Project
averaged GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
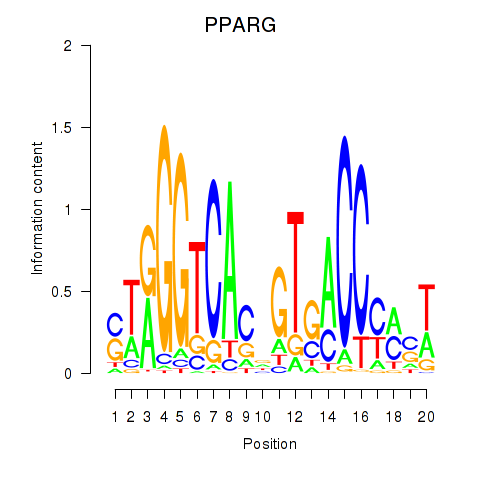
Results for PPARG
Z-value: 1.57
Transcription factors associated with PPARG
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PPARG
|
ENSG00000132170.15 | peroxisome proliferator activated receptor gamma |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PPARG | hg19_v2_chr3_+_12392971_12392983, hg19_v2_chr3_+_12329397_12329433, hg19_v2_chr3_+_12329358_12329393, hg19_v2_chr3_+_12330560_12330579 | -0.01 | 9.1e-01 | Click! |
Activity profile of PPARG motif
Sorted Z-values of PPARG motif
Network of associatons between targets according to the STRING database.
First level regulatory network of PPARG
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.2 | 36.5 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 4.0 | 16.1 | GO:1903781 | positive regulation of cardiac conduction(GO:1903781) positive regulation of atrial cardiac muscle cell action potential(GO:1903949) positive regulation of regulation of vascular smooth muscle cell membrane depolarization(GO:1904199) regulation of vascular smooth muscle cell membrane depolarization(GO:1990736) |
| 3.6 | 14.5 | GO:0070434 | positive regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070426) positive regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070434) |
| 3.5 | 49.5 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 3.5 | 17.5 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 2.9 | 8.6 | GO:0052250 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 2.8 | 14.1 | GO:0070295 | renal water absorption(GO:0070295) |
| 2.5 | 7.4 | GO:0032804 | negative regulation of low-density lipoprotein particle receptor catabolic process(GO:0032804) |
| 2.5 | 7.4 | GO:0043095 | regulation of GTP cyclohydrolase I activity(GO:0043095) negative regulation of GTP cyclohydrolase I activity(GO:0043105) |
| 2.4 | 9.4 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 2.0 | 6.1 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 1.9 | 7.5 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 1.8 | 7.4 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 1.7 | 6.9 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 1.7 | 5.0 | GO:0051295 | establishment of meiotic spindle localization(GO:0051295) |
| 1.7 | 24.9 | GO:0015671 | oxygen transport(GO:0015671) |
| 1.6 | 8.2 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 1.6 | 4.9 | GO:0002818 | intracellular defense response(GO:0002818) |
| 1.6 | 4.8 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
| 1.6 | 12.6 | GO:0046618 | drug export(GO:0046618) |
| 1.5 | 4.6 | GO:0055073 | cadmium ion homeostasis(GO:0055073) |
| 1.5 | 9.1 | GO:0070221 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 1.5 | 11.9 | GO:0007506 | gonadal mesoderm development(GO:0007506) |
| 1.4 | 8.6 | GO:0031437 | regulation of mRNA cleavage(GO:0031437) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) |
| 1.4 | 4.2 | GO:0001300 | chronological cell aging(GO:0001300) |
| 1.4 | 5.6 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 1.3 | 6.7 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 1.2 | 3.6 | GO:1905224 | clathrin-coated pit assembly(GO:1905224) |
| 1.2 | 4.7 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) |
| 1.2 | 4.6 | GO:0002774 | Fc receptor mediated inhibitory signaling pathway(GO:0002774) |
| 1.2 | 7.0 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 1.2 | 5.8 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 1.1 | 3.3 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 1.1 | 5.5 | GO:0002159 | desmosome assembly(GO:0002159) endothelial cell-cell adhesion(GO:0071603) |
| 1.1 | 6.3 | GO:0051708 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 1.0 | 4.1 | GO:0003095 | pressure natriuresis(GO:0003095) |
| 1.0 | 4.1 | GO:0002353 | regulation of thyroid hormone mediated signaling pathway(GO:0002155) kinin cascade(GO:0002254) plasma kallikrein-kinin cascade(GO:0002353) |
| 1.0 | 3.1 | GO:0019556 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 1.0 | 2.0 | GO:0097212 | lysosomal membrane organization(GO:0097212) |
| 1.0 | 71.7 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 1.0 | 5.0 | GO:0097267 | omega-hydroxylase P450 pathway(GO:0097267) |
| 1.0 | 10.7 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 1.0 | 4.8 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.9 | 3.8 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.9 | 3.7 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 0.9 | 14.3 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.9 | 13.7 | GO:0017085 | response to insecticide(GO:0017085) |
| 0.8 | 5.9 | GO:0097396 | response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 0.8 | 5.0 | GO:0070945 | neutrophil mediated killing of gram-negative bacterium(GO:0070945) |
| 0.8 | 2.5 | GO:2000395 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) ubiquitin-dependent endocytosis(GO:0070086) regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.8 | 14.1 | GO:2001204 | regulation of osteoclast development(GO:2001204) |
| 0.8 | 6.4 | GO:0070235 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.8 | 4.7 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.8 | 6.2 | GO:0035897 | proteolysis in other organism(GO:0035897) |
| 0.8 | 2.3 | GO:2000360 | negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.7 | 8.2 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.7 | 2.2 | GO:2000410 | tolerance induction to self antigen(GO:0002513) regulation of thymocyte migration(GO:2000410) |
| 0.7 | 2.2 | GO:0090675 | intermicrovillar adhesion(GO:0090675) |
| 0.7 | 8.9 | GO:0035826 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.7 | 6.7 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.6 | 10.9 | GO:0031953 | negative regulation of protein autophosphorylation(GO:0031953) |
| 0.6 | 3.8 | GO:0031335 | regulation of sulfur amino acid metabolic process(GO:0031335) regulation of homocysteine metabolic process(GO:0050666) |
| 0.6 | 6.9 | GO:0051712 | positive regulation of killing of cells of other organism(GO:0051712) |
| 0.6 | 4.3 | GO:0045591 | positive regulation of regulatory T cell differentiation(GO:0045591) |
| 0.6 | 6.7 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.5 | 2.1 | GO:0002329 | pre-B cell differentiation(GO:0002329) pre-B cell allelic exclusion(GO:0002331) |
| 0.5 | 5.0 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.5 | 2.0 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.5 | 2.9 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.5 | 1.5 | GO:0006522 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) |
| 0.5 | 13.4 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.5 | 1.4 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.4 | 1.3 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.4 | 2.6 | GO:1901098 | regulation of autophagosome maturation(GO:1901096) positive regulation of autophagosome maturation(GO:1901098) |
| 0.4 | 9.9 | GO:0046697 | decidualization(GO:0046697) |
| 0.4 | 6.0 | GO:1901750 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.4 | 6.4 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.4 | 7.1 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 0.4 | 8.3 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.4 | 2.4 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.4 | 10.8 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.4 | 6.0 | GO:0001893 | maternal placenta development(GO:0001893) |
| 0.4 | 1.1 | GO:1902463 | protein localization to cell leading edge(GO:1902463) |
| 0.4 | 5.6 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.4 | 1.4 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.4 | 3.9 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) |
| 0.3 | 1.4 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.3 | 1.0 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.3 | 1.3 | GO:0060023 | soft palate development(GO:0060023) |
| 0.3 | 1.0 | GO:0098759 | response to interleukin-8(GO:0098758) cellular response to interleukin-8(GO:0098759) |
| 0.3 | 1.0 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 0.3 | 1.0 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.3 | 0.9 | GO:0050894 | determination of affect(GO:0050894) |
| 0.3 | 7.8 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.3 | 0.9 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.3 | 1.8 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 0.3 | 1.2 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.3 | 0.9 | GO:2000824 | negative regulation of androgen receptor activity(GO:2000824) |
| 0.3 | 1.5 | GO:0044026 | DNA hypermethylation(GO:0044026) |
| 0.3 | 5.1 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.3 | 3.3 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.3 | 1.9 | GO:0060312 | regulation of blood vessel remodeling(GO:0060312) |
| 0.3 | 3.5 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.3 | 1.0 | GO:0021784 | postganglionic parasympathetic fiber development(GO:0021784) |
| 0.3 | 1.3 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.3 | 2.1 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.3 | 0.8 | GO:0097676 | histone H3-K36 dimethylation(GO:0097676) |
| 0.3 | 2.0 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.2 | 6.1 | GO:0002691 | regulation of cellular extravasation(GO:0002691) |
| 0.2 | 3.4 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.2 | 2.7 | GO:0070234 | positive regulation of T cell apoptotic process(GO:0070234) |
| 0.2 | 5.7 | GO:0097503 | sialylation(GO:0097503) |
| 0.2 | 1.7 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.2 | 6.5 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.2 | 1.4 | GO:1904783 | positive regulation of NMDA glutamate receptor activity(GO:1904783) |
| 0.2 | 1.2 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.2 | 4.2 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.2 | 0.9 | GO:1901091 | regulation of protein tetramerization(GO:1901090) negative regulation of protein tetramerization(GO:1901091) regulation of protein homotetramerization(GO:1901093) negative regulation of protein homotetramerization(GO:1901094) |
| 0.2 | 1.3 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.2 | 5.6 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.2 | 0.5 | GO:0010615 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.2 | 2.5 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.2 | 1.9 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.2 | 1.7 | GO:0048875 | chemical homeostasis within a tissue(GO:0048875) |
| 0.2 | 27.0 | GO:0002223 | stimulatory C-type lectin receptor signaling pathway(GO:0002223) |
| 0.2 | 3.7 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.2 | 1.4 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.2 | 1.1 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) regulation of microtubule binding(GO:1904526) |
| 0.1 | 1.3 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 0.4 | GO:0070781 | response to biotin(GO:0070781) |
| 0.1 | 4.5 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.1 | 21.1 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 0.8 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.1 | 1.8 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.1 | 10.5 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.1 | 1.2 | GO:0090344 | negative regulation of cell aging(GO:0090344) |
| 0.1 | 2.4 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.1 | 0.6 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.1 | 4.0 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.1 | 1.7 | GO:0043068 | positive regulation of apoptotic process(GO:0043065) positive regulation of programmed cell death(GO:0043068) |
| 0.1 | 1.8 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 | 5.1 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.1 | 4.4 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 1.9 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 | 0.4 | GO:0010820 | positive regulation of T cell chemotaxis(GO:0010820) |
| 0.1 | 0.5 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.1 | 1.5 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.1 | 9.0 | GO:0045995 | regulation of embryonic development(GO:0045995) |
| 0.1 | 0.3 | GO:0098838 | methotrexate transport(GO:0051958) reduced folate transmembrane transport(GO:0098838) |
| 0.1 | 2.4 | GO:0019835 | cytolysis(GO:0019835) |
| 0.1 | 1.2 | GO:0051683 | establishment of Golgi localization(GO:0051683) |
| 0.1 | 0.3 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.1 | 1.3 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.1 | 0.8 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.1 | 2.4 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.1 | 2.7 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.1 | 4.0 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.1 | 0.4 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.1 | 8.6 | GO:0048839 | inner ear development(GO:0048839) |
| 0.1 | 0.9 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.1 | 0.7 | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.1 | 1.7 | GO:0048255 | mRNA stabilization(GO:0048255) |
| 0.0 | 2.5 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 6.9 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.0 | 0.5 | GO:0007320 | insemination(GO:0007320) |
| 0.0 | 2.2 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.0 | 1.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.4 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.2 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.2 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 1.7 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.5 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 1.2 | GO:0042733 | embryonic digit morphogenesis(GO:0042733) |
| 0.0 | 2.7 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 1.2 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 1.1 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 1.3 | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.0 | 0.4 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.0 | 0.6 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 0.5 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 1.8 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 3.1 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 4.6 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.2 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 1.4 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 3.1 | GO:0007517 | muscle organ development(GO:0007517) |
| 0.0 | 0.0 | GO:0032759 | TRAIL production(GO:0032639) regulation of TRAIL production(GO:0032679) positive regulation of TRAIL production(GO:0032759) |
| 0.0 | 0.6 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.0 | 0.6 | GO:0031670 | cellular response to nutrient(GO:0031670) |
| 0.0 | 1.3 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 1.4 | GO:2001252 | positive regulation of chromosome organization(GO:2001252) |
| 0.0 | 1.6 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.0 | 1.0 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 0.2 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.9 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 2.1 | GO:0090305 | nucleic acid phosphodiester bond hydrolysis(GO:0090305) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.5 | GO:0036028 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 1.8 | 24.9 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 1.7 | 5.0 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 1.5 | 7.4 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 1.4 | 13.0 | GO:0044194 | cytolytic granule(GO:0044194) |
| 1.4 | 4.3 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 1.4 | 9.9 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 1.4 | 4.2 | GO:0072563 | endothelial microparticle(GO:0072563) |
| 1.4 | 69.8 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 1.4 | 5.6 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 1.2 | 5.8 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 1.1 | 6.7 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.9 | 19.7 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.7 | 8.9 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.7 | 5.5 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.6 | 16.1 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.5 | 3.9 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.5 | 3.9 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.5 | 1.4 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.4 | 2.2 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.4 | 13.7 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.4 | 2.7 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.4 | 6.7 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.4 | 2.5 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.4 | 5.0 | GO:0001673 | nuclear nucleosome(GO:0000788) male germ cell nucleus(GO:0001673) |
| 0.3 | 4.5 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.3 | 1.7 | GO:0071546 | pi-body(GO:0071546) |
| 0.3 | 2.2 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.3 | 3.8 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.3 | 1.2 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.3 | 5.1 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.2 | 1.7 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.2 | 1.2 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.2 | 1.9 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.2 | 6.4 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.2 | 6.3 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.2 | 1.0 | GO:0032449 | CBM complex(GO:0032449) |
| 0.2 | 18.0 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.2 | 6.9 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.2 | 32.5 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.2 | 15.2 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.2 | 8.2 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.2 | 3.2 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.2 | 0.6 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.2 | 0.9 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.1 | 0.9 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.1 | 1.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 1.3 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 2.1 | GO:0034518 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.1 | 3.7 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.1 | 1.3 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 6.0 | GO:0031672 | A band(GO:0031672) |
| 0.1 | 3.3 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 4.1 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 6.0 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 10.9 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 3.8 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 4.7 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 1.1 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.1 | 2.3 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.1 | 0.6 | GO:0098645 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.1 | 1.4 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.1 | 0.5 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 22.3 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 4.4 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 2.1 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 0.9 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 21.2 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.1 | 1.7 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 0.4 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.1 | 1.5 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 2.0 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 1.0 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 4.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.1 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 5.1 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.9 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 1.7 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 2.0 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 8.1 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 54.2 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 4.5 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 4.8 | GO:0098802 | plasma membrane receptor complex(GO:0098802) |
| 0.0 | 3.0 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 2.7 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.8 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.7 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.9 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.3 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.2 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.0 | 2.6 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 88.3 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 0.7 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 2.3 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.0 | 1.3 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 4.3 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 0.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 1.3 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 3.7 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 3.5 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.1 | 36.5 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 3.5 | 14.1 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 2.7 | 8.2 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 2.5 | 12.6 | GO:0015307 | drug:proton antiporter activity(GO:0015307) |
| 2.5 | 7.4 | GO:0044549 | GTP cyclohydrolase binding(GO:0044549) |
| 2.0 | 6.1 | GO:0050135 | NAD+ nucleosidase activity(GO:0003953) NAD(P)+ nucleosidase activity(GO:0050135) |
| 1.9 | 24.9 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 1.8 | 7.4 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 1.6 | 8.2 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 1.6 | 4.8 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 1.6 | 4.7 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 1.5 | 4.6 | GO:0051139 | metal ion:proton antiporter activity(GO:0051139) |
| 1.4 | 8.6 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 1.4 | 4.2 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) |
| 1.4 | 5.6 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 1.3 | 14.5 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 1.3 | 3.8 | GO:0004119 | cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) |
| 1.3 | 69.8 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 1.2 | 7.4 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 1.1 | 9.1 | GO:0016713 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 1.1 | 6.7 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 1.1 | 3.2 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 1.0 | 17.5 | GO:0003680 | AT DNA binding(GO:0003680) |
| 1.0 | 8.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 1.0 | 4.0 | GO:0008431 | vitamin E binding(GO:0008431) |
| 1.0 | 6.9 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.9 | 3.7 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.8 | 5.9 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.8 | 7.5 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.8 | 5.8 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.8 | 4.9 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.8 | 3.1 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.7 | 6.4 | GO:0043426 | MRF binding(GO:0043426) |
| 0.7 | 2.0 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.6 | 5.0 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.6 | 2.5 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.6 | 1.9 | GO:0047150 | betaine-homocysteine S-methyltransferase activity(GO:0047150) |
| 0.6 | 5.5 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.6 | 2.4 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.6 | 4.6 | GO:0032396 | inhibitory MHC class I receptor activity(GO:0032396) |
| 0.6 | 2.3 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.5 | 4.4 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.5 | 2.2 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.5 | 4.2 | GO:0005534 | galactose binding(GO:0005534) |
| 0.5 | 2.0 | GO:0071253 | connexin binding(GO:0071253) |
| 0.5 | 12.6 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.5 | 1.4 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.4 | 7.8 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.4 | 6.0 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.4 | 2.5 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.4 | 16.1 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.4 | 2.0 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.4 | 1.2 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.4 | 2.6 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.4 | 5.6 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.4 | 8.1 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.4 | 9.9 | GO:0001848 | complement binding(GO:0001848) |
| 0.4 | 11.4 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.3 | 1.4 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.3 | 1.0 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.3 | 6.0 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.3 | 8.1 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.3 | 0.9 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.3 | 3.3 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.3 | 5.1 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.3 | 2.0 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.3 | 1.4 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.3 | 6.2 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.3 | 12.3 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.3 | 6.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.3 | 5.0 | GO:0043495 | protein anchor(GO:0043495) |
| 0.3 | 4.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.2 | 3.7 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.2 | 1.3 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.2 | 1.2 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.2 | 2.3 | GO:0004568 | chitinase activity(GO:0004568) chitin binding(GO:0008061) |
| 0.2 | 1.2 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.2 | 1.8 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.2 | 0.9 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.2 | 1.1 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.2 | 3.8 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.2 | 1.9 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.2 | 29.6 | GO:0005178 | integrin binding(GO:0005178) |
| 0.2 | 2.8 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.2 | 0.6 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 0.6 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 1.0 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.1 | 2.6 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.1 | 1.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 3.7 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.1 | 22.8 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 1.4 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 1.4 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 1.5 | GO:0019841 | retinol binding(GO:0019841) |
| 0.1 | 0.9 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.1 | 9.9 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 9.5 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 3.4 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.1 | 11.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 5.4 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 1.9 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.1 | 9.5 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 10.5 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 1.7 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 2.1 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.1 | 0.3 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.1 | 1.7 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 2.1 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.1 | 0.9 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 2.7 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.1 | 0.8 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 2.1 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.1 | 2.7 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 0.9 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.1 | 3.6 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 19.8 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 2.1 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.7 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.1 | 14.1 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.1 | 7.2 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 10.9 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.1 | 0.3 | GO:0015350 | reduced folate carrier activity(GO:0008518) methotrexate transporter activity(GO:0015350) |
| 0.1 | 2.2 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 11.9 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 1.2 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 13.2 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.1 | 1.5 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 8.9 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 3.4 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 8.3 | GO:0001076 | transcription factor activity, RNA polymerase II transcription factor binding(GO:0001076) |
| 0.0 | 1.1 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.0 | 0.4 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.8 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 1.2 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 4.2 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 0.6 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.5 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.5 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.0 | 1.3 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 5.8 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 2.2 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 2.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.8 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 3.4 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.6 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 2.4 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.3 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 1.2 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.5 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 1.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.9 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.4 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.9 | GO:0060089 | receptor activity(GO:0004872) molecular transducer activity(GO:0060089) |
| 0.0 | 0.3 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.4 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 2.1 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 1.0 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 27.8 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.5 | 6.6 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.3 | 6.7 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.3 | 8.1 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.2 | 10.9 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.2 | 5.5 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.2 | 6.2 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.2 | 5.6 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.2 | 3.7 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.2 | 10.2 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.2 | 11.1 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.2 | 4.8 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.2 | 9.1 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.2 | 8.2 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.2 | 2.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 1.9 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.1 | 7.7 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 2.9 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.1 | 3.6 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.1 | 2.0 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.1 | 2.9 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 26.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 4.2 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 2.0 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 27.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 4.9 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 6.9 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.1 | 2.3 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 11.8 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.1 | 7.5 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 6.4 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 1.9 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.1 | 4.4 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.1 | 12.1 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 1.0 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 2.0 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 2.2 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 5.0 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 4.0 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 1.0 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 2.0 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 7.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.5 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 1.2 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.3 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.9 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.6 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.5 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 1.1 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 16.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 1.0 | 14.1 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.7 | 9.9 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.6 | 58.2 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.6 | 12.6 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.5 | 14.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.4 | 5.8 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.4 | 7.8 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.3 | 6.5 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.3 | 7.4 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.3 | 10.7 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.3 | 5.9 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.3 | 5.6 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.2 | 6.7 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.2 | 8.2 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.2 | 5.1 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.2 | 2.0 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.2 | 3.2 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.2 | 5.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.2 | 1.4 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.2 | 4.2 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.2 | 11.5 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.2 | 8.0 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.2 | 15.8 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.2 | 3.7 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 1.9 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 17.8 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 3.3 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 9.3 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 7.3 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 4.6 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 2.7 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 3.1 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 4.1 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 3.8 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 5.5 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 1.8 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 2.7 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 2.0 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.1 | 1.0 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.1 | 8.9 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 1.1 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 3.0 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 1.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 3.6 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 1.7 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 1.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.9 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.8 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.5 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.5 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 6.1 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 1.7 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 1.4 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.5 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 2.0 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 2.1 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 1.9 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 2.3 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.3 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 1.1 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.9 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.2 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 1.5 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |