Project
averaged GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
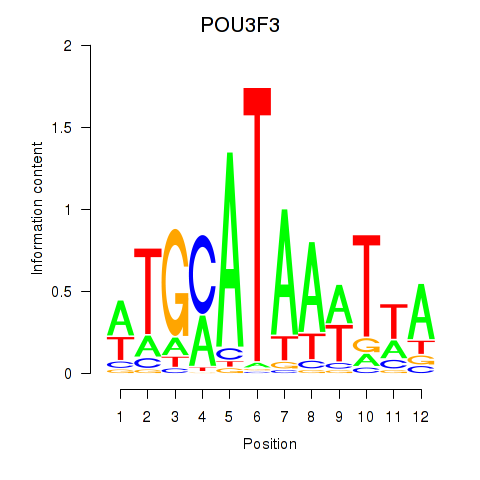
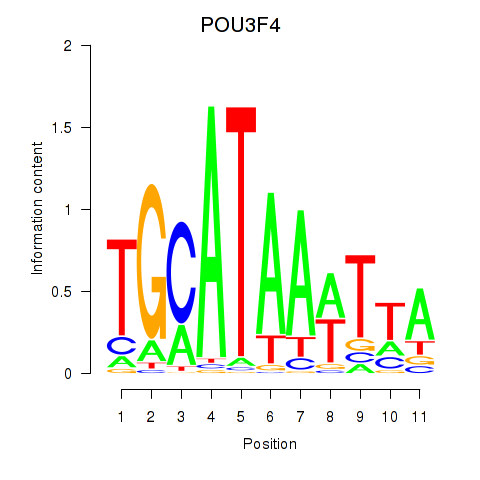
Results for POU3F3_POU3F4
Z-value: 1.69
Transcription factors associated with POU3F3_POU3F4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
POU3F3
|
ENSG00000198914.2 | POU class 3 homeobox 3 |
|
POU3F4
|
ENSG00000196767.4 | POU class 3 homeobox 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| POU3F3 | hg19_v2_chr2_+_105471969_105471969 | 0.38 | 7.2e-09 | Click! |
| POU3F4 | hg19_v2_chrX_+_82763265_82763283 | 0.35 | 1.2e-07 | Click! |
Activity profile of POU3F3_POU3F4 motif
Sorted Z-values of POU3F3_POU3F4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_158142750 | 71.76 |
ENST00000505888.1
ENST00000449365.1 |
GRIA2
|
glutamate receptor, ionotropic, AMPA 2 |
| chr4_+_158141899 | 66.41 |
ENST00000264426.9
ENST00000506284.1 |
GRIA2
|
glutamate receptor, ionotropic, AMPA 2 |
| chr15_-_45670924 | 66.02 |
ENST00000396659.3
|
GATM
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr3_-_58613323 | 50.77 |
ENST00000474531.1
ENST00000465970.1 |
FAM107A
|
family with sequence similarity 107, member A |
| chr5_+_140762268 | 44.45 |
ENST00000518325.1
|
PCDHGA7
|
protocadherin gamma subfamily A, 7 |
| chr6_-_46459675 | 40.38 |
ENST00000306764.7
|
RCAN2
|
regulator of calcineurin 2 |
| chr14_-_60097297 | 34.73 |
ENST00000395090.1
|
RTN1
|
reticulon 1 |
| chr14_-_21270561 | 32.56 |
ENST00000412779.2
|
RNASE1
|
ribonuclease, RNase A family, 1 (pancreatic) |
| chr14_-_21270995 | 29.58 |
ENST00000555698.1
ENST00000397970.4 ENST00000340900.3 |
RNASE1
|
ribonuclease, RNase A family, 1 (pancreatic) |
| chr5_-_11588907 | 28.42 |
ENST00000513598.1
ENST00000503622.1 |
CTNND2
|
catenin (cadherin-associated protein), delta 2 |
| chr3_+_35722487 | 28.38 |
ENST00000441454.1
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr3_+_158787041 | 26.61 |
ENST00000471575.1
ENST00000476809.1 ENST00000485419.1 |
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr16_-_21314360 | 24.61 |
ENST00000219599.3
ENST00000576703.1 |
CRYM
|
crystallin, mu |
| chr14_+_101297740 | 24.54 |
ENST00000555928.1
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr4_+_158141843 | 23.19 |
ENST00000509417.1
ENST00000296526.7 |
GRIA2
|
glutamate receptor, ionotropic, AMPA 2 |
| chr5_+_140753444 | 22.24 |
ENST00000517434.1
|
PCDHGA6
|
protocadherin gamma subfamily A, 6 |
| chr5_-_42811986 | 21.97 |
ENST00000511224.1
ENST00000507920.1 ENST00000510965.1 |
SEPP1
|
selenoprotein P, plasma, 1 |
| chr5_-_11589131 | 21.88 |
ENST00000511377.1
|
CTNND2
|
catenin (cadherin-associated protein), delta 2 |
| chr17_-_66951474 | 21.38 |
ENST00000269080.2
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr5_-_42812143 | 21.07 |
ENST00000514985.1
|
SEPP1
|
selenoprotein P, plasma, 1 |
| chr17_+_3379284 | 20.03 |
ENST00000263080.2
|
ASPA
|
aspartoacylase |
| chr5_+_36608422 | 18.54 |
ENST00000381918.3
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr9_+_137967366 | 17.77 |
ENST00000252854.4
|
OLFM1
|
olfactomedin 1 |
| chr18_-_52989217 | 17.10 |
ENST00000570287.2
|
TCF4
|
transcription factor 4 |
| chr10_-_75226166 | 17.06 |
ENST00000544628.1
|
PPP3CB
|
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr4_-_87281224 | 16.01 |
ENST00000395169.3
ENST00000395161.2 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr11_-_2906979 | 15.50 |
ENST00000380725.1
ENST00000313407.6 ENST00000430149.2 ENST00000440480.2 ENST00000414822.3 |
CDKN1C
|
cyclin-dependent kinase inhibitor 1C (p57, Kip2) |
| chr2_-_101034070 | 14.39 |
ENST00000264249.3
|
CHST10
|
carbohydrate sulfotransferase 10 |
| chr7_-_100026280 | 14.31 |
ENST00000360951.4
ENST00000398027.2 ENST00000324725.6 ENST00000472716.1 |
ZCWPW1
|
zinc finger, CW type with PWWP domain 1 |
| chr8_-_120685608 | 13.45 |
ENST00000427067.2
|
ENPP2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr2_-_175711133 | 13.41 |
ENST00000409597.1
ENST00000413882.1 |
CHN1
|
chimerin 1 |
| chr5_-_146461027 | 12.93 |
ENST00000394410.2
ENST00000508267.1 ENST00000504198.1 |
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta |
| chr1_+_28099683 | 12.81 |
ENST00000373943.4
|
STX12
|
syntaxin 12 |
| chr5_+_125758813 | 12.78 |
ENST00000285689.3
ENST00000515200.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr2_-_55237484 | 11.84 |
ENST00000394609.2
|
RTN4
|
reticulon 4 |
| chr6_-_52859046 | 11.42 |
ENST00000457564.1
ENST00000541324.1 ENST00000370960.1 |
GSTA4
|
glutathione S-transferase alpha 4 |
| chr5_+_125758865 | 11.22 |
ENST00000542322.1
ENST00000544396.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr8_-_91095099 | 10.89 |
ENST00000265431.3
|
CALB1
|
calbindin 1, 28kDa |
| chr12_-_111926342 | 10.34 |
ENST00000389154.3
|
ATXN2
|
ataxin 2 |
| chr7_-_14880892 | 10.24 |
ENST00000406247.3
ENST00000399322.3 ENST00000258767.5 |
DGKB
|
diacylglycerol kinase, beta 90kDa |
| chr3_-_127455200 | 9.99 |
ENST00000398101.3
|
MGLL
|
monoglyceride lipase |
| chr2_+_239335449 | 9.73 |
ENST00000264607.4
|
ASB1
|
ankyrin repeat and SOCS box containing 1 |
| chr19_-_18632861 | 9.28 |
ENST00000262809.4
|
ELL
|
elongation factor RNA polymerase II |
| chr3_-_33686743 | 9.24 |
ENST00000333778.6
ENST00000539981.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr18_-_52989525 | 9.19 |
ENST00000457482.3
|
TCF4
|
transcription factor 4 |
| chr5_-_160279207 | 8.99 |
ENST00000327245.5
|
ATP10B
|
ATPase, class V, type 10B |
| chr22_-_37213554 | 8.47 |
ENST00000443735.1
|
PVALB
|
parvalbumin |
| chr19_-_44123734 | 8.30 |
ENST00000598676.1
|
ZNF428
|
zinc finger protein 428 |
| chr17_+_66255310 | 8.16 |
ENST00000448504.2
|
ARSG
|
arylsulfatase G |
| chr12_-_91574142 | 8.00 |
ENST00000547937.1
|
DCN
|
decorin |
| chr1_+_19923454 | 7.84 |
ENST00000602662.1
ENST00000602293.1 ENST00000322753.6 |
MINOS1-NBL1
MINOS1
|
MINOS1-NBL1 readthrough mitochondrial inner membrane organizing system 1 |
| chr5_+_140710061 | 7.83 |
ENST00000517417.1
ENST00000378105.3 |
PCDHGA1
|
protocadherin gamma subfamily A, 1 |
| chr1_+_104198377 | 7.78 |
ENST00000370083.4
|
AMY1A
|
amylase, alpha 1A (salivary) |
| chr2_-_166930131 | 7.78 |
ENST00000303395.4
ENST00000409050.1 ENST00000423058.2 ENST00000375405.3 |
SCN1A
|
sodium channel, voltage-gated, type I, alpha subunit |
| chr12_-_91573249 | 7.63 |
ENST00000550099.1
ENST00000546391.1 ENST00000551354.1 |
DCN
|
decorin |
| chr9_+_137967268 | 7.56 |
ENST00000371799.4
ENST00000277415.11 |
OLFM1
|
olfactomedin 1 |
| chr17_-_50236039 | 7.39 |
ENST00000451037.2
|
CA10
|
carbonic anhydrase X |
| chr15_-_90892669 | 7.19 |
ENST00000412799.2
|
GABARAPL3
|
GABA(A) receptors associated protein like 3, pseudogene |
| chr3_-_185826855 | 7.16 |
ENST00000306376.5
|
ETV5
|
ets variant 5 |
| chr5_+_141488070 | 6.69 |
ENST00000253814.4
|
NDFIP1
|
Nedd4 family interacting protein 1 |
| chr17_-_37844267 | 6.67 |
ENST00000579146.1
ENST00000378011.4 ENST00000429199.2 ENST00000300658.4 |
PGAP3
|
post-GPI attachment to proteins 3 |
| chr12_-_91573316 | 6.43 |
ENST00000393155.1
|
DCN
|
decorin |
| chr19_-_44124019 | 6.31 |
ENST00000300811.3
|
ZNF428
|
zinc finger protein 428 |
| chr5_+_129083772 | 6.14 |
ENST00000564719.1
|
KIAA1024L
|
KIAA1024-like |
| chr1_-_1297157 | 6.09 |
ENST00000477278.2
|
MXRA8
|
matrix-remodelling associated 8 |
| chr2_+_189839046 | 6.03 |
ENST00000304636.3
ENST00000317840.5 |
COL3A1
|
collagen, type III, alpha 1 |
| chr12_-_91573132 | 6.03 |
ENST00000550563.1
ENST00000546370.1 |
DCN
|
decorin |
| chr1_+_50574585 | 5.95 |
ENST00000371824.1
ENST00000371823.4 |
ELAVL4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr11_-_57004658 | 5.80 |
ENST00000606794.1
|
APLNR
|
apelin receptor |
| chr3_+_53528659 | 5.55 |
ENST00000350061.5
|
CACNA1D
|
calcium channel, voltage-dependent, L type, alpha 1D subunit |
| chr7_+_116593433 | 5.53 |
ENST00000323984.3
ENST00000393449.1 |
ST7
|
suppression of tumorigenicity 7 |
| chr5_-_78809950 | 5.41 |
ENST00000334082.6
|
HOMER1
|
homer homolog 1 (Drosophila) |
| chr11_+_2405833 | 5.22 |
ENST00000527343.1
ENST00000464784.2 |
CD81
|
CD81 molecule |
| chrX_+_65382433 | 5.12 |
ENST00000374727.3
|
HEPH
|
hephaestin |
| chr2_+_166095898 | 5.06 |
ENST00000424833.1
ENST00000375437.2 ENST00000357398.3 |
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit |
| chr8_-_67090825 | 4.81 |
ENST00000276571.3
|
CRH
|
corticotropin releasing hormone |
| chr17_-_61996136 | 4.77 |
ENST00000342364.4
|
GH1
|
growth hormone 1 |
| chr5_+_140782351 | 4.74 |
ENST00000573521.1
|
PCDHGA9
|
protocadherin gamma subfamily A, 9 |
| chr4_+_62067860 | 4.74 |
ENST00000514591.1
|
LPHN3
|
latrophilin 3 |
| chr15_+_74422585 | 4.74 |
ENST00000561740.1
ENST00000435464.1 ENST00000453268.2 |
ISLR2
|
immunoglobulin superfamily containing leucine-rich repeat 2 |
| chrX_+_57618269 | 4.60 |
ENST00000374888.1
|
ZXDB
|
zinc finger, X-linked, duplicated B |
| chr10_-_90712520 | 4.56 |
ENST00000224784.6
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta |
| chr13_+_52586517 | 4.33 |
ENST00000523764.1
ENST00000521508.1 |
ALG11
|
ALG11, alpha-1,2-mannosyltransferase |
| chr18_-_33647487 | 4.31 |
ENST00000590898.1
ENST00000357384.4 ENST00000319040.6 ENST00000588737.1 ENST00000399022.4 |
RPRD1A
|
regulation of nuclear pre-mRNA domain containing 1A |
| chr11_-_95657231 | 4.20 |
ENST00000409459.1
ENST00000352297.7 ENST00000393223.3 ENST00000346299.5 |
MTMR2
|
myotubularin related protein 2 |
| chr11_+_8040739 | 4.18 |
ENST00000534099.1
|
TUB
|
tubby bipartite transcription factor |
| chr13_-_26795840 | 4.17 |
ENST00000381570.3
ENST00000399762.2 ENST00000346166.3 |
RNF6
|
ring finger protein (C3H2C3 type) 6 |
| chr1_+_101702417 | 4.09 |
ENST00000305352.6
|
S1PR1
|
sphingosine-1-phosphate receptor 1 |
| chr16_-_70835034 | 4.05 |
ENST00000261776.5
|
VAC14
|
Vac14 homolog (S. cerevisiae) |
| chr12_+_41086297 | 4.00 |
ENST00000551295.2
|
CNTN1
|
contactin 1 |
| chr4_+_88754113 | 3.90 |
ENST00000560249.1
ENST00000540395.1 ENST00000511670.1 ENST00000361056.3 |
MEPE
|
matrix extracellular phosphoglycoprotein |
| chr9_-_15472730 | 3.88 |
ENST00000481862.1
|
PSIP1
|
PC4 and SFRS1 interacting protein 1 |
| chr21_-_27423339 | 3.73 |
ENST00000415997.1
|
APP
|
amyloid beta (A4) precursor protein |
| chr19_+_36486078 | 3.61 |
ENST00000378887.2
|
SDHAF1
|
succinate dehydrogenase complex assembly factor 1 |
| chrX_+_134975858 | 3.61 |
ENST00000537770.1
|
SAGE1
|
sarcoma antigen 1 |
| chrX_+_134975753 | 3.55 |
ENST00000535938.1
|
SAGE1
|
sarcoma antigen 1 |
| chr4_-_102268484 | 3.51 |
ENST00000394853.4
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr6_-_11779403 | 3.50 |
ENST00000414691.3
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr15_-_30113676 | 3.40 |
ENST00000400011.2
|
TJP1
|
tight junction protein 1 |
| chr1_-_216978709 | 3.32 |
ENST00000360012.3
|
ESRRG
|
estrogen-related receptor gamma |
| chr7_+_119913688 | 3.28 |
ENST00000331113.4
|
KCND2
|
potassium voltage-gated channel, Shal-related subfamily, member 2 |
| chr17_-_26220366 | 3.26 |
ENST00000460380.2
ENST00000508862.1 ENST00000379102.3 ENST00000582441.1 |
LYRM9
RP1-66C13.4
|
LYR motif containing 9 Uncharacterized protein |
| chr12_+_110011571 | 3.11 |
ENST00000539696.1
ENST00000228510.3 ENST00000392727.3 |
MVK
|
mevalonate kinase |
| chr12_-_102224457 | 3.09 |
ENST00000549165.1
ENST00000549940.1 ENST00000392919.4 |
GNPTAB
|
N-acetylglucosamine-1-phosphate transferase, alpha and beta subunits |
| chr7_+_29237354 | 3.07 |
ENST00000546235.1
|
CHN2
|
chimerin 2 |
| chr1_-_104238912 | 3.06 |
ENST00000330330.5
|
AMY1B
|
amylase, alpha 1B (salivary) |
| chr14_+_100485712 | 3.06 |
ENST00000544450.2
|
EVL
|
Enah/Vasp-like |
| chr1_-_193155729 | 2.98 |
ENST00000367434.4
|
B3GALT2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
| chr2_-_183387430 | 2.95 |
ENST00000410103.1
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr14_+_70918874 | 2.94 |
ENST00000603540.1
|
ADAM21
|
ADAM metallopeptidase domain 21 |
| chr6_+_29429217 | 2.93 |
ENST00000396792.2
|
OR2H1
|
olfactory receptor, family 2, subfamily H, member 1 |
| chr15_-_74658519 | 2.90 |
ENST00000450547.1
ENST00000358632.4 |
CYP11A1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr10_-_49813090 | 2.90 |
ENST00000249601.4
|
ARHGAP22
|
Rho GTPase activating protein 22 |
| chr4_+_186990298 | 2.84 |
ENST00000296795.3
ENST00000513189.1 |
TLR3
|
toll-like receptor 3 |
| chr4_-_102268628 | 2.84 |
ENST00000323055.6
ENST00000512215.1 ENST00000394854.3 |
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr16_+_31128978 | 2.82 |
ENST00000448516.2
ENST00000219797.4 |
KAT8
|
K(lysine) acetyltransferase 8 |
| chr15_-_74658493 | 2.82 |
ENST00000419019.2
ENST00000569662.1 |
CYP11A1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chrX_-_55020511 | 2.82 |
ENST00000375006.3
ENST00000374992.2 |
PFKFB1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr7_+_134528635 | 2.80 |
ENST00000445569.2
|
CALD1
|
caldesmon 1 |
| chr9_+_21967137 | 2.77 |
ENST00000441769.2
|
C9orf53
|
chromosome 9 open reading frame 53 |
| chr1_-_104239076 | 2.73 |
ENST00000370080.3
|
AMY1B
|
amylase, alpha 1B (salivary) |
| chr5_+_74807886 | 2.68 |
ENST00000514296.1
|
POLK
|
polymerase (DNA directed) kappa |
| chr1_-_62190793 | 2.64 |
ENST00000371177.2
ENST00000606498.1 |
TM2D1
|
TM2 domain containing 1 |
| chrX_-_112084043 | 2.61 |
ENST00000304758.1
|
AMOT
|
angiomotin |
| chr6_-_49712147 | 2.61 |
ENST00000433368.2
ENST00000354620.4 |
CRISP3
|
cysteine-rich secretory protein 3 |
| chr16_-_279405 | 2.60 |
ENST00000430864.1
ENST00000293872.8 ENST00000337351.4 ENST00000397783.1 |
LUC7L
|
LUC7-like (S. cerevisiae) |
| chr2_-_183387064 | 2.56 |
ENST00000536095.1
ENST00000331935.6 ENST00000358139.2 ENST00000456212.1 |
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr13_+_109248500 | 2.55 |
ENST00000356711.2
|
MYO16
|
myosin XVI |
| chr9_-_14180778 | 2.51 |
ENST00000380924.1
ENST00000543693.1 |
NFIB
|
nuclear factor I/B |
| chr17_-_61996160 | 2.50 |
ENST00000458650.2
ENST00000351388.4 ENST00000323322.5 |
GH1
|
growth hormone 1 |
| chr3_+_148508845 | 2.47 |
ENST00000491148.1
|
CPB1
|
carboxypeptidase B1 (tissue) |
| chr1_-_48866517 | 2.46 |
ENST00000371841.1
|
SPATA6
|
spermatogenesis associated 6 |
| chr6_+_44355257 | 2.41 |
ENST00000371477.3
|
CDC5L
|
cell division cycle 5-like |
| chrX_+_54834004 | 2.41 |
ENST00000375068.1
|
MAGED2
|
melanoma antigen family D, 2 |
| chr19_-_18508396 | 2.40 |
ENST00000595840.1
ENST00000339007.3 |
LRRC25
|
leucine rich repeat containing 25 |
| chr12_-_42631529 | 2.40 |
ENST00000548917.1
|
YAF2
|
YY1 associated factor 2 |
| chr4_+_175839551 | 2.36 |
ENST00000404450.4
ENST00000514159.1 |
ADAM29
|
ADAM metallopeptidase domain 29 |
| chr1_+_104293028 | 2.35 |
ENST00000370079.3
|
AMY1C
|
amylase, alpha 1C (salivary) |
| chr1_-_67142710 | 2.27 |
ENST00000502413.2
|
AL139147.1
|
Uncharacterized protein |
| chr12_-_71148357 | 2.26 |
ENST00000378778.1
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr17_-_61996192 | 2.23 |
ENST00000392824.4
|
CSHL1
|
chorionic somatomammotropin hormone-like 1 |
| chr5_+_140474181 | 2.16 |
ENST00000194155.4
|
PCDHB2
|
protocadherin beta 2 |
| chr5_+_36152179 | 2.15 |
ENST00000508514.1
ENST00000513151.1 ENST00000546211.1 |
SKP2
|
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr14_-_80697396 | 2.11 |
ENST00000557010.1
|
DIO2
|
deiodinase, iodothyronine, type II |
| chr2_+_210444142 | 2.11 |
ENST00000360351.4
ENST00000361559.4 |
MAP2
|
microtubule-associated protein 2 |
| chr11_+_55029628 | 2.11 |
ENST00000417545.2
|
TRIM48
|
tripartite motif containing 48 |
| chr2_-_228244013 | 2.10 |
ENST00000304568.3
|
TM4SF20
|
transmembrane 4 L six family member 20 |
| chr5_-_160973649 | 2.09 |
ENST00000393959.1
ENST00000517547.1 |
GABRB2
|
gamma-aminobutyric acid (GABA) A receptor, beta 2 |
| chr4_-_70826725 | 2.01 |
ENST00000353151.3
|
CSN2
|
casein beta |
| chr1_-_152332480 | 2.00 |
ENST00000388718.5
|
FLG2
|
filaggrin family member 2 |
| chr7_-_140482926 | 2.00 |
ENST00000496384.2
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B |
| chr15_+_63569785 | 1.97 |
ENST00000380343.4
ENST00000560353.1 |
APH1B
|
APH1B gamma secretase subunit |
| chr11_+_28129795 | 1.92 |
ENST00000406787.3
ENST00000342303.5 ENST00000403099.1 ENST00000407364.3 |
METTL15
|
methyltransferase like 15 |
| chr6_+_131894284 | 1.91 |
ENST00000368087.3
ENST00000356962.2 |
ARG1
|
arginase 1 |
| chr22_-_41032668 | 1.87 |
ENST00000355630.3
ENST00000396617.3 ENST00000402042.1 |
MKL1
|
megakaryoblastic leukemia (translocation) 1 |
| chr11_-_62369291 | 1.83 |
ENST00000278823.2
|
MTA2
|
metastasis associated 1 family, member 2 |
| chr12_-_56753858 | 1.82 |
ENST00000314128.4
ENST00000557235.1 ENST00000418572.2 |
STAT2
|
signal transducer and activator of transcription 2, 113kDa |
| chr7_+_106505912 | 1.82 |
ENST00000359195.3
|
PIK3CG
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
| chr1_-_89664595 | 1.82 |
ENST00000355754.6
|
GBP4
|
guanylate binding protein 4 |
| chr14_+_22337014 | 1.78 |
ENST00000390436.2
|
TRAV13-1
|
T cell receptor alpha variable 13-1 |
| chr6_+_158733692 | 1.77 |
ENST00000367094.2
ENST00000367097.3 |
TULP4
|
tubby like protein 4 |
| chr3_-_52931557 | 1.77 |
ENST00000504329.1
ENST00000355083.5 |
TMEM110-MUSTN1
TMEM110
|
TMEM110-MUSTN1 readthrough transmembrane protein 110 |
| chr9_+_34458771 | 1.77 |
ENST00000437363.1
ENST00000242317.4 |
DNAI1
|
dynein, axonemal, intermediate chain 1 |
| chrX_-_32173579 | 1.75 |
ENST00000359836.1
ENST00000343523.2 ENST00000378707.3 ENST00000541735.1 ENST00000474231.1 |
DMD
|
dystrophin |
| chr10_+_114710425 | 1.75 |
ENST00000352065.5
ENST00000369395.1 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr8_-_17942432 | 1.74 |
ENST00000381733.4
ENST00000314146.10 |
ASAH1
|
N-acylsphingosine amidohydrolase (acid ceramidase) 1 |
| chr17_-_39093672 | 1.73 |
ENST00000209718.3
ENST00000436344.3 ENST00000485751.1 |
KRT23
|
keratin 23 (histone deacetylase inducible) |
| chr6_-_11779014 | 1.73 |
ENST00000229583.5
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr1_-_179112189 | 1.72 |
ENST00000512653.1
ENST00000344730.3 |
ABL2
|
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr12_-_10324716 | 1.69 |
ENST00000545927.1
ENST00000432556.2 ENST00000309539.3 ENST00000544577.1 |
OLR1
|
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chr12_-_71148413 | 1.69 |
ENST00000440835.2
ENST00000549308.1 ENST00000550661.1 |
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr17_-_61951090 | 1.68 |
ENST00000345366.7
ENST00000392886.2 ENST00000336844.5 ENST00000560142.1 |
CSH2
|
chorionic somatomammotropin hormone 2 |
| chr2_-_113993020 | 1.66 |
ENST00000465084.1
|
PAX8
|
paired box 8 |
| chr15_+_71228826 | 1.65 |
ENST00000558456.1
ENST00000560158.2 ENST00000558808.1 ENST00000559806.1 ENST00000559069.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr6_-_49712123 | 1.64 |
ENST00000263045.4
|
CRISP3
|
cysteine-rich secretory protein 3 |
| chr8_-_93029865 | 1.63 |
ENST00000422361.2
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr3_+_189507523 | 1.58 |
ENST00000437221.1
ENST00000392463.2 ENST00000392461.3 ENST00000449992.1 ENST00000456148.1 |
TP63
|
tumor protein p63 |
| chr17_-_3301704 | 1.53 |
ENST00000322608.2
|
OR1E1
|
olfactory receptor, family 1, subfamily E, member 1 |
| chr7_-_36406750 | 1.49 |
ENST00000453212.1
ENST00000415803.2 ENST00000440378.1 ENST00000431396.1 ENST00000317020.6 ENST00000436884.1 |
KIAA0895
|
KIAA0895 |
| chr1_+_160336851 | 1.48 |
ENST00000302101.5
|
NHLH1
|
nescient helix loop helix 1 |
| chr17_+_17206635 | 1.47 |
ENST00000389022.4
|
NT5M
|
5',3'-nucleotidase, mitochondrial |
| chr8_-_38008783 | 1.45 |
ENST00000276449.4
|
STAR
|
steroidogenic acute regulatory protein |
| chr8_-_41166953 | 1.43 |
ENST00000220772.3
|
SFRP1
|
secreted frizzled-related protein 1 |
| chr12_-_91505608 | 1.42 |
ENST00000266718.4
|
LUM
|
lumican |
| chr5_-_133702761 | 1.38 |
ENST00000521118.1
ENST00000265334.4 ENST00000435211.1 |
CDKL3
|
cyclin-dependent kinase-like 3 |
| chr5_-_111312622 | 1.37 |
ENST00000395634.3
|
NREP
|
neuronal regeneration related protein |
| chr1_+_104159999 | 1.37 |
ENST00000414303.2
ENST00000423678.1 |
AMY2A
|
amylase, alpha 2A (pancreatic) |
| chrX_+_23352133 | 1.36 |
ENST00000379361.4
|
PTCHD1
|
patched domain containing 1 |
| chr4_+_175839506 | 1.34 |
ENST00000505141.1
ENST00000359240.3 ENST00000445694.1 |
ADAM29
|
ADAM metallopeptidase domain 29 |
| chr12_-_102874416 | 1.29 |
ENST00000392904.1
ENST00000337514.6 |
IGF1
|
insulin-like growth factor 1 (somatomedin C) |
| chrX_+_96138907 | 1.28 |
ENST00000373040.3
|
RPA4
|
replication protein A4, 30kDa |
| chr19_-_8373173 | 1.27 |
ENST00000537716.2
ENST00000301458.5 |
CD320
|
CD320 molecule |
| chr22_-_39268308 | 1.25 |
ENST00000407418.3
|
CBX6
|
chromobox homolog 6 |
| chr5_+_175288631 | 1.24 |
ENST00000509837.1
|
CPLX2
|
complexin 2 |
| chr11_+_124824000 | 1.21 |
ENST00000529051.1
ENST00000344762.5 |
CCDC15
|
coiled-coil domain containing 15 |
| chr12_-_7848364 | 1.21 |
ENST00000329913.3
|
GDF3
|
growth differentiation factor 3 |
| chr1_-_100231349 | 1.21 |
ENST00000287474.5
ENST00000414213.1 |
FRRS1
|
ferric-chelate reductase 1 |
| chr3_+_121902511 | 1.20 |
ENST00000490131.1
|
CASR
|
calcium-sensing receptor |
| chr15_+_59279851 | 1.17 |
ENST00000348370.4
ENST00000434298.1 ENST00000559160.1 |
RNF111
|
ring finger protein 111 |
| chr2_+_201170596 | 1.16 |
ENST00000439084.1
ENST00000409718.1 |
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of POU3F3_POU3F4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.7 | 20.0 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 6.0 | 66.0 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 3.4 | 165.1 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 2.8 | 25.3 | GO:0003190 | atrioventricular valve formation(GO:0003190) |
| 2.6 | 18.5 | GO:0006537 | glutamate biosynthetic process(GO:0006537) gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 2.3 | 28.1 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 2.3 | 9.3 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 2.2 | 24.6 | GO:0006554 | lysine catabolic process(GO:0006554) |
| 2.2 | 6.7 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 2.2 | 15.5 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 2.1 | 6.3 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 2.0 | 4.1 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) |
| 2.0 | 16.0 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 1.9 | 21.4 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 1.8 | 5.4 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 1.6 | 4.8 | GO:0043400 | cortisol secretion(GO:0043400) positive regulation of corticotropin secretion(GO:0051461) regulation of cortisol secretion(GO:0051462) negative regulation of glucagon secretion(GO:0070093) positive regulation of corticosterone secretion(GO:2000854) |
| 1.4 | 4.3 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 1.4 | 7.2 | GO:0042078 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 1.4 | 10.0 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 1.4 | 17.1 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 1.4 | 4.2 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 1.3 | 11.8 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 1.3 | 5.2 | GO:0043126 | regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 1.3 | 12.5 | GO:0072221 | distal convoluted tubule development(GO:0072025) metanephric distal convoluted tubule development(GO:0072221) |
| 1.2 | 14.5 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 1.2 | 9.2 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 1.1 | 3.4 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 1.0 | 3.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 1.0 | 2.9 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.9 | 7.3 | GO:0010536 | positive regulation of activation of Janus kinase activity(GO:0010536) |
| 0.9 | 13.4 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.8 | 56.3 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.8 | 5.6 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.8 | 3.9 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.8 | 8.4 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.8 | 4.6 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.8 | 6.0 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.7 | 50.0 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.7 | 2.8 | GO:0034344 | microglial cell activation involved in immune response(GO:0002282) type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.7 | 2.0 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.6 | 1.2 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.6 | 1.8 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.6 | 12.9 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.5 | 1.0 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.5 | 5.8 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.5 | 1.4 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) |
| 0.5 | 76.3 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.5 | 1.4 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.5 | 2.8 | GO:0033133 | fructose 2,6-bisphosphate metabolic process(GO:0006003) positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.5 | 2.3 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.5 | 20.3 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.4 | 7.5 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.4 | 6.1 | GO:0060856 | establishment of blood-brain barrier(GO:0060856) |
| 0.4 | 3.9 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.4 | 7.2 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.4 | 5.1 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.4 | 4.2 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to photoreceptor outer segment(GO:1903546) |
| 0.4 | 13.4 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.4 | 2.0 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.4 | 9.7 | GO:0030539 | male genitalia development(GO:0030539) |
| 0.4 | 1.9 | GO:1902023 | L-arginine import(GO:0043091) arginine import(GO:0090467) L-arginine transport(GO:1902023) |
| 0.4 | 2.5 | GO:2000795 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.3 | 9.0 | GO:0045332 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.3 | 2.4 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.3 | 11.4 | GO:1901687 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.3 | 3.0 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.3 | 1.8 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) secretory granule localization(GO:0032252) |
| 0.3 | 12.8 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.3 | 10.2 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.3 | 1.9 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.3 | 4.3 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.2 | 1.9 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.2 | 3.3 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.2 | 1.8 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.2 | 0.4 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) |
| 0.2 | 5.7 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.2 | 1.4 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.2 | 2.6 | GO:0042074 | cell migration involved in gastrulation(GO:0042074) |
| 0.2 | 1.4 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.2 | 2.1 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.2 | 0.6 | GO:0061743 | motor learning(GO:0061743) |
| 0.2 | 2.8 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.2 | 0.5 | GO:2000768 | positive regulation of epithelial cell differentiation involved in kidney development(GO:2000698) positive regulation of nephron tubule epithelial cell differentiation(GO:2000768) |
| 0.2 | 3.3 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.2 | 0.7 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.2 | 0.9 | GO:0032096 | negative regulation of response to food(GO:0032096) negative regulation of appetite(GO:0032099) |
| 0.2 | 4.3 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 0.9 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.1 | 1.8 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 1.5 | GO:0046078 | dUMP metabolic process(GO:0046078) |
| 0.1 | 1.2 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 | 0.7 | GO:0051006 | positive regulation of lipoprotein lipase activity(GO:0051006) |
| 0.1 | 1.3 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.1 | 24.7 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.1 | 1.1 | GO:0048505 | positive regulation of activin receptor signaling pathway(GO:0032927) regulation of development, heterochronic(GO:0040034) regulation of timing of cell differentiation(GO:0048505) |
| 0.1 | 1.4 | GO:0021794 | thalamus development(GO:0021794) |
| 0.1 | 2.5 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.1 | 56.9 | GO:0001558 | regulation of cell growth(GO:0001558) |
| 0.1 | 1.2 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.1 | 2.2 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.1 | 0.5 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.1 | 1.1 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.1 | 2.7 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.1 | 2.1 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.1 | 8.5 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.1 | 2.6 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 0.8 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 0.6 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.1 | 1.8 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.1 | 1.7 | GO:0061436 | regulation of water loss via skin(GO:0033561) establishment of skin barrier(GO:0061436) |
| 0.1 | 23.3 | GO:0045666 | positive regulation of neuron differentiation(GO:0045666) |
| 0.1 | 4.1 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.1 | 0.8 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 1.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 2.9 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.1 | 2.1 | GO:0050770 | regulation of axonogenesis(GO:0050770) |
| 0.1 | 1.0 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.1 | 0.5 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.1 | 3.1 | GO:0010633 | negative regulation of epithelial cell migration(GO:0010633) |
| 0.1 | 0.9 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 0.2 | GO:0060423 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) positive regulation of branching involved in lung morphogenesis(GO:0061047) |
| 0.1 | 1.8 | GO:0010762 | regulation of fibroblast migration(GO:0010762) |
| 0.1 | 1.0 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.1 | 1.8 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.1 | 2.6 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 1.1 | GO:0032026 | response to magnesium ion(GO:0032026) |
| 0.0 | 0.3 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.0 | 3.6 | GO:0033108 | mitochondrial respiratory chain complex assembly(GO:0033108) |
| 0.0 | 0.7 | GO:0097242 | beta-amyloid clearance(GO:0097242) |
| 0.0 | 6.5 | GO:0016051 | carbohydrate biosynthetic process(GO:0016051) |
| 0.0 | 0.4 | GO:0071732 | cellular response to nitric oxide(GO:0071732) |
| 0.0 | 1.3 | GO:0033260 | DNA replication initiation(GO:0006270) nuclear DNA replication(GO:0033260) |
| 0.0 | 3.8 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 1.0 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 1.4 | GO:0021549 | cerebellum development(GO:0021549) |
| 0.0 | 0.4 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.1 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.2 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) negative regulation of centrosome cycle(GO:0046606) |
| 0.0 | 0.1 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.2 | GO:0034142 | toll-like receptor 4 signaling pathway(GO:0034142) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 32.3 | 161.3 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 2.6 | 23.4 | GO:0005955 | calcineurin complex(GO:0005955) |
| 2.4 | 7.3 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 2.0 | 28.1 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 1.2 | 25.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 1.1 | 4.6 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 1.1 | 7.8 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.9 | 9.2 | GO:0045180 | basal cortex(GO:0045180) |
| 0.8 | 12.9 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.7 | 67.5 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.7 | 12.8 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.7 | 13.6 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.6 | 7.5 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.6 | 2.5 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.6 | 1.8 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.6 | 4.8 | GO:0043196 | varicosity(GO:0043196) |
| 0.6 | 4.1 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.5 | 13.2 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.4 | 5.6 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.4 | 23.4 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.4 | 3.7 | GO:0097449 | astrocyte projection(GO:0097449) growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.4 | 12.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.4 | 2.8 | GO:0072487 | MSL complex(GO:0072487) |
| 0.4 | 2.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.3 | 52.3 | GO:0043204 | perikaryon(GO:0043204) |
| 0.3 | 41.4 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.2 | 3.7 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.2 | 20.6 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.2 | 1.8 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.2 | 2.5 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.2 | 0.8 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.2 | 1.8 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.2 | 2.8 | GO:0030478 | actin cap(GO:0030478) |
| 0.2 | 10.3 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 2.8 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.1 | 5.4 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 1.1 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.1 | 3.4 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 2.6 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.1 | 0.5 | GO:0071439 | CORVET complex(GO:0033263) clathrin complex(GO:0071439) |
| 0.1 | 43.0 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 1.0 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 7.7 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.1 | 2.1 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 29.8 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.1 | 13.9 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.1 | 1.8 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 0.6 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 0.7 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.1 | 1.7 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.6 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.5 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 2.0 | GO:0044297 | cell body(GO:0044297) |
| 0.0 | 8.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 4.2 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 3.9 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 3.7 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.0 | 0.9 | GO:0012505 | endomembrane system(GO:0012505) |
| 0.0 | 1.6 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.7 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 1.6 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 2.8 | GO:0061695 | transferase complex, transferring phosphorus-containing groups(GO:0061695) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.1 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 1.0 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 6.1 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 6.7 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 1.1 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 9.2 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 2.3 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 2.7 | GO:0043005 | neuron projection(GO:0043005) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 13.4 | 161.3 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 10.4 | 62.1 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 6.7 | 40.4 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 6.2 | 24.6 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 3.4 | 13.4 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 2.9 | 20.0 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 2.7 | 21.4 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 2.7 | 16.0 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 2.6 | 18.5 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 2.6 | 66.0 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 2.4 | 7.3 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 2.3 | 23.4 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 2.3 | 9.1 | GO:0016160 | amylase activity(GO:0016160) |
| 2.0 | 26.3 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 1.9 | 5.7 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) |
| 1.8 | 10.9 | GO:0005499 | vitamin D binding(GO:0005499) |
| 1.4 | 4.3 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 1.1 | 7.5 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 1.0 | 3.0 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.9 | 15.5 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.8 | 3.9 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.8 | 2.3 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.8 | 10.0 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.6 | 1.9 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.6 | 1.3 | GO:0015235 | cobalamin transporter activity(GO:0015235) |
| 0.6 | 2.4 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.6 | 8.2 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.6 | 10.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.6 | 2.8 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.6 | 5.6 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.5 | 13.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.5 | 12.9 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.5 | 4.3 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.5 | 3.3 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.4 | 4.8 | GO:0051430 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.4 | 2.6 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.4 | 5.2 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.4 | 1.7 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.4 | 6.0 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.4 | 3.3 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.4 | 28.1 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.4 | 1.2 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.4 | 9.0 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.4 | 3.7 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.4 | 2.6 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.4 | 5.4 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.4 | 2.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.3 | 4.1 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) sphingolipid binding(GO:0046625) |
| 0.3 | 15.2 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.3 | 39.0 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.3 | 4.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.3 | 3.9 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.3 | 2.1 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.3 | 12.8 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.2 | 1.7 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.2 | 2.8 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.2 | 11.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.2 | 14.4 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.2 | 1.1 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.2 | 2.5 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.2 | 1.2 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.2 | 6.1 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.2 | 3.9 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.2 | 0.9 | GO:0008430 | selenium binding(GO:0008430) |
| 0.1 | 0.7 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 6.4 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 0.7 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.1 | 1.9 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 0.5 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.1 | 1.5 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 12.9 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.1 | 30.1 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.1 | 1.9 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 0.8 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.1 | 2.7 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 1.8 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 1.4 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.1 | 0.6 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 1.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 2.5 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 52.1 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.1 | 1.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.9 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 2.6 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 1.0 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 1.1 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 1.8 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 3.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.2 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 1.6 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 5.9 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 1.4 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 1.4 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.4 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 1.2 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 1.2 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.6 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 1.1 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.2 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.9 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.4 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 1.3 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 1.3 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.5 | GO:0042805 | actinin binding(GO:0042805) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 161.3 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.9 | 49.5 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.5 | 25.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.4 | 17.7 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.2 | 16.5 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.2 | 24.4 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.2 | 12.9 | PID ATR PATHWAY | ATR signaling pathway |
| 0.2 | 17.7 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.2 | 4.1 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 1.8 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 2.0 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 1.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 4.0 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 3.4 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 5.1 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 5.2 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.1 | 1.8 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 2.8 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.1 | 1.4 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 1.1 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.5 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 1.1 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 1.9 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 2.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 2.1 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 3.0 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 1.9 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 1.2 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 3.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.7 | 161.3 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 1.6 | 17.3 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 1.0 | 28.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.8 | 8.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.8 | 16.0 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.7 | 8.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.6 | 21.4 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.5 | 26.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.5 | 2.0 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.5 | 20.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.5 | 12.8 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.4 | 3.9 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.4 | 7.3 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.4 | 20.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.3 | 11.4 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.3 | 18.5 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.3 | 6.1 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.3 | 7.5 | REACTOME DAG AND IP3 SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.2 | 2.8 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.2 | 2.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.2 | 3.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.2 | 8.7 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.2 | 3.7 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.2 | 51.4 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.2 | 4.3 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.2 | 5.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 4.3 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.2 | 2.0 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.1 | 5.0 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 7.2 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 15.5 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 5.7 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 7.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 1.8 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 11.8 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.1 | 2.5 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 1.5 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 6.0 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 1.1 | REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | Genes involved in SCF-beta-TrCP mediated degradation of Emi1 |
| 0.1 | 1.1 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.1 | 4.1 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 4.4 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 2.1 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 1.4 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 1.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 1.8 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.1 | 1.7 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 1.0 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 1.8 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.1 | 5.2 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 5.1 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.5 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 1.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 1.8 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 2.8 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.0 | 5.6 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 1.3 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |