Project
averaged GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
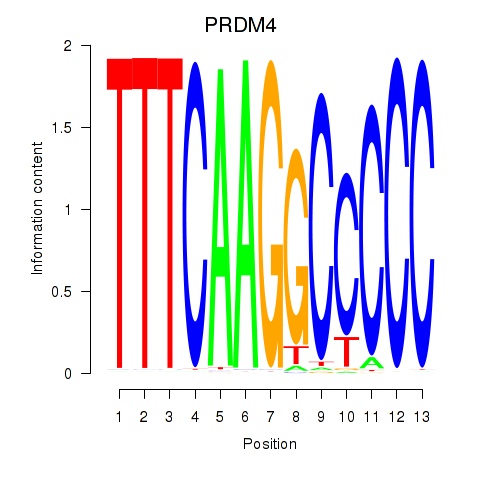
Results for PRDM4
Z-value: 0.65
Transcription factors associated with PRDM4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PRDM4
|
ENSG00000110851.7 | PR/SET domain 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PRDM4 | hg19_v2_chr12_-_108154925_108154945 | -0.01 | 8.9e-01 | Click! |
Activity profile of PRDM4 motif
Sorted Z-values of PRDM4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_105953246 | 11.28 |
ENST00000392531.3
|
CRIP1
|
cysteine-rich protein 1 (intestinal) |
| chr14_+_105953204 | 11.17 |
ENST00000409393.2
|
CRIP1
|
cysteine-rich protein 1 (intestinal) |
| chr12_-_58240470 | 11.01 |
ENST00000548823.1
ENST00000398073.2 |
CTDSP2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr19_+_42724423 | 9.90 |
ENST00000301215.3
ENST00000597945.1 |
ZNF526
|
zinc finger protein 526 |
| chr2_+_219264466 | 5.31 |
ENST00000273062.2
|
CTDSP1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1 |
| chr7_+_143079000 | 4.95 |
ENST00000392910.2
|
ZYX
|
zyxin |
| chr22_-_39636914 | 4.81 |
ENST00000381551.4
|
PDGFB
|
platelet-derived growth factor beta polypeptide |
| chr19_+_41117770 | 4.65 |
ENST00000601032.1
|
LTBP4
|
latent transforming growth factor beta binding protein 4 |
| chr6_-_24721054 | 4.27 |
ENST00000378119.4
|
C6orf62
|
chromosome 6 open reading frame 62 |
| chr2_+_234296792 | 3.87 |
ENST00000409813.3
|
DGKD
|
diacylglycerol kinase, delta 130kDa |
| chr7_+_143078652 | 3.55 |
ENST00000354434.4
ENST00000449423.2 |
ZYX
|
zyxin |
| chr19_+_50180317 | 3.37 |
ENST00000534465.1
|
PRMT1
|
protein arginine methyltransferase 1 |
| chr12_+_104682496 | 3.30 |
ENST00000378070.4
|
TXNRD1
|
thioredoxin reductase 1 |
| chr3_+_148709128 | 3.23 |
ENST00000345003.4
ENST00000296048.6 ENST00000483267.1 |
GYG1
|
glycogenin 1 |
| chr3_+_148709310 | 3.12 |
ENST00000484197.1
ENST00000492285.2 ENST00000461191.1 |
GYG1
|
glycogenin 1 |
| chr12_+_69979446 | 3.12 |
ENST00000543146.2
|
CCT2
|
chaperonin containing TCP1, subunit 2 (beta) |
| chr15_-_66797172 | 2.88 |
ENST00000569438.1
ENST00000569696.1 ENST00000307961.6 |
RPL4
|
ribosomal protein L4 |
| chr9_-_70488865 | 2.79 |
ENST00000377392.5
|
CBWD5
|
COBW domain containing 5 |
| chr10_-_105677886 | 2.78 |
ENST00000224950.3
|
OBFC1
|
oligonucleotide/oligosaccharide-binding fold containing 1 |
| chr10_-_105212141 | 2.58 |
ENST00000369788.3
|
CALHM2
|
calcium homeostasis modulator 2 |
| chr7_-_23571586 | 2.37 |
ENST00000538367.1
ENST00000392502.4 ENST00000297071.4 |
TRA2A
|
transformer 2 alpha homolog (Drosophila) |
| chr10_+_88728189 | 2.35 |
ENST00000416348.1
|
ADIRF
|
adipogenesis regulatory factor |
| chr1_+_55107449 | 2.28 |
ENST00000421030.2
ENST00000545244.1 ENST00000339553.5 ENST00000409996.1 ENST00000454855.2 |
MROH7
|
maestro heat-like repeat family member 7 |
| chr20_-_61569296 | 2.21 |
ENST00000370371.4
|
DIDO1
|
death inducer-obliterator 1 |
| chr3_+_16926441 | 2.21 |
ENST00000418129.2
ENST00000396755.2 |
PLCL2
|
phospholipase C-like 2 |
| chr20_+_61569463 | 2.20 |
ENST00000266069.3
|
GID8
|
GID complex subunit 8 |
| chr11_+_58910295 | 2.05 |
ENST00000420244.1
|
FAM111A
|
family with sequence similarity 111, member A |
| chr17_+_42634844 | 1.83 |
ENST00000315323.3
|
FZD2
|
frizzled family receptor 2 |
| chr17_+_7788104 | 1.78 |
ENST00000380358.4
|
CHD3
|
chromodomain helicase DNA binding protein 3 |
| chr6_-_32160622 | 1.77 |
ENST00000487761.1
ENST00000375040.3 |
GPSM3
|
G-protein signaling modulator 3 |
| chr11_+_47279504 | 1.74 |
ENST00000441012.2
ENST00000437276.1 ENST00000436029.1 ENST00000467728.1 ENST00000405853.3 |
NR1H3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr4_+_667307 | 1.59 |
ENST00000506838.1
|
MYL5
|
myosin, light chain 5, regulatory |
| chr9_+_96338647 | 1.51 |
ENST00000359246.4
|
PHF2
|
PHD finger protein 2 |
| chr4_-_83295103 | 1.48 |
ENST00000313899.7
ENST00000352301.4 ENST00000509107.1 ENST00000353341.4 ENST00000541060.1 |
HNRNPD
|
heterogeneous nuclear ribonucleoprotein D (AU-rich element RNA binding protein 1, 37kDa) |
| chr15_+_45315302 | 1.38 |
ENST00000267814.9
|
SORD
|
sorbitol dehydrogenase |
| chr5_-_180688105 | 1.32 |
ENST00000327767.4
|
TRIM52
|
tripartite motif containing 52 |
| chr7_+_90893783 | 1.30 |
ENST00000287934.2
|
FZD1
|
frizzled family receptor 1 |
| chr1_+_172502336 | 1.24 |
ENST00000263688.3
|
SUCO
|
SUN domain containing ossification factor |
| chr10_+_123923105 | 1.22 |
ENST00000368999.1
|
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr16_+_15068955 | 1.13 |
ENST00000396410.4
ENST00000569715.1 ENST00000450288.2 |
PDXDC1
|
pyridoxal-dependent decarboxylase domain containing 1 |
| chr15_+_66797627 | 0.99 |
ENST00000565627.1
ENST00000564179.1 |
ZWILCH
|
zwilch kinetochore protein |
| chr1_+_43232913 | 0.82 |
ENST00000372525.5
ENST00000536543.1 |
C1orf50
|
chromosome 1 open reading frame 50 |
| chr5_-_34916871 | 0.75 |
ENST00000382038.2
|
RAD1
|
RAD1 homolog (S. pombe) |
| chr20_-_61569227 | 0.66 |
ENST00000266070.4
ENST00000395335.2 ENST00000266071.5 |
DIDO1
|
death inducer-obliterator 1 |
| chr11_+_19138670 | 0.65 |
ENST00000446113.2
ENST00000399351.3 |
ZDHHC13
|
zinc finger, DHHC-type containing 13 |
| chr1_-_43232649 | 0.64 |
ENST00000372526.2
ENST00000236040.4 ENST00000296388.5 ENST00000397054.3 |
LEPRE1
|
leucine proline-enriched proteoglycan (leprecan) 1 |
| chr4_+_667686 | 0.60 |
ENST00000505477.1
|
MYL5
|
myosin, light chain 5, regulatory |
| chr2_-_8723918 | 0.60 |
ENST00000454224.1
|
AC011747.4
|
AC011747.4 |
| chr13_+_52436111 | 0.60 |
ENST00000242819.4
|
CCDC70
|
coiled-coil domain containing 70 |
| chrX_-_73072534 | 0.52 |
ENST00000429829.1
|
XIST
|
X inactive specific transcript (non-protein coding) |
| chr14_+_32030582 | 0.52 |
ENST00000550649.1
ENST00000281081.7 |
NUBPL
|
nucleotide binding protein-like |
| chr1_+_27114418 | 0.49 |
ENST00000078527.4
|
PIGV
|
phosphatidylinositol glycan anchor biosynthesis, class V |
| chr19_-_11639910 | 0.41 |
ENST00000588998.1
ENST00000586149.1 |
ECSIT
|
ECSIT signalling integrator |
| chr5_-_102455801 | 0.35 |
ENST00000508629.1
ENST00000399004.2 |
GIN1
|
gypsy retrotransposon integrase 1 |
| chr7_-_38398721 | 0.29 |
ENST00000390346.2
|
TRGV3
|
T cell receptor gamma variable 3 |
| chr5_+_159343688 | 0.25 |
ENST00000306675.3
|
ADRA1B
|
adrenoceptor alpha 1B |
| chr12_+_15475331 | 0.23 |
ENST00000281171.4
|
PTPRO
|
protein tyrosine phosphatase, receptor type, O |
| chr12_+_15475462 | 0.19 |
ENST00000543886.1
ENST00000348962.2 |
PTPRO
|
protein tyrosine phosphatase, receptor type, O |
| chrX_+_15808569 | 0.15 |
ENST00000380308.3
ENST00000307771.7 |
ZRSR2
|
zinc finger (CCCH type), RNA-binding motif and serine/arginine rich 2 |
| chr1_+_27114589 | 0.12 |
ENST00000431541.1
ENST00000449950.2 ENST00000374145.1 |
PIGV
|
phosphatidylinositol glycan anchor biosynthesis, class V |
| chr14_+_24025194 | 0.09 |
ENST00000404535.3
ENST00000288014.6 |
THTPA
|
thiamine triphosphatase |
Network of associatons between targets according to the STRING database.
First level regulatory network of PRDM4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.5 | 22.5 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 1.2 | 4.8 | GO:1900241 | metanephric glomerular mesangial cell development(GO:0072255) reversible differentiation(GO:0090677) cell dedifferentiation involved in phenotypic switching(GO:0090678) positive regulation of phenotypic switching(GO:1900241) regulation of vascular smooth muscle cell dedifferentiation(GO:1905174) positive regulation of vascular smooth muscle cell dedifferentiation(GO:1905176) vascular smooth muscle cell dedifferentiation(GO:1990936) |
| 1.0 | 3.1 | GO:0051086 | chaperone mediated protein folding independent of cofactor(GO:0051086) |
| 0.7 | 2.2 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 0.6 | 1.7 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.6 | 3.4 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.5 | 1.5 | GO:0061188 | negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.5 | 1.5 | GO:1901355 | response to rapamycin(GO:1901355) |
| 0.5 | 1.4 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.4 | 1.3 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.4 | 2.3 | GO:0072719 | cellular response to cisplatin(GO:0072719) |
| 0.4 | 3.3 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.3 | 1.8 | GO:0003150 | muscular septum morphogenesis(GO:0003150) |
| 0.2 | 0.8 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.2 | 3.9 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.2 | 4.6 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.2 | 6.4 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.1 | 0.4 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.1 | 1.8 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.1 | 15.1 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.1 | 0.3 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.1 | 8.5 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 1.2 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.0 | 0.7 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.0 | 0.6 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.4 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 2.9 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 2.1 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.1 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.0 | 1.8 | GO:0032508 | DNA duplex unwinding(GO:0032508) |
| 0.0 | 0.5 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 1.0 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.2 | 3.1 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.2 | 0.8 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.2 | 3.4 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 0.6 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.1 | 1.3 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.1 | 8.5 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 2.2 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 1.8 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 1.2 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 4.8 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 2.9 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 5.4 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 1.8 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 3.1 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.7 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.0 | 1.5 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 16.3 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 1.6 | 6.4 | GO:0008466 | glycogenin glucosyltransferase activity(GO:0008466) |
| 1.4 | 23.9 | GO:0003680 | AT DNA binding(GO:0003680) |
| 1.1 | 3.3 | GO:0098626 | methylselenol reductase activity(GO:0098625) methylseleninic acid reductase activity(GO:0098626) |
| 0.6 | 3.4 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.5 | 4.8 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.4 | 1.7 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.2 | 3.9 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) diacylglycerol binding(GO:0019992) |
| 0.2 | 4.6 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.2 | 0.6 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.1 | 3.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.8 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.1 | 2.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 3.1 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 0.3 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 1.5 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 1.8 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.7 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.4 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 1.8 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.1 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 2.2 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 1.1 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 1.4 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 2.9 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.5 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 16.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.2 | 8.5 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.2 | 4.8 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.1 | 3.1 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.8 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 4.6 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.8 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 2.2 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.4 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 8.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 3.1 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.1 | 3.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 3.9 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 2.9 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 4.8 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 3.1 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 2.9 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 1.5 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.8 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.0 | 1.0 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |