Project
averaged GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
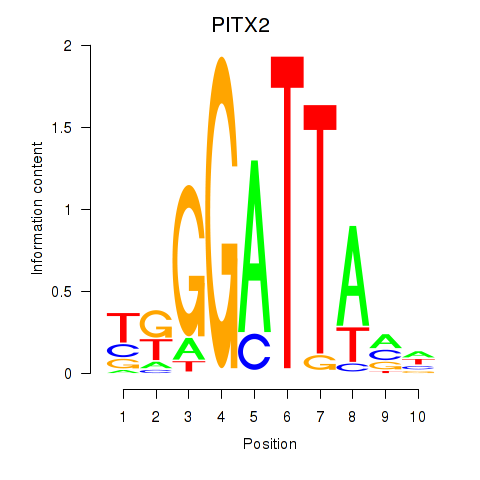
Results for PITX2
Z-value: 0.47
Transcription factors associated with PITX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PITX2
|
ENSG00000164093.11 | paired like homeodomain 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PITX2 | hg19_v2_chr4_-_111544254_111544270, hg19_v2_chr4_-_111563076_111563172, hg19_v2_chr4_-_111558135_111558198 | 0.15 | 2.5e-02 | Click! |
Activity profile of PITX2 motif
Sorted Z-values of PITX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_5248294 | 17.07 |
ENST00000335295.4
|
HBB
|
hemoglobin, beta |
| chr8_-_27462822 | 11.24 |
ENST00000522098.1
|
CLU
|
clusterin |
| chr13_-_36429763 | 8.27 |
ENST00000379893.1
|
DCLK1
|
doublecortin-like kinase 1 |
| chrX_-_54384425 | 4.73 |
ENST00000375169.3
ENST00000354646.2 |
WNK3
|
WNK lysine deficient protein kinase 3 |
| chr17_-_40021656 | 3.86 |
ENST00000319121.3
|
KLHL11
|
kelch-like family member 11 |
| chrX_+_15767971 | 3.74 |
ENST00000479740.1
ENST00000454127.2 |
CA5B
|
carbonic anhydrase VB, mitochondrial |
| chr5_-_64920115 | 3.46 |
ENST00000381018.3
ENST00000274327.7 |
TRIM23
|
tripartite motif containing 23 |
| chr16_-_5116025 | 3.06 |
ENST00000472572.3
ENST00000315997.5 ENST00000422873.1 ENST00000350219.4 |
C16orf89
|
chromosome 16 open reading frame 89 |
| chr5_+_64920543 | 2.30 |
ENST00000399438.3
ENST00000510585.2 |
TRAPPC13
CTC-534A2.2
|
trafficking protein particle complex 13 CDNA FLJ26957 fis, clone SLV00486; Uncharacterized protein |
| chr2_+_120770645 | 2.25 |
ENST00000443902.2
|
EPB41L5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr19_+_2476116 | 2.24 |
ENST00000215631.4
ENST00000587345.1 |
GADD45B
|
growth arrest and DNA-damage-inducible, beta |
| chr2_+_120770581 | 2.12 |
ENST00000263713.5
|
EPB41L5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr1_+_22303503 | 1.88 |
ENST00000337107.6
|
CELA3B
|
chymotrypsin-like elastase family, member 3B |
| chr10_-_61900762 | 1.87 |
ENST00000355288.2
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr14_+_75988851 | 1.76 |
ENST00000555504.1
|
BATF
|
basic leucine zipper transcription factor, ATF-like |
| chr6_-_53013620 | 1.74 |
ENST00000259803.7
|
GCM1
|
glial cells missing homolog 1 (Drosophila) |
| chr1_-_31230650 | 1.47 |
ENST00000294507.3
|
LAPTM5
|
lysosomal protein transmembrane 5 |
| chr16_-_68014732 | 1.46 |
ENST00000268793.4
|
DPEP3
|
dipeptidase 3 |
| chr12_+_7941989 | 1.41 |
ENST00000229307.4
|
NANOG
|
Nanog homeobox |
| chr9_+_8858102 | 1.40 |
ENST00000447950.1
ENST00000430766.1 |
RP11-75C9.1
|
RP11-75C9.1 |
| chr15_-_89764929 | 1.39 |
ENST00000268125.5
|
RLBP1
|
retinaldehyde binding protein 1 |
| chr9_+_34458771 | 1.35 |
ENST00000437363.1
ENST00000242317.4 |
DNAI1
|
dynein, axonemal, intermediate chain 1 |
| chr7_-_128415844 | 1.24 |
ENST00000249389.2
|
OPN1SW
|
opsin 1 (cone pigments), short-wave-sensitive |
| chr9_-_95166841 | 1.06 |
ENST00000262551.4
|
OGN
|
osteoglycin |
| chr1_+_92417716 | 1.06 |
ENST00000402388.1
|
BRDT
|
bromodomain, testis-specific |
| chrX_-_6146876 | 1.05 |
ENST00000381095.3
|
NLGN4X
|
neuroligin 4, X-linked |
| chr11_+_61522844 | 1.05 |
ENST00000265460.5
|
MYRF
|
myelin regulatory factor |
| chr6_+_35310391 | 1.01 |
ENST00000337400.2
ENST00000311565.4 ENST00000540939.1 |
PPARD
|
peroxisome proliferator-activated receptor delta |
| chr11_-_36619771 | 0.93 |
ENST00000311485.3
ENST00000527033.1 ENST00000532616.1 |
RAG2
|
recombination activating gene 2 |
| chrX_-_48931648 | 0.92 |
ENST00000376386.3
ENST00000376390.4 |
PRAF2
|
PRA1 domain family, member 2 |
| chr21_-_19775973 | 0.88 |
ENST00000284885.3
|
TMPRSS15
|
transmembrane protease, serine 15 |
| chr8_-_133123406 | 0.87 |
ENST00000434736.2
|
HHLA1
|
HERV-H LTR-associating 1 |
| chr6_+_35310312 | 0.86 |
ENST00000448077.2
ENST00000360694.3 ENST00000418635.2 ENST00000444397.1 |
PPARD
|
peroxisome proliferator-activated receptor delta |
| chr21_-_34915147 | 0.78 |
ENST00000381831.3
ENST00000381839.3 |
GART
|
phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase |
| chrX_+_107069063 | 0.74 |
ENST00000262843.6
|
MID2
|
midline 2 |
| chr1_-_165414414 | 0.70 |
ENST00000359842.5
|
RXRG
|
retinoid X receptor, gamma |
| chr4_+_70916119 | 0.70 |
ENST00000246896.3
ENST00000511674.1 |
HTN1
|
histatin 1 |
| chr4_-_76912070 | 0.68 |
ENST00000395711.4
ENST00000356260.5 |
SDAD1
|
SDA1 domain containing 1 |
| chr18_-_3880051 | 0.67 |
ENST00000584874.1
|
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1 |
| chr4_-_68620053 | 0.65 |
ENST00000420975.2
ENST00000226413.4 |
GNRHR
|
gonadotropin-releasing hormone receptor |
| chr1_+_22328144 | 0.64 |
ENST00000290122.3
ENST00000374663.1 |
CELA3A
|
chymotrypsin-like elastase family, member 3A |
| chr1_+_2407754 | 0.51 |
ENST00000419816.2
ENST00000378486.3 ENST00000378488.3 ENST00000288766.5 |
PLCH2
|
phospholipase C, eta 2 |
| chr13_+_73632897 | 0.35 |
ENST00000377687.4
|
KLF5
|
Kruppel-like factor 5 (intestinal) |
| chr4_+_126237554 | 0.32 |
ENST00000394329.3
|
FAT4
|
FAT atypical cadherin 4 |
| chr9_-_110251836 | 0.22 |
ENST00000374672.4
|
KLF4
|
Kruppel-like factor 4 (gut) |
| chr17_+_79495397 | 0.15 |
ENST00000417245.2
ENST00000334850.7 |
FSCN2
|
fascin homolog 2, actin-bundling protein, retinal (Strongylocentrotus purpuratus) |
| chr1_+_201979645 | 0.13 |
ENST00000367284.5
ENST00000367283.3 |
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr12_-_323689 | 0.08 |
ENST00000428720.1
|
SLC6A12
|
solute carrier family 6 (neurotransmitter transporter), member 12 |
| chr14_+_75988768 | 0.06 |
ENST00000286639.6
|
BATF
|
basic leucine zipper transcription factor, ATF-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of PITX2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.3 | 17.1 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 1.9 | 11.2 | GO:0061517 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 1.5 | 4.4 | GO:0003383 | apical constriction(GO:0003383) mesoderm migration involved in gastrulation(GO:0007509) |
| 1.2 | 4.7 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.6 | 1.7 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.5 | 1.9 | GO:1900827 | positive regulation of cell communication by electrical coupling(GO:0010650) maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.4 | 1.1 | GO:0051039 | histone displacement(GO:0001207) regulation of transcription involved in meiotic cell cycle(GO:0051037) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.3 | 8.3 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.3 | 3.3 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.3 | 1.1 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.2 | 1.4 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.2 | 0.9 | GO:0002329 | pre-B cell differentiation(GO:0002329) pre-B cell allelic exclusion(GO:0002331) |
| 0.2 | 1.8 | GO:0003360 | brainstem development(GO:0003360) |
| 0.2 | 0.6 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.1 | 1.3 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 0.3 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.1 | 2.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 1.8 | GO:0072540 | T-helper 17 cell lineage commitment(GO:0072540) |
| 0.1 | 0.7 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.1 | 1.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 0.2 | GO:0071409 | negative regulation of muscle hyperplasia(GO:0014740) cellular response to cycloheximide(GO:0071409) |
| 0.1 | 3.7 | GO:0006730 | one-carbon metabolic process(GO:0006730) bicarbonate transport(GO:0015701) |
| 0.1 | 0.7 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 1.1 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.4 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 1.5 | GO:0007140 | male meiosis(GO:0007140) |
| 0.0 | 0.9 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.0 | 0.7 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.7 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 | 2.5 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 1.5 | GO:1990823 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.3 | 17.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.7 | 11.2 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) neurofibrillary tangle(GO:0097418) |
| 0.2 | 1.3 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 1.9 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 3.9 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.2 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 4.4 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 9.8 | GO:0099572 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 4.7 | GO:0005923 | bicellular tight junction(GO:0005923) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 17.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.6 | 11.2 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.4 | 4.7 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.3 | 1.4 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.2 | 3.7 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.2 | 0.7 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.2 | 0.8 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.1 | 1.2 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 1.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 1.9 | GO:0036041 | long-chain fatty acid binding(GO:0036041) |
| 0.1 | 1.3 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 1.5 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 3.5 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.2 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.0 | 2.3 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.6 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.9 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 8.3 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.0 | 6.2 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 1.1 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.5 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.0 | 1.7 | GO:0042826 | histone deacetylase binding(GO:0042826) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 11.0 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 2.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 2.2 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 3.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.7 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 1.2 | REACTOME OPSINS | Genes involved in Opsins |
| 0.1 | 17.1 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 11.2 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 1.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 1.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.8 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.6 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 2.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |