Project
averaged GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
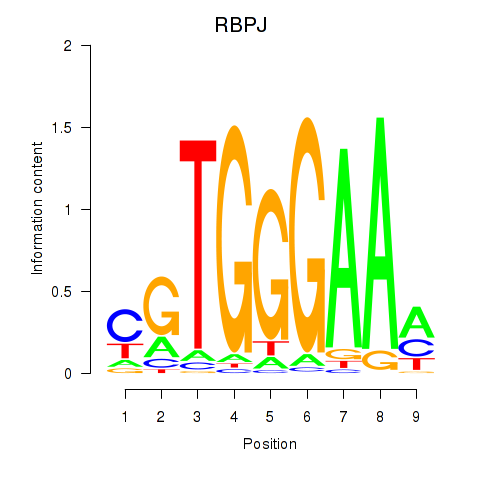
Results for RBPJ
Z-value: 1.10
Transcription factors associated with RBPJ
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RBPJ
|
ENSG00000168214.16 | recombination signal binding protein for immunoglobulin kappa J region |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RBPJ | hg19_v2_chr4_+_26322185_26322334 | -0.32 | 1.3e-06 | Click! |
Activity profile of RBPJ motif
Sorted Z-values of RBPJ motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_80680078 | 18.81 |
ENST00000337919.5
ENST00000354724.3 |
HEY1
|
hes-related family bHLH transcription factor with YRPW motif 1 |
| chr17_-_56606664 | 16.34 |
ENST00000580844.1
|
SEPT4
|
septin 4 |
| chr17_-_56606705 | 16.10 |
ENST00000317268.3
|
SEPT4
|
septin 4 |
| chr17_-_56606639 | 15.48 |
ENST00000579371.1
|
SEPT4
|
septin 4 |
| chr3_+_32726774 | 15.15 |
ENST00000538368.1
|
CNOT10
|
CCR4-NOT transcription complex, subunit 10 |
| chr16_-_70719925 | 14.83 |
ENST00000338779.6
|
MTSS1L
|
metastasis suppressor 1-like |
| chr4_-_176733897 | 13.01 |
ENST00000393658.2
|
GPM6A
|
glycoprotein M6A |
| chr19_+_50706866 | 12.89 |
ENST00000440075.2
ENST00000376970.2 ENST00000425460.1 ENST00000599920.1 ENST00000601313.1 |
MYH14
|
myosin, heavy chain 14, non-muscle |
| chr12_-_16759711 | 12.09 |
ENST00000447609.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr4_-_73935409 | 11.76 |
ENST00000507544.2
ENST00000295890.4 |
COX18
|
COX18 cytochrome C oxidase assembly factor |
| chr8_-_22089845 | 11.64 |
ENST00000454243.2
|
PHYHIP
|
phytanoyl-CoA 2-hydroxylase interacting protein |
| chrX_-_153236819 | 11.49 |
ENST00000354233.3
|
HCFC1
|
host cell factor C1 (VP16-accessory protein) |
| chr6_+_126070726 | 11.28 |
ENST00000368364.3
|
HEY2
|
hes-related family bHLH transcription factor with YRPW motif 2 |
| chr8_-_22089533 | 11.25 |
ENST00000321613.3
|
PHYHIP
|
phytanoyl-CoA 2-hydroxylase interacting protein |
| chr19_-_39390440 | 11.14 |
ENST00000249396.7
ENST00000414941.1 ENST00000392081.2 |
SIRT2
|
sirtuin 2 |
| chr17_-_34257731 | 10.64 |
ENST00000431884.2
ENST00000425909.3 ENST00000394528.3 ENST00000430160.2 |
RDM1
|
RAD52 motif 1 |
| chr5_-_139943830 | 10.63 |
ENST00000412920.3
ENST00000511201.2 ENST00000356738.2 ENST00000354402.5 ENST00000358580.5 ENST00000508496.2 |
APBB3
|
amyloid beta (A4) precursor protein-binding, family B, member 3 |
| chr12_+_4918342 | 10.59 |
ENST00000280684.3
ENST00000433855.1 |
KCNA6
|
potassium voltage-gated channel, shaker-related subfamily, member 6 |
| chr19_-_39390350 | 9.93 |
ENST00000447739.1
ENST00000358931.5 ENST00000407552.1 |
SIRT2
|
sirtuin 2 |
| chr19_-_55690758 | 9.77 |
ENST00000590851.1
|
SYT5
|
synaptotagmin V |
| chr19_-_46296011 | 9.14 |
ENST00000377735.3
ENST00000270223.6 |
DMWD
|
dystrophia myotonica, WD repeat containing |
| chr12_+_12938541 | 9.10 |
ENST00000356591.4
|
APOLD1
|
apolipoprotein L domain containing 1 |
| chr16_-_30102547 | 8.82 |
ENST00000279386.2
|
TBX6
|
T-box 6 |
| chr1_+_36690011 | 8.74 |
ENST00000354618.5
ENST00000469141.2 ENST00000478853.1 |
THRAP3
|
thyroid hormone receptor associated protein 3 |
| chr10_-_38146510 | 8.44 |
ENST00000395867.3
|
ZNF248
|
zinc finger protein 248 |
| chr19_+_56652686 | 8.21 |
ENST00000592949.1
|
ZNF444
|
zinc finger protein 444 |
| chr1_+_15272271 | 8.20 |
ENST00000400797.3
|
KAZN
|
kazrin, periplakin interacting protein |
| chr9_-_35665165 | 8.09 |
ENST00000343259.3
ENST00000378387.3 |
ARHGEF39
|
Rho guanine nucleotide exchange factor (GEF) 39 |
| chr1_-_223536679 | 7.68 |
ENST00000608996.1
|
SUSD4
|
sushi domain containing 4 |
| chr14_+_29234870 | 7.47 |
ENST00000382535.3
|
FOXG1
|
forkhead box G1 |
| chr19_-_36870087 | 7.34 |
ENST00000270001.7
|
ZFP14
|
ZFP14 zinc finger protein |
| chr14_-_53258314 | 7.12 |
ENST00000216410.3
ENST00000557604.1 |
GNPNAT1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chr22_-_38245304 | 7.07 |
ENST00000609454.1
|
ANKRD54
|
ankyrin repeat domain 54 |
| chr3_+_142442841 | 7.01 |
ENST00000476941.1
ENST00000273482.6 |
TRPC1
|
transient receptor potential cation channel, subfamily C, member 1 |
| chr2_+_219824357 | 6.96 |
ENST00000302625.4
|
CDK5R2
|
cyclin-dependent kinase 5, regulatory subunit 2 (p39) |
| chr11_+_112832202 | 6.82 |
ENST00000534015.1
|
NCAM1
|
neural cell adhesion molecule 1 |
| chrX_-_40594755 | 6.74 |
ENST00000324817.1
|
MED14
|
mediator complex subunit 14 |
| chr1_-_146644036 | 6.73 |
ENST00000425272.2
|
PRKAB2
|
protein kinase, AMP-activated, beta 2 non-catalytic subunit |
| chr5_-_137514617 | 6.69 |
ENST00000254900.5
|
BRD8
|
bromodomain containing 8 |
| chr18_-_52989217 | 6.64 |
ENST00000570287.2
|
TCF4
|
transcription factor 4 |
| chrX_-_153236620 | 6.63 |
ENST00000369984.4
|
HCFC1
|
host cell factor C1 (VP16-accessory protein) |
| chr14_-_53258180 | 6.56 |
ENST00000554230.1
|
GNPNAT1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chr5_-_115872142 | 6.51 |
ENST00000510263.1
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr10_-_51371321 | 6.46 |
ENST00000602930.1
|
AGAP8
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 8 |
| chr17_+_2699697 | 6.24 |
ENST00000254695.8
ENST00000366401.4 ENST00000542807.1 |
RAP1GAP2
|
RAP1 GTPase activating protein 2 |
| chr1_-_223537401 | 6.21 |
ENST00000343846.3
ENST00000454695.2 ENST00000484758.2 |
SUSD4
|
sushi domain containing 4 |
| chr19_+_56652556 | 6.08 |
ENST00000337080.3
|
ZNF444
|
zinc finger protein 444 |
| chr10_+_112836779 | 5.91 |
ENST00000280155.2
|
ADRA2A
|
adrenoceptor alpha 2A |
| chrX_+_122993827 | 5.75 |
ENST00000371199.3
|
XIAP
|
X-linked inhibitor of apoptosis |
| chr5_+_178368186 | 5.69 |
ENST00000320129.3
ENST00000519564.1 |
ZNF454
|
zinc finger protein 454 |
| chr11_+_112832090 | 5.66 |
ENST00000533760.1
|
NCAM1
|
neural cell adhesion molecule 1 |
| chr17_-_71410794 | 5.65 |
ENST00000424778.1
|
SDK2
|
sidekick cell adhesion molecule 2 |
| chr12_+_175930 | 5.65 |
ENST00000538872.1
ENST00000326261.4 |
IQSEC3
|
IQ motif and Sec7 domain 3 |
| chr19_+_42746927 | 5.46 |
ENST00000378108.1
|
AC006486.1
|
AC006486.1 |
| chr13_-_25496926 | 5.34 |
ENST00000545981.1
ENST00000381884.4 |
CENPJ
|
centromere protein J |
| chr11_-_790060 | 5.31 |
ENST00000330106.4
|
CEND1
|
cell cycle exit and neuronal differentiation 1 |
| chrX_+_16141667 | 5.13 |
ENST00000380289.2
|
GRPR
|
gastrin-releasing peptide receptor |
| chr14_-_93214915 | 5.10 |
ENST00000553918.1
ENST00000555699.1 ENST00000553802.1 ENST00000554397.1 ENST00000554919.1 ENST00000554080.1 ENST00000553371.1 |
LGMN
|
legumain |
| chr12_-_49259643 | 5.07 |
ENST00000309739.5
|
RND1
|
Rho family GTPase 1 |
| chr11_-_32452357 | 5.02 |
ENST00000379079.2
ENST00000530998.1 |
WT1
|
Wilms tumor 1 |
| chr10_+_74927875 | 4.76 |
ENST00000242505.6
|
FAM149B1
|
family with sequence similarity 149, member B1 |
| chr17_+_29421987 | 4.75 |
ENST00000431387.4
|
NF1
|
neurofibromin 1 |
| chr3_+_171561127 | 4.69 |
ENST00000334567.5
ENST00000450693.1 |
TMEM212
|
transmembrane protein 212 |
| chr11_-_64512273 | 4.56 |
ENST00000377497.3
ENST00000377487.1 ENST00000430645.1 |
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr14_-_93214988 | 4.50 |
ENST00000557434.1
ENST00000393218.2 ENST00000334869.4 |
LGMN
|
legumain |
| chr1_+_50571949 | 4.34 |
ENST00000357083.4
|
ELAVL4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr20_+_43211149 | 4.34 |
ENST00000372886.1
|
PKIG
|
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr9_+_102584128 | 4.34 |
ENST00000338488.4
ENST00000395097.2 |
NR4A3
|
nuclear receptor subfamily 4, group A, member 3 |
| chr5_+_140792614 | 4.26 |
ENST00000398610.2
|
PCDHGA10
|
protocadherin gamma subfamily A, 10 |
| chr5_+_178322893 | 4.13 |
ENST00000361362.2
ENST00000520660.1 ENST00000520805.1 |
ZFP2
|
ZFP2 zinc finger protein |
| chr2_+_3642545 | 4.07 |
ENST00000382062.2
ENST00000236693.7 ENST00000349077.4 |
COLEC11
|
collectin sub-family member 11 |
| chr16_-_10276611 | 4.04 |
ENST00000396573.2
|
GRIN2A
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2A |
| chr3_+_46449049 | 4.02 |
ENST00000357392.4
ENST00000400880.3 ENST00000433848.1 |
CCRL2
|
chemokine (C-C motif) receptor-like 2 |
| chr1_-_146644122 | 4.00 |
ENST00000254101.3
|
PRKAB2
|
protein kinase, AMP-activated, beta 2 non-catalytic subunit |
| chr1_+_43855560 | 3.90 |
ENST00000562955.1
|
SZT2
|
seizure threshold 2 homolog (mouse) |
| chr10_+_118305435 | 3.80 |
ENST00000369221.2
|
PNLIP
|
pancreatic lipase |
| chr10_+_124320156 | 3.73 |
ENST00000338354.3
ENST00000344338.3 ENST00000330163.4 ENST00000368909.3 ENST00000368955.3 ENST00000368956.2 |
DMBT1
|
deleted in malignant brain tumors 1 |
| chr9_-_130712995 | 3.70 |
ENST00000373084.4
|
FAM102A
|
family with sequence similarity 102, member A |
| chr2_-_129076151 | 3.57 |
ENST00000259241.6
|
HS6ST1
|
heparan sulfate 6-O-sulfotransferase 1 |
| chr1_-_150980828 | 3.49 |
ENST00000361936.5
ENST00000361738.6 |
FAM63A
|
family with sequence similarity 63, member A |
| chr3_-_183735731 | 3.48 |
ENST00000334444.6
|
ABCC5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr7_+_76090993 | 3.47 |
ENST00000425780.1
ENST00000456590.1 ENST00000451769.1 ENST00000324432.5 ENST00000307569.8 ENST00000457529.1 ENST00000446600.1 ENST00000413936.2 ENST00000423646.1 ENST00000438930.1 ENST00000430490.2 |
DTX2
|
deltex homolog 2 (Drosophila) |
| chr3_+_155860751 | 3.44 |
ENST00000471742.1
|
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr11_+_118478313 | 3.34 |
ENST00000356063.5
|
PHLDB1
|
pleckstrin homology-like domain, family B, member 1 |
| chr1_-_112531777 | 3.26 |
ENST00000315987.2
ENST00000302127.4 |
KCND3
|
potassium voltage-gated channel, Shal-related subfamily, member 3 |
| chr17_-_39780819 | 3.23 |
ENST00000311208.8
|
KRT17
|
keratin 17 |
| chr3_+_159570722 | 3.23 |
ENST00000482804.1
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr10_+_124320195 | 3.12 |
ENST00000359586.6
|
DMBT1
|
deleted in malignant brain tumors 1 |
| chr16_+_23313591 | 3.09 |
ENST00000343070.2
|
SCNN1B
|
sodium channel, non-voltage-gated 1, beta subunit |
| chr17_+_73452545 | 3.05 |
ENST00000314256.7
|
KIAA0195
|
KIAA0195 |
| chr12_-_4758159 | 3.04 |
ENST00000545990.2
|
AKAP3
|
A kinase (PRKA) anchor protein 3 |
| chr14_+_22337014 | 2.99 |
ENST00000390436.2
|
TRAV13-1
|
T cell receptor alpha variable 13-1 |
| chr2_+_48796120 | 2.98 |
ENST00000394754.1
|
STON1-GTF2A1L
|
STON1-GTF2A1L readthrough |
| chr11_+_64052692 | 2.95 |
ENST00000377702.4
|
GPR137
|
G protein-coupled receptor 137 |
| chr16_-_53737795 | 2.93 |
ENST00000262135.4
ENST00000564374.1 ENST00000566096.1 |
RPGRIP1L
|
RPGRIP1-like |
| chr14_-_53417732 | 2.93 |
ENST00000399304.3
ENST00000395631.2 ENST00000341590.3 ENST00000343279.4 |
FERMT2
|
fermitin family member 2 |
| chr9_-_98269481 | 2.90 |
ENST00000418258.1
ENST00000553011.1 ENST00000551845.1 |
PTCH1
|
patched 1 |
| chr11_+_66512089 | 2.83 |
ENST00000524551.1
ENST00000525908.1 ENST00000360962.4 ENST00000346672.4 ENST00000527634.1 ENST00000540737.1 |
C11orf80
|
chromosome 11 open reading frame 80 |
| chr16_-_53737722 | 2.73 |
ENST00000569716.1
ENST00000562588.1 ENST00000562230.1 ENST00000379925.3 ENST00000563746.1 ENST00000568653.3 |
RPGRIP1L
|
RPGRIP1-like |
| chr21_+_38792602 | 2.61 |
ENST00000398960.2
ENST00000398956.2 |
DYRK1A
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A |
| chr13_-_74708372 | 2.61 |
ENST00000377666.4
|
KLF12
|
Kruppel-like factor 12 |
| chrX_-_49121165 | 2.57 |
ENST00000376207.4
ENST00000376199.2 |
FOXP3
|
forkhead box P3 |
| chr7_-_120498357 | 2.55 |
ENST00000415871.1
ENST00000222747.3 ENST00000430985.1 |
TSPAN12
|
tetraspanin 12 |
| chr19_+_50528971 | 2.51 |
ENST00000598809.1
ENST00000595661.1 ENST00000391821.2 |
ZNF473
|
zinc finger protein 473 |
| chr17_-_16118835 | 2.48 |
ENST00000582357.1
ENST00000436828.1 ENST00000411510.1 ENST00000268712.3 |
NCOR1
|
nuclear receptor corepressor 1 |
| chr6_-_76203345 | 2.44 |
ENST00000393004.2
|
FILIP1
|
filamin A interacting protein 1 |
| chr11_+_66512303 | 2.39 |
ENST00000532565.2
|
C11orf80
|
chromosome 11 open reading frame 80 |
| chr11_-_64512469 | 2.35 |
ENST00000377485.1
|
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr21_-_31538971 | 2.33 |
ENST00000286808.3
|
CLDN17
|
claudin 17 |
| chr10_-_97200772 | 2.16 |
ENST00000371241.1
ENST00000354106.3 ENST00000371239.1 ENST00000361941.3 ENST00000277982.5 ENST00000371245.3 |
SORBS1
|
sorbin and SH3 domain containing 1 |
| chr7_+_90338712 | 2.08 |
ENST00000265741.3
ENST00000406263.1 |
CDK14
|
cyclin-dependent kinase 14 |
| chr19_+_39390587 | 2.07 |
ENST00000572515.1
ENST00000392079.3 ENST00000575359.1 ENST00000313582.5 |
NFKBIB
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr22_+_40297105 | 2.04 |
ENST00000540310.1
|
GRAP2
|
GRB2-related adaptor protein 2 |
| chr20_+_30063067 | 2.02 |
ENST00000201979.2
|
REM1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr3_+_193853927 | 2.02 |
ENST00000232424.3
|
HES1
|
hes family bHLH transcription factor 1 |
| chr9_-_14722715 | 1.94 |
ENST00000380911.3
|
CER1
|
cerberus 1, DAN family BMP antagonist |
| chr19_+_50529212 | 1.84 |
ENST00000270617.3
ENST00000445728.3 ENST00000601364.1 |
ZNF473
|
zinc finger protein 473 |
| chr1_-_8000872 | 1.80 |
ENST00000377507.3
|
TNFRSF9
|
tumor necrosis factor receptor superfamily, member 9 |
| chr12_-_7848364 | 1.72 |
ENST00000329913.3
|
GDF3
|
growth differentiation factor 3 |
| chr2_+_204193101 | 1.63 |
ENST00000430418.1
ENST00000424558.1 ENST00000261016.6 |
ABI2
|
abl-interactor 2 |
| chr19_+_39390320 | 1.60 |
ENST00000576510.1
|
NFKBIB
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr7_+_90339169 | 1.58 |
ENST00000436577.2
|
CDK14
|
cyclin-dependent kinase 14 |
| chr11_-_64512803 | 1.55 |
ENST00000377489.1
ENST00000354024.3 |
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr18_+_10526008 | 1.51 |
ENST00000542979.1
ENST00000322897.6 |
NAPG
|
N-ethylmaleimide-sensitive factor attachment protein, gamma |
| chr1_-_40105617 | 1.50 |
ENST00000372852.3
|
HEYL
|
hes-related family bHLH transcription factor with YRPW motif-like |
| chr17_+_29421900 | 1.48 |
ENST00000358273.4
ENST00000356175.3 |
NF1
|
neurofibromin 1 |
| chr17_+_38474489 | 1.46 |
ENST00000394089.2
ENST00000425707.3 |
RARA
|
retinoic acid receptor, alpha |
| chr6_-_94129244 | 1.45 |
ENST00000369303.4
ENST00000369297.1 |
EPHA7
|
EPH receptor A7 |
| chr8_+_107738240 | 1.43 |
ENST00000449762.2
ENST00000297447.6 |
OXR1
|
oxidation resistance 1 |
| chr10_-_33623310 | 1.39 |
ENST00000395995.1
ENST00000374823.5 ENST00000374821.5 ENST00000374816.3 |
NRP1
|
neuropilin 1 |
| chr7_-_94953878 | 1.39 |
ENST00000222381.3
|
PON1
|
paraoxonase 1 |
| chr9_-_35691017 | 1.34 |
ENST00000378292.3
|
TPM2
|
tropomyosin 2 (beta) |
| chr1_+_147374915 | 1.32 |
ENST00000240986.4
|
GJA8
|
gap junction protein, alpha 8, 50kDa |
| chr7_-_27169801 | 1.28 |
ENST00000511914.1
|
HOXA4
|
homeobox A4 |
| chr2_-_79315112 | 1.21 |
ENST00000305089.3
|
REG1B
|
regenerating islet-derived 1 beta |
| chr22_+_40297079 | 1.16 |
ENST00000344138.4
ENST00000543252.1 |
GRAP2
|
GRB2-related adaptor protein 2 |
| chr11_+_64053005 | 1.12 |
ENST00000538032.1
|
GPR137
|
G protein-coupled receptor 137 |
| chr3_-_87325728 | 1.00 |
ENST00000350375.2
|
POU1F1
|
POU class 1 homeobox 1 |
| chr1_-_85358850 | 0.98 |
ENST00000370611.3
|
LPAR3
|
lysophosphatidic acid receptor 3 |
| chr7_+_100187196 | 0.87 |
ENST00000468962.1
ENST00000427939.2 |
FBXO24
|
F-box protein 24 |
| chr11_-_8959758 | 0.85 |
ENST00000531618.1
|
ASCL3
|
achaete-scute family bHLH transcription factor 3 |
| chr5_+_132009675 | 0.74 |
ENST00000231449.2
ENST00000350025.2 |
IL4
|
interleukin 4 |
| chr11_+_64052944 | 0.71 |
ENST00000535675.1
ENST00000543383.1 |
GPR137
|
G protein-coupled receptor 137 |
| chr12_+_6309963 | 0.68 |
ENST00000382515.2
|
CD9
|
CD9 molecule |
| chr11_-_83393457 | 0.63 |
ENST00000404783.3
|
DLG2
|
discs, large homolog 2 (Drosophila) |
| chr4_+_26321284 | 0.58 |
ENST00000506956.1
ENST00000512671.1 ENST00000345843.3 ENST00000342295.1 |
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr11_-_83393429 | 0.54 |
ENST00000426717.2
|
DLG2
|
discs, large homolog 2 (Drosophila) |
| chr1_-_43855479 | 0.51 |
ENST00000290663.6
ENST00000372457.4 |
MED8
|
mediator complex subunit 8 |
| chr9_+_706842 | 0.49 |
ENST00000382293.3
|
KANK1
|
KN motif and ankyrin repeat domains 1 |
| chr20_+_44637526 | 0.49 |
ENST00000372330.3
|
MMP9
|
matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) |
| chr19_+_44100544 | 0.44 |
ENST00000391965.2
ENST00000525771.1 |
ZNF576
|
zinc finger protein 576 |
| chr19_-_46088068 | 0.35 |
ENST00000263275.4
ENST00000323060.3 |
OPA3
|
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia) |
| chr15_-_70390191 | 0.33 |
ENST00000559191.1
|
TLE3
|
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) |
| chr15_+_40697988 | 0.18 |
ENST00000487418.2
ENST00000479013.2 |
IVD
|
isovaleryl-CoA dehydrogenase |
| chr10_-_44144292 | 0.17 |
ENST00000374433.2
|
ZNF32
|
zinc finger protein 32 |
| chrX_+_19362011 | 0.16 |
ENST00000379806.5
ENST00000545074.1 ENST00000540249.1 ENST00000423505.1 ENST00000417819.1 ENST00000422285.2 ENST00000355808.5 ENST00000379805.3 |
PDHA1
|
pyruvate dehydrogenase (lipoamide) alpha 1 |
| chr22_-_31885727 | 0.09 |
ENST00000330125.5
ENST00000344710.5 ENST00000397518.1 |
EIF4ENIF1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr13_-_38443860 | 0.06 |
ENST00000426868.2
ENST00000379681.3 ENST00000338947.5 ENST00000355779.2 ENST00000358477.2 ENST00000379673.2 |
TRPC4
|
transient receptor potential cation channel, subfamily C, member 4 |
| chrX_+_152907913 | 0.03 |
ENST00000370167.4
|
DUSP9
|
dual specificity phosphatase 9 |
Network of associatons between targets according to the STRING database.
First level regulatory network of RBPJ
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.6 | 47.9 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 7.5 | 30.1 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 7.0 | 21.1 | GO:2000777 | positive regulation of oocyte maturation(GO:1900195) negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 4.5 | 18.1 | GO:0019046 | release from viral latency(GO:0019046) |
| 3.2 | 9.6 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 2.7 | 13.7 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 2.3 | 7.0 | GO:0021722 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 2.1 | 6.2 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 2.0 | 12.1 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 1.9 | 5.8 | GO:1902528 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 1.5 | 5.9 | GO:0035625 | negative regulation of epinephrine secretion(GO:0032811) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 1.5 | 8.8 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 1.4 | 4.3 | GO:0032765 | positive regulation of mast cell cytokine production(GO:0032765) |
| 1.3 | 5.1 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 1.3 | 5.0 | GO:0072299 | posterior mesonephric tubule development(GO:0072166) negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) |
| 1.1 | 5.3 | GO:0061511 | centriole elongation(GO:0061511) |
| 1.1 | 5.3 | GO:0021699 | cerebellum maturation(GO:0021590) cerebellar cortex maturation(GO:0021699) |
| 1.0 | 2.9 | GO:0021997 | neural plate axis specification(GO:0021997) |
| 0.9 | 4.4 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.9 | 1.7 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.9 | 2.6 | GO:0032829 | regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032829) positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032831) negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.8 | 9.1 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.7 | 6.9 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.7 | 2.0 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.7 | 2.0 | GO:0021558 | midbrain-hindbrain boundary morphogenesis(GO:0021555) trochlear nerve development(GO:0021558) regulation of timing of neuron differentiation(GO:0060164) |
| 0.6 | 11.8 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.6 | 3.9 | GO:0021540 | corpus callosum morphogenesis(GO:0021540) |
| 0.5 | 6.5 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.5 | 10.7 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.5 | 1.5 | GO:2000824 | negative regulation of androgen receptor activity(GO:2000824) |
| 0.5 | 1.9 | GO:1900145 | regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900175) |
| 0.5 | 5.1 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.4 | 4.0 | GO:0042428 | serotonin metabolic process(GO:0042428) |
| 0.4 | 5.7 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.4 | 13.2 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.4 | 2.5 | GO:0072367 | regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) |
| 0.4 | 8.7 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.4 | 1.5 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.3 | 1.4 | GO:0034444 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 0.3 | 15.3 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.3 | 5.7 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.3 | 4.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.3 | 3.3 | GO:1904261 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.3 | 1.8 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.3 | 6.1 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.3 | 3.4 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.3 | 5.7 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.3 | 10.1 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.3 | 4.3 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.3 | 15.1 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.3 | 3.8 | GO:0061365 | positive regulation of triglyceride lipase activity(GO:0061365) |
| 0.2 | 1.0 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 0.2 | 3.5 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.2 | 0.7 | GO:2000320 | negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.2 | 9.8 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.2 | 4.2 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.2 | 1.6 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.2 | 3.3 | GO:0097623 | potassium ion export across plasma membrane(GO:0097623) |
| 0.2 | 3.2 | GO:0051798 | positive regulation of hair follicle development(GO:0051798) |
| 0.2 | 7.1 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.1 | 1.5 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.1 | 2.6 | GO:0090312 | positive regulation of protein deacetylation(GO:0090312) |
| 0.1 | 2.6 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 3.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 2.9 | GO:0033622 | integrin activation(GO:0033622) |
| 0.1 | 1.4 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.1 | 13.9 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.1 | 1.0 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 3.0 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.1 | 2.2 | GO:0045725 | positive regulation of glycogen biosynthetic process(GO:0045725) |
| 0.1 | 2.3 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 0.5 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
| 0.1 | 10.1 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.1 | 5.8 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.1 | 7.6 | GO:0070268 | cornification(GO:0070268) |
| 0.1 | 0.7 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.1 | 3.1 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.1 | 8.1 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 10.6 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 1.2 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 4.7 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 4.0 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 3.2 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.0 | 6.2 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.0 | 4.3 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.3 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 1.5 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 3.7 | GO:0071222 | cellular response to lipopolysaccharide(GO:0071222) |
| 0.0 | 0.2 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.2 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 1.3 | GO:0033275 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 47.9 | GO:0097227 | sperm annulus(GO:0097227) |
| 2.6 | 12.9 | GO:0097513 | myosin II filament(GO:0097513) |
| 2.3 | 21.1 | GO:0072687 | meiotic spindle(GO:0072687) |
| 1.9 | 5.7 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 1.7 | 7.0 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 1.6 | 9.8 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 1.4 | 18.1 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 1.4 | 9.6 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 1.3 | 6.7 | GO:0070847 | core mediator complex(GO:0070847) |
| 1.2 | 3.7 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 1.1 | 5.3 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.9 | 15.1 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.7 | 15.9 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.6 | 3.9 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.6 | 13.8 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.5 | 10.7 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.5 | 3.4 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.5 | 5.7 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.4 | 2.2 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.4 | 11.8 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.4 | 6.9 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.3 | 6.2 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.3 | 3.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.3 | 8.7 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.3 | 3.0 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.3 | 8.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.2 | 15.0 | GO:0015030 | Cajal body(GO:0015030) |
| 0.2 | 3.4 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 14.8 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 15.0 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.1 | 1.6 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 3.1 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.1 | 1.3 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 4.3 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 0.6 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 2.9 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 13.7 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 1.4 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.1 | 1.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 8.9 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 10.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 4.0 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 4.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 1.5 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 2.3 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.7 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 3.2 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 9.7 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 0.2 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 25.5 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 2.1 | GO:0031965 | nuclear membrane(GO:0031965) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.9 | 31.6 | GO:0035939 | microsatellite binding(GO:0035939) |
| 7.0 | 21.1 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 2.3 | 7.0 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 2.0 | 5.9 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 1.5 | 18.1 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 1.1 | 3.4 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 1.0 | 5.0 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 1.0 | 15.5 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.8 | 10.7 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.7 | 3.3 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.6 | 3.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.6 | 2.9 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.6 | 12.9 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.5 | 6.6 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.5 | 6.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.5 | 6.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.5 | 8.5 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.5 | 1.4 | GO:0102007 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.4 | 10.6 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.3 | 24.6 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.3 | 3.5 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.3 | 4.3 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.3 | 1.9 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.3 | 6.9 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.2 | 1.5 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.2 | 1.5 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.2 | 3.6 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.2 | 2.6 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.2 | 3.1 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.2 | 5.7 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.2 | 4.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.2 | 4.1 | GO:0005537 | mannose binding(GO:0005537) |
| 0.2 | 10.9 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.2 | 5.8 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.2 | 55.0 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.2 | 13.1 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.1 | 10.6 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 5.1 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.1 | 3.8 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 2.6 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.1 | 3.4 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.1 | 2.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 14.2 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.1 | 4.3 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 2.9 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 1.0 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 4.0 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 1.3 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 11.9 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.1 | 3.7 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 8.1 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 6.9 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 1.6 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 1.8 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 0.6 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 2.2 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.2 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 3.2 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 7.1 | GO:0019887 | protein kinase regulator activity(GO:0019887) |
| 0.0 | 1.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 5.7 | GO:0001664 | G-protein coupled receptor binding(GO:0001664) |
| 0.0 | 1.7 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 2.0 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 2.3 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 1.5 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 9.1 | GO:0008289 | lipid binding(GO:0008289) |
| 0.0 | 1.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 2.8 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.2 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 21.1 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.5 | 29.5 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.3 | 14.7 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.2 | 7.0 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 3.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 4.3 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 2.9 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 10.5 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 3.7 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.1 | 3.3 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 11.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 4.4 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 19.8 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.5 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 1.5 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 2.2 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 1.8 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.7 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 1.9 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.6 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 1.0 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 4.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.3 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 12.1 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.7 | 9.6 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.5 | 10.7 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.4 | 31.8 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.4 | 15.1 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.4 | 18.0 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.4 | 17.3 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.3 | 14.7 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.3 | 4.4 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.3 | 6.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.2 | 5.9 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.2 | 3.5 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.1 | 3.7 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.1 | 9.8 | REACTOME REGULATION OF INSULIN SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.1 | 7.2 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 3.6 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 3.8 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.1 | 2.6 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 4.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 1.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 3.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 2.3 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 3.2 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 1.9 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 2.9 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 5.2 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.6 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |