Project
averaged GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
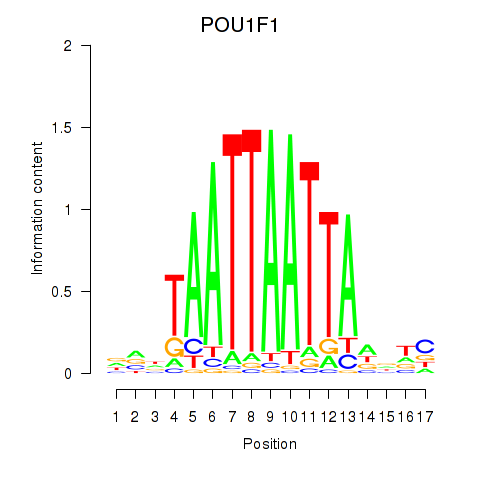
Results for POU1F1
Z-value: 0.87
Transcription factors associated with POU1F1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
POU1F1
|
ENSG00000064835.6 | POU class 1 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| POU1F1 | hg19_v2_chr3_-_87325612_87325654 | 0.27 | 6.8e-05 | Click! |
Activity profile of POU1F1 motif
Sorted Z-values of POU1F1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_140762268 | 18.09 |
ENST00000518325.1
|
PCDHGA7
|
protocadherin gamma subfamily A, 7 |
| chr6_-_52859046 | 14.49 |
ENST00000457564.1
ENST00000541324.1 ENST00000370960.1 |
GSTA4
|
glutathione S-transferase alpha 4 |
| chrX_-_13835147 | 12.75 |
ENST00000493677.1
ENST00000355135.2 |
GPM6B
|
glycoprotein M6B |
| chr17_+_1674982 | 11.73 |
ENST00000572048.1
ENST00000573763.1 |
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr6_-_52859968 | 10.29 |
ENST00000370959.1
|
GSTA4
|
glutathione S-transferase alpha 4 |
| chr12_-_15038779 | 8.79 |
ENST00000228938.5
ENST00000539261.1 |
MGP
|
matrix Gla protein |
| chr21_-_27423339 | 8.09 |
ENST00000415997.1
|
APP
|
amyloid beta (A4) precursor protein |
| chr19_+_54466179 | 8.06 |
ENST00000270458.2
|
CACNG8
|
calcium channel, voltage-dependent, gamma subunit 8 |
| chr8_+_30244580 | 7.17 |
ENST00000523115.1
ENST00000519647.1 |
RBPMS
|
RNA binding protein with multiple splicing |
| chr15_+_84115868 | 6.81 |
ENST00000427482.2
|
SH3GL3
|
SH3-domain GRB2-like 3 |
| chr11_+_12399071 | 6.59 |
ENST00000539723.1
ENST00000550549.1 |
PARVA
|
parvin, alpha |
| chr11_-_117186946 | 6.51 |
ENST00000313005.6
ENST00000528053.1 |
BACE1
|
beta-site APP-cleaving enzyme 1 |
| chr7_-_45960850 | 6.50 |
ENST00000381083.4
ENST00000381086.5 ENST00000275521.6 |
IGFBP3
|
insulin-like growth factor binding protein 3 |
| chr11_-_128894053 | 6.19 |
ENST00000392657.3
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr9_-_21351377 | 6.16 |
ENST00000380210.1
|
IFNA6
|
interferon, alpha 6 |
| chr11_+_2405833 | 5.74 |
ENST00000527343.1
ENST00000464784.2 |
CD81
|
CD81 molecule |
| chr19_-_51920952 | 5.66 |
ENST00000356298.5
ENST00000339313.5 ENST00000529627.1 ENST00000439889.2 ENST00000353836.5 ENST00000432469.2 |
SIGLEC10
|
sialic acid binding Ig-like lectin 10 |
| chr4_-_46911248 | 5.51 |
ENST00000355591.3
ENST00000505102.1 |
COX7B2
|
cytochrome c oxidase subunit VIIb2 |
| chr8_-_30670384 | 5.35 |
ENST00000221138.4
ENST00000518243.1 |
PPP2CB
|
protein phosphatase 2, catalytic subunit, beta isozyme |
| chr1_+_233086326 | 5.23 |
ENST00000366628.5
ENST00000366627.4 |
NTPCR
|
nucleoside-triphosphatase, cancer-related |
| chr6_-_110501200 | 5.14 |
ENST00000392586.1
ENST00000419252.1 ENST00000392589.1 ENST00000392588.1 ENST00000359451.2 |
WASF1
|
WAS protein family, member 1 |
| chr5_+_173472607 | 5.11 |
ENST00000303177.3
ENST00000519867.1 |
NSG2
|
Neuron-specific protein family member 2 |
| chr1_-_13390765 | 5.03 |
ENST00000357367.2
|
PRAMEF8
|
PRAME family member 8 |
| chr5_+_129083772 | 4.96 |
ENST00000564719.1
|
KIAA1024L
|
KIAA1024-like |
| chr10_+_64133934 | 4.89 |
ENST00000395254.3
ENST00000395255.3 ENST00000410046.3 |
ZNF365
|
zinc finger protein 365 |
| chr7_+_134528635 | 4.81 |
ENST00000445569.2
|
CALD1
|
caldesmon 1 |
| chr2_-_152118352 | 4.81 |
ENST00000331426.5
|
RBM43
|
RNA binding motif protein 43 |
| chr7_+_130126012 | 4.64 |
ENST00000341441.5
|
MEST
|
mesoderm specific transcript |
| chr7_+_130126165 | 4.63 |
ENST00000427521.1
ENST00000416162.2 ENST00000378576.4 |
MEST
|
mesoderm specific transcript |
| chr4_+_48833234 | 4.56 |
ENST00000510824.1
ENST00000425583.2 |
OCIAD1
|
OCIA domain containing 1 |
| chr8_+_98900132 | 4.45 |
ENST00000520016.1
|
MATN2
|
matrilin 2 |
| chr3_-_48130707 | 4.32 |
ENST00000360240.6
ENST00000383737.4 |
MAP4
|
microtubule-associated protein 4 |
| chr4_-_153601136 | 4.22 |
ENST00000504064.1
ENST00000304385.3 |
TMEM154
|
transmembrane protein 154 |
| chr5_+_81575281 | 4.19 |
ENST00000380167.4
|
ATP6AP1L
|
ATPase, H+ transporting, lysosomal accessory protein 1-like |
| chrX_-_102941596 | 3.99 |
ENST00000441076.2
ENST00000422355.1 ENST00000442614.1 ENST00000422154.2 ENST00000451301.1 |
MORF4L2
|
mortality factor 4 like 2 |
| chr5_+_95066823 | 3.98 |
ENST00000506817.1
ENST00000379982.3 |
RHOBTB3
|
Rho-related BTB domain containing 3 |
| chr5_-_125930929 | 3.84 |
ENST00000553117.1
ENST00000447989.2 ENST00000409134.3 |
ALDH7A1
|
aldehyde dehydrogenase 7 family, member A1 |
| chr12_-_25150373 | 3.82 |
ENST00000549828.1
|
C12orf77
|
chromosome 12 open reading frame 77 |
| chr6_+_26087509 | 3.82 |
ENST00000397022.3
ENST00000353147.5 ENST00000352392.4 ENST00000349999.4 ENST00000317896.7 ENST00000357618.5 ENST00000470149.1 ENST00000336625.8 ENST00000461397.1 ENST00000488199.1 |
HFE
|
hemochromatosis |
| chr4_-_185275104 | 3.69 |
ENST00000317596.3
|
RP11-290F5.2
|
RP11-290F5.2 |
| chr15_-_56757329 | 3.66 |
ENST00000260453.3
|
MNS1
|
meiosis-specific nuclear structural 1 |
| chr5_-_55412774 | 3.65 |
ENST00000434982.2
|
ANKRD55
|
ankyrin repeat domain 55 |
| chr16_-_66764119 | 3.62 |
ENST00000569320.1
|
DYNC1LI2
|
dynein, cytoplasmic 1, light intermediate chain 2 |
| chr10_-_28571015 | 3.61 |
ENST00000375719.3
ENST00000375732.1 |
MPP7
|
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chrX_+_43515467 | 3.61 |
ENST00000338702.3
ENST00000542639.1 |
MAOA
|
monoamine oxidase A |
| chr12_-_118796910 | 3.60 |
ENST00000541186.1
ENST00000539872.1 |
TAOK3
|
TAO kinase 3 |
| chr1_-_182921119 | 3.49 |
ENST00000423786.1
|
SHCBP1L
|
SHC SH2-domain binding protein 1-like |
| chr2_-_190044480 | 3.38 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chrX_-_102942961 | 3.31 |
ENST00000434230.1
ENST00000418819.1 ENST00000360458.1 |
MORF4L2
|
mortality factor 4 like 2 |
| chr8_-_38008783 | 3.19 |
ENST00000276449.4
|
STAR
|
steroidogenic acute regulatory protein |
| chr3_+_38537960 | 3.17 |
ENST00000453767.1
|
EXOG
|
endo/exonuclease (5'-3'), endonuclease G-like |
| chr19_+_19144384 | 3.16 |
ENST00000392335.2
ENST00000535612.1 ENST00000537263.1 ENST00000540707.1 ENST00000541725.1 ENST00000269932.6 ENST00000546344.1 ENST00000540792.1 ENST00000536098.1 ENST00000541898.1 ENST00000543877.1 |
ARMC6
|
armadillo repeat containing 6 |
| chr10_+_91152303 | 3.11 |
ENST00000371804.3
|
IFIT1
|
interferon-induced protein with tetratricopeptide repeats 1 |
| chrX_-_102943022 | 3.10 |
ENST00000433176.2
|
MORF4L2
|
mortality factor 4 like 2 |
| chr21_-_43735628 | 3.07 |
ENST00000291525.10
ENST00000518498.1 |
TFF3
|
trefoil factor 3 (intestinal) |
| chr3_-_194072019 | 3.03 |
ENST00000429275.1
ENST00000323830.3 |
CPN2
|
carboxypeptidase N, polypeptide 2 |
| chr9_-_215744 | 3.02 |
ENST00000382387.2
|
C9orf66
|
chromosome 9 open reading frame 66 |
| chr6_+_26087646 | 2.99 |
ENST00000309234.6
|
HFE
|
hemochromatosis |
| chr4_+_169552748 | 2.97 |
ENST00000504519.1
ENST00000512127.1 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr10_+_18549645 | 2.96 |
ENST00000396576.2
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr16_+_16434185 | 2.96 |
ENST00000524823.2
|
AC138969.4
|
Protein PKD1P1 |
| chr16_-_66584059 | 2.89 |
ENST00000417693.3
ENST00000544898.1 ENST00000569718.1 ENST00000527284.1 ENST00000299697.7 ENST00000451102.2 |
TK2
|
thymidine kinase 2, mitochondrial |
| chr11_-_327537 | 2.89 |
ENST00000602735.1
|
IFITM3
|
interferon induced transmembrane protein 3 |
| chr4_+_3344141 | 2.89 |
ENST00000306648.7
|
RGS12
|
regulator of G-protein signaling 12 |
| chr1_+_77333117 | 2.87 |
ENST00000477717.1
|
ST6GALNAC5
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 |
| chr18_+_21269404 | 2.82 |
ENST00000313654.9
|
LAMA3
|
laminin, alpha 3 |
| chr8_-_141774467 | 2.81 |
ENST00000520151.1
ENST00000519024.1 ENST00000519465.1 |
PTK2
|
protein tyrosine kinase 2 |
| chr6_+_114178512 | 2.73 |
ENST00000368635.4
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate |
| chr5_+_140588269 | 2.73 |
ENST00000541609.1
ENST00000239450.2 |
PCDHB12
|
protocadherin beta 12 |
| chr4_-_122854612 | 2.66 |
ENST00000264811.5
|
TRPC3
|
transient receptor potential cation channel, subfamily C, member 3 |
| chr16_-_28937027 | 2.65 |
ENST00000358201.4
|
RABEP2
|
rabaptin, RAB GTPase binding effector protein 2 |
| chr18_-_52989217 | 2.65 |
ENST00000570287.2
|
TCF4
|
transcription factor 4 |
| chr7_-_140482926 | 2.56 |
ENST00000496384.2
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B |
| chr2_+_210517895 | 2.55 |
ENST00000447185.1
|
MAP2
|
microtubule-associated protein 2 |
| chr3_-_160823158 | 2.54 |
ENST00000392779.2
ENST00000392780.1 ENST00000494173.1 |
B3GALNT1
|
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chr4_+_88754113 | 2.51 |
ENST00000560249.1
ENST00000540395.1 ENST00000511670.1 ENST00000361056.3 |
MEPE
|
matrix extracellular phosphoglycoprotein |
| chr14_-_90420862 | 2.47 |
ENST00000556005.1
ENST00000555872.1 |
EFCAB11
|
EF-hand calcium binding domain 11 |
| chr10_+_53806501 | 2.47 |
ENST00000373975.2
|
PRKG1
|
protein kinase, cGMP-dependent, type I |
| chr22_-_32767017 | 2.46 |
ENST00000400234.1
|
RFPL3S
|
RFPL3 antisense |
| chr3_+_195447738 | 2.45 |
ENST00000447234.2
ENST00000320736.6 ENST00000436408.1 |
MUC20
|
mucin 20, cell surface associated |
| chr7_+_129932974 | 2.44 |
ENST00000445470.2
ENST00000222482.4 ENST00000492072.1 ENST00000473956.1 ENST00000493259.1 ENST00000486598.1 |
CPA4
|
carboxypeptidase A4 |
| chr11_-_63376013 | 2.42 |
ENST00000540943.1
|
PLA2G16
|
phospholipase A2, group XVI |
| chr8_-_110986918 | 2.42 |
ENST00000297404.1
|
KCNV1
|
potassium channel, subfamily V, member 1 |
| chr14_-_80697396 | 2.41 |
ENST00000557010.1
|
DIO2
|
deiodinase, iodothyronine, type II |
| chr8_+_70404996 | 2.39 |
ENST00000402687.4
ENST00000419716.3 |
SULF1
|
sulfatase 1 |
| chr16_-_18462221 | 2.37 |
ENST00000528301.1
|
RP11-1212A22.4
|
LOC339047 protein; Nuclear pore complex-interacting protein family member A3; Nuclear pore complex-interacting protein family member A5; Protein PKD1P1 |
| chr6_+_29555683 | 2.33 |
ENST00000383640.2
|
OR2H2
|
olfactory receptor, family 2, subfamily H, member 2 |
| chr1_+_117963209 | 2.32 |
ENST00000449370.2
|
MAN1A2
|
mannosidase, alpha, class 1A, member 2 |
| chr19_-_56826157 | 2.31 |
ENST00000592509.1
ENST00000592679.1 ENST00000588442.1 ENST00000593106.1 ENST00000587492.1 ENST00000254165.3 |
ZSCAN5A
|
zinc finger and SCAN domain containing 5A |
| chr12_-_71148357 | 2.31 |
ENST00000378778.1
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr3_-_160823040 | 2.30 |
ENST00000484127.1
ENST00000492353.1 ENST00000473142.1 ENST00000468268.1 ENST00000460353.1 ENST00000320474.4 ENST00000392781.2 |
B3GALNT1
|
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chr18_+_21269556 | 2.25 |
ENST00000399516.3
|
LAMA3
|
laminin, alpha 3 |
| chr7_+_138943265 | 2.24 |
ENST00000483726.1
|
UBN2
|
ubinuclein 2 |
| chr2_-_166930131 | 2.23 |
ENST00000303395.4
ENST00000409050.1 ENST00000423058.2 ENST00000375405.3 |
SCN1A
|
sodium channel, voltage-gated, type I, alpha subunit |
| chr5_+_140552218 | 2.14 |
ENST00000231137.3
|
PCDHB7
|
protocadherin beta 7 |
| chr8_-_49833978 | 2.13 |
ENST00000020945.1
|
SNAI2
|
snail family zinc finger 2 |
| chr11_+_122753391 | 2.11 |
ENST00000307257.6
ENST00000227349.2 |
C11orf63
|
chromosome 11 open reading frame 63 |
| chr10_+_17270214 | 2.09 |
ENST00000544301.1
|
VIM
|
vimentin |
| chr6_+_29429217 | 2.09 |
ENST00000396792.2
|
OR2H1
|
olfactory receptor, family 2, subfamily H, member 1 |
| chr1_-_109584608 | 2.05 |
ENST00000400794.3
ENST00000528747.1 ENST00000369962.3 ENST00000361054.3 |
WDR47
|
WD repeat domain 47 |
| chr5_-_111754948 | 2.05 |
ENST00000261486.5
|
EPB41L4A
|
erythrocyte membrane protein band 4.1 like 4A |
| chr9_+_111624577 | 2.04 |
ENST00000333999.3
|
ACTL7A
|
actin-like 7A |
| chr5_+_119867159 | 2.01 |
ENST00000505123.1
|
PRR16
|
proline rich 16 |
| chr17_+_66255310 | 1.98 |
ENST00000448504.2
|
ARSG
|
arylsulfatase G |
| chr12_+_26348246 | 1.97 |
ENST00000422622.2
|
SSPN
|
sarcospan |
| chr17_+_29664830 | 1.97 |
ENST00000444181.2
ENST00000417592.2 |
NF1
|
neurofibromin 1 |
| chr1_+_119957554 | 1.96 |
ENST00000543831.1
ENST00000433745.1 ENST00000369416.3 |
HSD3B2
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2 |
| chr12_-_71148413 | 1.92 |
ENST00000440835.2
ENST00000549308.1 ENST00000550661.1 |
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr19_-_4535233 | 1.91 |
ENST00000381848.3
ENST00000588887.1 ENST00000586133.1 |
PLIN5
|
perilipin 5 |
| chr1_+_197237352 | 1.89 |
ENST00000538660.1
ENST00000367400.3 ENST00000367399.2 |
CRB1
|
crumbs homolog 1 (Drosophila) |
| chr18_-_64271363 | 1.88 |
ENST00000262150.2
|
CDH19
|
cadherin 19, type 2 |
| chr4_+_186990298 | 1.86 |
ENST00000296795.3
ENST00000513189.1 |
TLR3
|
toll-like receptor 3 |
| chr17_-_4938712 | 1.85 |
ENST00000254853.5
ENST00000424747.1 |
SLC52A1
|
solute carrier family 52 (riboflavin transporter), member 1 |
| chr2_+_11696464 | 1.84 |
ENST00000234142.5
|
GREB1
|
growth regulation by estrogen in breast cancer 1 |
| chr3_+_130650738 | 1.79 |
ENST00000504612.1
|
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chrX_+_154113317 | 1.77 |
ENST00000354461.2
|
H2AFB1
|
H2A histone family, member B1 |
| chr1_+_81771806 | 1.77 |
ENST00000370721.1
ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2
|
latrophilin 2 |
| chrX_+_153533275 | 1.76 |
ENST00000426989.1
ENST00000426203.1 ENST00000369912.2 |
TKTL1
|
transketolase-like 1 |
| chr17_+_25799008 | 1.72 |
ENST00000583370.1
ENST00000398988.3 ENST00000268763.6 |
KSR1
|
kinase suppressor of ras 1 |
| chr8_-_91095099 | 1.71 |
ENST00000265431.3
|
CALB1
|
calbindin 1, 28kDa |
| chr2_+_161993465 | 1.71 |
ENST00000457476.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr16_+_19535235 | 1.70 |
ENST00000565376.2
ENST00000396208.2 |
CCP110
|
centriolar coiled coil protein 110kDa |
| chr4_+_71263599 | 1.69 |
ENST00000399575.2
|
PROL1
|
proline rich, lacrimal 1 |
| chr16_-_66583701 | 1.68 |
ENST00000527800.1
ENST00000525974.1 ENST00000563369.2 |
TK2
|
thymidine kinase 2, mitochondrial |
| chr4_+_37455536 | 1.65 |
ENST00000381980.4
ENST00000508175.1 |
C4orf19
|
chromosome 4 open reading frame 19 |
| chr12_+_26348429 | 1.65 |
ENST00000242729.2
|
SSPN
|
sarcospan |
| chr22_+_23487513 | 1.65 |
ENST00000263116.2
ENST00000341989.4 |
RAB36
|
RAB36, member RAS oncogene family |
| chr12_-_112123524 | 1.60 |
ENST00000327551.6
|
BRAP
|
BRCA1 associated protein |
| chr7_-_138666053 | 1.59 |
ENST00000440172.1
ENST00000422774.1 |
KIAA1549
|
KIAA1549 |
| chr19_-_22193731 | 1.58 |
ENST00000601773.1
ENST00000397126.4 ENST00000601993.1 ENST00000599916.1 |
ZNF208
|
zinc finger protein 208 |
| chr14_-_36990354 | 1.57 |
ENST00000518149.1
|
NKX2-1
|
NK2 homeobox 1 |
| chr8_-_49834299 | 1.57 |
ENST00000396822.1
|
SNAI2
|
snail family zinc finger 2 |
| chr9_-_33473882 | 1.57 |
ENST00000455041.2
ENST00000353159.2 ENST00000297990.4 ENST00000379471.2 |
NOL6
|
nucleolar protein 6 (RNA-associated) |
| chr19_+_9296279 | 1.55 |
ENST00000344248.2
|
OR7D2
|
olfactory receptor, family 7, subfamily D, member 2 |
| chr17_-_2996290 | 1.53 |
ENST00000331459.1
|
OR1D2
|
olfactory receptor, family 1, subfamily D, member 2 |
| chr1_-_67266939 | 1.51 |
ENST00000304526.2
|
INSL5
|
insulin-like 5 |
| chr2_-_26700900 | 1.50 |
ENST00000338581.6
ENST00000339598.3 ENST00000402415.3 |
OTOF
|
otoferlin |
| chr15_+_75970672 | 1.49 |
ENST00000435356.1
|
AC105020.1
|
Uncharacterized protein; cDNA FLJ12988 fis, clone NT2RP3000080 |
| chr7_+_107224364 | 1.49 |
ENST00000491150.1
|
BCAP29
|
B-cell receptor-associated protein 29 |
| chr9_-_21239978 | 1.42 |
ENST00000380222.2
|
IFNA14
|
interferon, alpha 14 |
| chr20_+_55904815 | 1.41 |
ENST00000371263.3
ENST00000345868.4 ENST00000371260.4 ENST00000418127.1 |
SPO11
|
SPO11 meiotic protein covalently bound to DSB |
| chr7_-_16872932 | 1.37 |
ENST00000419572.2
ENST00000412973.1 |
AGR2
|
anterior gradient 2 |
| chr2_+_103035102 | 1.37 |
ENST00000264260.2
|
IL18RAP
|
interleukin 18 receptor accessory protein |
| chr4_+_175839551 | 1.37 |
ENST00000404450.4
ENST00000514159.1 |
ADAM29
|
ADAM metallopeptidase domain 29 |
| chrX_-_71458802 | 1.37 |
ENST00000373657.1
ENST00000334463.3 |
ERCC6L
|
excision repair cross-complementing rodent repair deficiency, complementation group 6-like |
| chr2_-_169887827 | 1.37 |
ENST00000263817.6
|
ABCB11
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11 |
| chr1_+_162039558 | 1.35 |
ENST00000530878.1
ENST00000361897.5 |
NOS1AP
|
nitric oxide synthase 1 (neuronal) adaptor protein |
| chr18_-_52989525 | 1.32 |
ENST00000457482.3
|
TCF4
|
transcription factor 4 |
| chr20_-_43589109 | 1.31 |
ENST00000372813.3
|
TOMM34
|
translocase of outer mitochondrial membrane 34 |
| chr12_-_58220078 | 1.29 |
ENST00000549039.1
|
CTDSP2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chrX_+_84258832 | 1.27 |
ENST00000373173.2
|
APOOL
|
apolipoprotein O-like |
| chr6_-_136788001 | 1.26 |
ENST00000544465.1
|
MAP7
|
microtubule-associated protein 7 |
| chr11_-_13517565 | 1.25 |
ENST00000282091.1
ENST00000529816.1 |
PTH
|
parathyroid hormone |
| chr2_+_67624430 | 1.21 |
ENST00000272342.5
|
ETAA1
|
Ewing tumor-associated antigen 1 |
| chr1_-_211307404 | 1.20 |
ENST00000367007.4
|
KCNH1
|
potassium voltage-gated channel, subfamily H (eag-related), member 1 |
| chr4_+_5527117 | 1.18 |
ENST00000505296.1
|
C4orf6
|
chromosome 4 open reading frame 6 |
| chr6_-_146057144 | 1.15 |
ENST00000367519.3
|
EPM2A
|
epilepsy, progressive myoclonus type 2A, Lafora disease (laforin) |
| chr9_+_103947311 | 1.14 |
ENST00000395056.2
|
LPPR1
|
Lipid phosphate phosphatase-related protein type 1 |
| chr4_+_175839506 | 1.13 |
ENST00000505141.1
ENST00000359240.3 ENST00000445694.1 |
ADAM29
|
ADAM metallopeptidase domain 29 |
| chr14_+_61654271 | 1.11 |
ENST00000555185.1
ENST00000557294.1 ENST00000556778.1 |
PRKCH
|
protein kinase C, eta |
| chrX_-_11308598 | 1.09 |
ENST00000380717.3
|
ARHGAP6
|
Rho GTPase activating protein 6 |
| chr1_-_45988542 | 1.08 |
ENST00000424390.1
|
PRDX1
|
peroxiredoxin 1 |
| chr5_+_140593509 | 1.08 |
ENST00000341948.4
|
PCDHB13
|
protocadherin beta 13 |
| chr10_-_52645379 | 1.03 |
ENST00000395489.2
|
A1CF
|
APOBEC1 complementation factor |
| chr9_-_99540328 | 1.03 |
ENST00000223428.4
ENST00000375231.1 ENST00000374641.3 |
ZNF510
|
zinc finger protein 510 |
| chr3_+_52245458 | 1.03 |
ENST00000459884.1
|
ALAS1
|
aminolevulinate, delta-, synthase 1 |
| chr11_-_102651343 | 1.00 |
ENST00000279441.4
ENST00000539681.1 |
MMP10
|
matrix metallopeptidase 10 (stromelysin 2) |
| chr1_-_53608289 | 0.98 |
ENST00000371491.4
|
SLC1A7
|
solute carrier family 1 (glutamate transporter), member 7 |
| chr22_-_43010928 | 0.95 |
ENST00000348657.2
ENST00000252115.5 |
POLDIP3
|
polymerase (DNA-directed), delta interacting protein 3 |
| chr1_+_87012753 | 0.95 |
ENST00000370563.3
|
CLCA4
|
chloride channel accessory 4 |
| chr10_+_57358750 | 0.95 |
ENST00000512524.2
|
MTRNR2L5
|
MT-RNR2-like 5 |
| chr4_+_5526883 | 0.91 |
ENST00000195455.2
|
C4orf6
|
chromosome 4 open reading frame 6 |
| chr1_-_53608249 | 0.90 |
ENST00000371494.4
|
SLC1A7
|
solute carrier family 1 (glutamate transporter), member 7 |
| chr12_+_26164645 | 0.89 |
ENST00000542004.1
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr12_-_52761262 | 0.87 |
ENST00000257901.3
|
KRT85
|
keratin 85 |
| chr5_+_137225158 | 0.87 |
ENST00000290431.5
|
PKD2L2
|
polycystic kidney disease 2-like 2 |
| chr16_-_70835034 | 0.86 |
ENST00000261776.5
|
VAC14
|
Vac14 homolog (S. cerevisiae) |
| chr4_-_46996424 | 0.84 |
ENST00000264318.3
|
GABRA4
|
gamma-aminobutyric acid (GABA) A receptor, alpha 4 |
| chr17_-_40950698 | 0.82 |
ENST00000328434.7
|
COA3
|
cytochrome c oxidase assembly factor 3 |
| chr2_+_152214098 | 0.80 |
ENST00000243347.3
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6 |
| chrX_+_83116142 | 0.78 |
ENST00000329312.4
|
CYLC1
|
cylicin, basic protein of sperm head cytoskeleton 1 |
| chr9_-_100684845 | 0.77 |
ENST00000375119.3
|
C9orf156
|
chromosome 9 open reading frame 156 |
| chr2_-_109605663 | 0.76 |
ENST00000409271.1
ENST00000258443.2 ENST00000376651.1 |
EDAR
|
ectodysplasin A receptor |
| chr15_+_67418047 | 0.76 |
ENST00000540846.2
|
SMAD3
|
SMAD family member 3 |
| chr7_+_99425633 | 0.74 |
ENST00000354829.2
ENST00000421837.2 ENST00000417625.1 ENST00000342499.4 ENST00000444905.1 ENST00000415413.1 ENST00000312017.5 ENST00000222382.5 |
CYP3A43
|
cytochrome P450, family 3, subfamily A, polypeptide 43 |
| chrX_-_154689596 | 0.72 |
ENST00000369444.2
|
H2AFB3
|
H2A histone family, member B3 |
| chr16_+_48278178 | 0.72 |
ENST00000285737.4
ENST00000535754.1 |
LONP2
|
lon peptidase 2, peroxisomal |
| chr19_-_43969796 | 0.71 |
ENST00000244333.3
|
LYPD3
|
LY6/PLAUR domain containing 3 |
| chr12_-_10978957 | 0.67 |
ENST00000240619.2
|
TAS2R10
|
taste receptor, type 2, member 10 |
| chr5_-_78809950 | 0.66 |
ENST00000334082.6
|
HOMER1
|
homer homolog 1 (Drosophila) |
| chrY_-_6740649 | 0.66 |
ENST00000383036.1
ENST00000383037.4 |
AMELY
|
amelogenin, Y-linked |
| chr1_+_11333245 | 0.65 |
ENST00000376810.5
|
UBIAD1
|
UbiA prenyltransferase domain containing 1 |
| chr3_-_46759314 | 0.62 |
ENST00000315170.7
|
PRSS50
|
protease, serine, 50 |
| chr6_-_49712147 | 0.60 |
ENST00000433368.2
ENST00000354620.4 |
CRISP3
|
cysteine-rich secretory protein 3 |
| chr3_+_193310918 | 0.60 |
ENST00000361908.3
ENST00000392438.3 ENST00000361510.2 ENST00000361715.2 ENST00000361828.2 ENST00000361150.2 |
OPA1
|
optic atrophy 1 (autosomal dominant) |
| chr4_-_120243545 | 0.59 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr18_-_13915530 | 0.58 |
ENST00000327606.3
|
MC2R
|
melanocortin 2 receptor (adrenocorticotropic hormone) |
| chr6_-_25874440 | 0.56 |
ENST00000361703.6
ENST00000397060.4 |
SLC17A3
|
solute carrier family 17 (organic anion transporter), member 3 |
| chr8_+_67687413 | 0.53 |
ENST00000521960.1
ENST00000522398.1 ENST00000522629.1 ENST00000520976.1 ENST00000396596.1 |
SGK3
|
serum/glucocorticoid regulated kinase family, member 3 |
| chr8_-_59412717 | 0.49 |
ENST00000301645.3
|
CYP7A1
|
cytochrome P450, family 7, subfamily A, polypeptide 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of POU1F1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 11.7 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 2.3 | 6.8 | GO:0002589 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) regulation of T cell antigen processing and presentation(GO:0002625) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) response to iron ion starvation(GO:1990641) |
| 1.6 | 8.1 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 1.4 | 5.7 | GO:0043128 | regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 1.3 | 12.7 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 1.2 | 3.7 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 1.1 | 4.6 | GO:0046104 | thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 1.1 | 3.2 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) |
| 1.0 | 3.1 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 1.0 | 3.8 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.8 | 3.4 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) |
| 0.7 | 24.8 | GO:1901685 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.7 | 4.3 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.7 | 4.2 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.7 | 2.0 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.6 | 1.9 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.6 | 5.1 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.5 | 3.8 | GO:0042426 | choline catabolic process(GO:0042426) |
| 0.5 | 4.3 | GO:1902514 | regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.5 | 3.6 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.5 | 2.6 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.5 | 6.6 | GO:0071670 | smooth muscle cell chemotaxis(GO:0071670) |
| 0.5 | 1.5 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.5 | 1.9 | GO:0034343 | microglial cell activation involved in immune response(GO:0002282) type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.5 | 1.9 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.4 | 2.7 | GO:0010615 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.4 | 1.3 | GO:1900158 | positive regulation of osteoclast proliferation(GO:0090290) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.4 | 2.4 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 0.4 | 2.3 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.4 | 7.6 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.4 | 7.2 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.4 | 7.7 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.3 | 9.1 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.3 | 2.4 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.3 | 9.6 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.3 | 1.6 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.3 | 2.8 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.3 | 6.5 | GO:0044342 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) type B pancreatic cell proliferation(GO:0044342) |
| 0.3 | 1.8 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.3 | 1.4 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.3 | 3.0 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.2 | 1.7 | GO:0072205 | metanephric collecting duct development(GO:0072205) |
| 0.2 | 3.6 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.2 | 0.7 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.2 | 2.4 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.2 | 4.6 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.2 | 2.2 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.2 | 1.4 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
| 0.2 | 1.4 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.2 | 2.1 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.2 | 0.9 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.2 | 0.9 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.2 | 0.4 | GO:0010841 | positive regulation of circadian sleep/wake cycle, wakefulness(GO:0010841) |
| 0.2 | 5.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.2 | 1.0 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.2 | 1.2 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.2 | 5.1 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.2 | 2.5 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.2 | 0.8 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.2 | 2.0 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.2 | 23.9 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.2 | 0.8 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.1 | 1.5 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 2.0 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.1 | 1.1 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.1 | 6.8 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 6.5 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) beta-amyloid metabolic process(GO:0050435) |
| 0.1 | 2.0 | GO:0006705 | mineralocorticoid biosynthetic process(GO:0006705) mineralocorticoid metabolic process(GO:0008212) |
| 0.1 | 0.7 | GO:0009233 | menaquinone metabolic process(GO:0009233) |
| 0.1 | 2.7 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 3.0 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 1.2 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.1 | 0.6 | GO:0014042 | positive regulation of neuron maturation(GO:0014042) |
| 0.1 | 2.5 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.1 | 4.2 | GO:0015988 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.1 | 1.4 | GO:0090220 | meiotic telomere clustering(GO:0045141) chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) |
| 0.1 | 2.9 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.1 | 0.8 | GO:2001023 | regulation of response to drug(GO:2001023) |
| 0.1 | 3.6 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.1 | 0.5 | GO:0050955 | thermoception(GO:0050955) |
| 0.1 | 4.4 | GO:0031102 | neuron projection regeneration(GO:0031102) |
| 0.1 | 1.9 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.1 | 5.4 | GO:0051058 | negative regulation of Ras protein signal transduction(GO:0046580) negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.1 | 2.4 | GO:0048012 | hepatocyte growth factor receptor signaling pathway(GO:0048012) |
| 0.1 | 0.6 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 0.8 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.1 | 9.3 | GO:0007498 | mesoderm development(GO:0007498) |
| 0.1 | 0.5 | GO:0070857 | regulation of bile acid biosynthetic process(GO:0070857) |
| 0.1 | 0.4 | GO:1901529 | regulation of anion channel activity(GO:0010359) positive regulation of anion channel activity(GO:1901529) |
| 0.1 | 4.4 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.1 | 1.2 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 1.0 | GO:0019216 | regulation of lipid metabolic process(GO:0019216) |
| 0.1 | 1.9 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.1 | 1.0 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.7 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.0 | 0.3 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 1.1 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) |
| 0.0 | 0.1 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.0 | 0.2 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.0 | 1.3 | GO:0006626 | protein targeting to mitochondrion(GO:0006626) |
| 0.0 | 2.5 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 1.3 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.4 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 1.1 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 0.7 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.0 | 1.8 | GO:0006007 | glucose catabolic process(GO:0006007) |
| 0.0 | 0.2 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.2 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.9 | GO:0050982 | detection of mechanical stimulus(GO:0050982) |
| 0.0 | 1.7 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 0.2 | GO:1904322 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 | 1.7 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 0.3 | GO:0048007 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 2.0 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 3.7 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.9 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.0 | 0.9 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.0 | 2.4 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.0 | 5.4 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.0 | 4.5 | GO:0065004 | protein-DNA complex assembly(GO:0065004) |
| 0.0 | 1.5 | GO:0016079 | synaptic vesicle exocytosis(GO:0016079) |
| 0.0 | 0.4 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 1.9 | GO:2001252 | positive regulation of chromosome organization(GO:2001252) |
| 0.0 | 6.1 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 1.3 | GO:0006970 | response to osmotic stress(GO:0006970) |
| 0.0 | 0.2 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 1.0 | GO:0030574 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 1.5 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 1.6 | GO:0007265 | Ras protein signal transduction(GO:0007265) |
| 0.0 | 0.5 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.0 | 0.4 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 1.5 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.7 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 6.8 | GO:1990357 | terminal web(GO:1990357) |
| 1.1 | 3.4 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 1.0 | 11.7 | GO:0043203 | axon hillock(GO:0043203) |
| 0.9 | 8.1 | GO:1990761 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.7 | 6.5 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.6 | 5.1 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.5 | 3.8 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.4 | 3.5 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.4 | 2.5 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.4 | 5.1 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.3 | 1.0 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.3 | 4.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.3 | 5.5 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.3 | 4.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.3 | 4.8 | GO:0030478 | actin cap(GO:0030478) |
| 0.3 | 4.8 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.3 | 3.6 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.3 | 9.6 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.3 | 8.1 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.3 | 0.8 | GO:0043159 | perinuclear theca(GO:0033011) cytoskeletal calyx(GO:0033150) acrosomal matrix(GO:0043159) |
| 0.2 | 5.1 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.2 | 7.2 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.2 | 2.0 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.2 | 1.4 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.2 | 1.3 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.2 | 5.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 2.2 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 4.0 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 1.5 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 0.9 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.1 | 3.9 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 4.6 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 2.0 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 5.7 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 0.3 | GO:0097679 | other organism cytoplasm(GO:0097679) |
| 0.1 | 1.9 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.1 | 0.8 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 2.4 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 8.0 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.1 | 0.4 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.1 | 1.0 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.1 | 3.0 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 4.5 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.1 | 21.4 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 4.0 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 6.9 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 1.9 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 7.9 | GO:0044309 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 0.8 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 2.9 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.7 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 12.7 | GO:0045121 | membrane raft(GO:0045121) |
| 0.0 | 0.3 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 2.6 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.7 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 3.1 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 3.0 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.9 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 2.3 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 1.2 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 1.6 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 1.5 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 1.7 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 3.0 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 2.8 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 1.4 | GO:0000228 | nuclear chromosome(GO:0000228) |
| 0.0 | 11.4 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 2.6 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 1.5 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 1.7 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.6 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.8 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 1.6 | GO:0031965 | nuclear membrane(GO:0031965) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 6.5 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 1.7 | 5.2 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 1.6 | 4.8 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 1.5 | 4.6 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 1.1 | 12.9 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 1.0 | 7.2 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 1.0 | 2.9 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.9 | 6.5 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.8 | 8.1 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.7 | 2.0 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) steroid delta-isomerase activity(GO:0004769) |
| 0.6 | 2.4 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.6 | 1.8 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.6 | 5.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.5 | 7.6 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.5 | 2.5 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.5 | 24.8 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.5 | 2.5 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.5 | 2.4 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.5 | 1.9 | GO:0035473 | lipase binding(GO:0035473) |
| 0.5 | 1.4 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.5 | 1.9 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.5 | 3.6 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.4 | 2.4 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.4 | 3.0 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.3 | 2.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.3 | 4.0 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.3 | 1.7 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.3 | 1.7 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.3 | 1.3 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.2 | 1.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.2 | 7.8 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.2 | 2.3 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.2 | 2.7 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.2 | 4.0 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.2 | 1.2 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.2 | 2.8 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.2 | 5.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 3.2 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.2 | 4.2 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.2 | 4.8 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.2 | 1.9 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.2 | 2.0 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.2 | 3.8 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.2 | 4.2 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.2 | 4.2 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.2 | 3.2 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.2 | 2.0 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.2 | 0.8 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.1 | 6.8 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 1.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 3.0 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.7 | GO:0016209 | antioxidant activity(GO:0016209) |
| 0.1 | 12.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 11.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 2.2 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 0.4 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.1 | 1.6 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.1 | 3.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 0.7 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 1.3 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.1 | 0.8 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.1 | 0.6 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.1 | 2.3 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.6 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.1 | 5.4 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 1.8 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 0.6 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 0.3 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.1 | 1.0 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 1.4 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 2.4 | GO:0008200 | ion channel inhibitor activity(GO:0008200) |
| 0.1 | 1.3 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 2.7 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.7 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.7 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 32.7 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.5 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.4 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 1.2 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 3.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 10.9 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.7 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 2.9 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.3 | GO:0030882 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.0 | 4.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.6 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.5 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 5.6 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 7.6 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 1.0 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.6 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.5 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.2 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 8.8 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 8.1 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 9.1 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.1 | 2.6 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 5.5 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.1 | 6.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 5.1 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 2.5 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 2.8 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 6.6 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 2.7 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.1 | 6.1 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 19.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 2.0 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 7.7 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 1.9 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 1.4 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 4.0 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 2.5 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 3.5 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.9 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 2.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.6 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.1 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 1.7 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 5.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.1 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 1.8 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.5 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 1.0 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.4 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.0 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 24.8 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.4 | 8.1 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.3 | 5.4 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.2 | 6.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 2.7 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.2 | 6.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.2 | 7.6 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.2 | 2.7 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.2 | 3.6 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.2 | 2.0 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.2 | 1.9 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.2 | 8.1 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.2 | 1.9 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.2 | 2.4 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.2 | 2.6 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.1 | 2.0 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 3.2 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 4.6 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 2.8 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.1 | 6.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 1.9 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.1 | 4.5 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 0.9 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 4.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 2.6 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 3.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 1.1 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 1.5 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 2.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 5.1 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 3.7 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 2.4 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 2.2 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.4 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 1.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.1 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 1.0 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.8 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.8 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.7 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 1.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 4.1 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.7 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.9 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |