.4 | GO:0030238 | male sex determination(GO:0030238) |
| 0.0 | 1.1 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.0 | 0.4 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.0 | GO:0035992 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.0 | 0.1 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.0 | 1.7 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.0 | 16.5 | GO:0043687 | post-translational protein modification(GO:0043687) |
| 0.0 | 0.3 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.2 | GO:0002003 | angiotensin maturation(GO:0002003) |
| 0.0 | 0.2 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.0 | 1.0 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.0 | 4.9 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 2.1 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.0 | 3.0 | GO:0016032 | viral process(GO:0016032) |
| 0.0 | 1.1 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) peptidyl-serine modification(GO:0018209) |
| 0.0 | 0.4 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.0 | 4.7 | GO:0007163 | establishment or maintenance of cell polarity(GO:0007163) |
| 0.0 | 1.2 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.0 | 0.7 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.2 | GO:0009744 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.0 | 2.7 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 1.0 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.2 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.0 | 0.5 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.5 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.0 | 0.4 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 0.8 | GO:0007050 | cell cycle arrest(GO:0007050) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.4 | GO:0020003 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 1.2 | 6.0 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 1.0 | 3.1 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.9 | 2.6 | GO:0098536 | deuterosome(GO:0098536) |
| 0.8 | 7.6 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.8 | 3.0 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.6 | 3.2 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.5 | 2.4 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.5 | 5.8 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.4 | 1.8 | GO:0071062 | rough endoplasmic reticulum lumen(GO:0048237) alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.4 | 4.4 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.4 | 7.4 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.2 | 1.9 | GO:0045179 | apical cortex(GO:0045179) |
| 0.2 | 1.1 | GO:0070081 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.2 | 2.2 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.2 | 1.2 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.2 | 7.0 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 0.5 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.2 | 1.9 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.2 | 0.8 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 3.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.6 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.1 | 0.9 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 0.7 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.1 | 4.3 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 0.7 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 8.1 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.1 | 3.4 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 4.0 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 3.3 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 5.6 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.1 | 2.9 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 0.9 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 2.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.1 | 1.7 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 0.5 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.1 | 1.9 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 1.8 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.1 | 0.5 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 5.7 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.1 | 0.5 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.1 | 0.4 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 9.1 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 1.1 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.8 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.5 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.8 | GO:0030128 | clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 11.6 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 2.3 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.6 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 2.2 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.4 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 1.3 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 4.2 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 6.2 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 2.0 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 3.9 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.0 | 0.4 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 2.5 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 1.1 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 3.0 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 5.3 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 4.7 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.4 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 4.2 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.3 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 3.0 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 1.9 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 1.4 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.0 | 4.8 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 1.0 | GO:0031902 | late endosome membrane(GO:0031902) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.4 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 1.2 | 6.0 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.9 | 3.6 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.7 | 3.7 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.6 | 5.2 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.6 | 6.0 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.6 | 3.5 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.6 | 2.9 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.6 | 2.9 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.6 | 5.5 | GO:0009374 | biotin carboxylase activity(GO:0004075) biotin binding(GO:0009374) |
| 0.5 | 1.6 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.5 | 2.6 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.5 | 2.3 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.5 | 5.2 | GO:0004568 | chitinase activity(GO:0004568) chitin binding(GO:0008061) |
| 0.5 | 3.6 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.5 | 3.2 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.4 | 4.9 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.4 | 1.7 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.4 | 2.0 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.4 | 3.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.4 | 1.5 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.4 | 3.0 | GO:0043559 | insulin binding(GO:0043559) |
| 0.4 | 1.1 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.3 | 1.0 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.3 | 1.9 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.3 | 21.1 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.3 | 5.1 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.3 | 2.5 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.3 | 1.6 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.3 | 1.4 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.3 | 2.2 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.3 | 7.4 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.2 | 0.7 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.2 | 1.2 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.2 | 2.4 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.2 | 6.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.2 | 0.8 | GO:0004102 | choline O-acetyltransferase activity(GO:0004102) |
| 0.2 | 0.6 | GO:0004603 | phenylethanolamine N-methyltransferase activity(GO:0004603) |
| 0.2 | 3.1 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.2 | 2.1 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.2 | 3.9 | GO:0004857 | enzyme inhibitor activity(GO:0004857) |
| 0.2 | 1.2 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.2 | 2.2 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.2 | 2.7 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.2 | 4.0 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.2 | 7.6 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 2.5 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.1 | 3.9 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.1 | 0.6 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 0.5 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.1 | 20.5 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 0.7 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.1 | 5.3 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 5.4 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 1.7 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 1.6 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.1 | 0.3 | GO:0008424 | glycoprotein 6-alpha-L-fucosyltransferase activity(GO:0008424) alpha-(1->6)-fucosyltransferase activity(GO:0046921) |
| 0.1 | 2.8 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.1 | 3.6 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 8.5 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 0.5 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.1 | 2.1 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.1 | 1.7 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 6.9 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 3.0 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.1 | 8.8 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 0.8 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 0.7 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 1.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 3.6 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.1 | 0.7 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.7 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 1.3 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.7 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.0 | 1.9 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 3.5 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 3.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.2 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 1.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.5 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.5 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.4 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 3.2 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 4.5 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.6 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.6 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 6.1 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.3 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.8 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 1.7 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.9 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 2.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 3.4 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 4.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 2.8 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.4 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 6.3 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 0.4 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.3 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.2 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.4 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.1 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.2 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 1.5 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.9 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.6 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.2 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 12.5 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.2 | 4.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 8.7 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 3.0 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 1.1 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 28.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 5.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 3.3 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 1.6 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.8 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 3.7 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.8 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 1.8 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 2.5 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.5 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.7 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.8 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.3 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 1.2 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.2 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.6 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 1.0 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 4.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.7 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.7 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 2.0 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.2 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.6 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 8.7 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.5 | 6.4 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.4 | 2.2 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.2 | 5.1 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.2 | 2.7 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.2 | 3.5 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.2 | 10.4 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 7.6 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 4.1 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.1 | 3.0 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 3.6 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 0.8 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.1 | 6.1 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 1.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 5.8 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.1 | 2.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 2.9 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 10.0 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 2.6 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 5.0 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 3.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 1.8 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 3.1 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 0.9 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 3.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 2.2 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 3.8 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 2.5 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 1.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 1.1 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 1.6 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.8 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.9 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.5 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 1.7 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 5.6 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.7 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.6 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 1.7 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 2.3 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 1.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 3.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.7 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 1.0 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |