Project
averaged GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for RUNX3_BCL11A
Z-value: 1.68
Transcription factors associated with RUNX3_BCL11A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
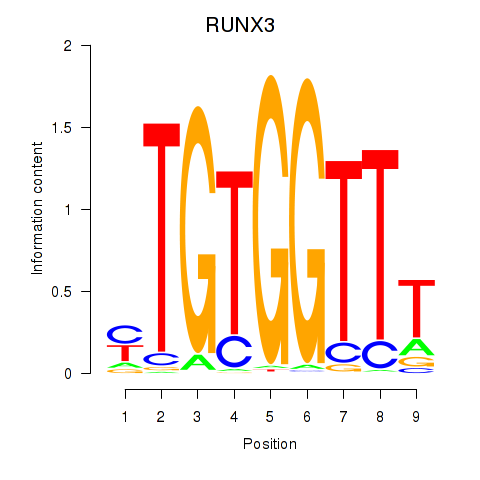
RUNX3
|
ENSG00000020633.14 | RUNX family transcription factor 3 |
|
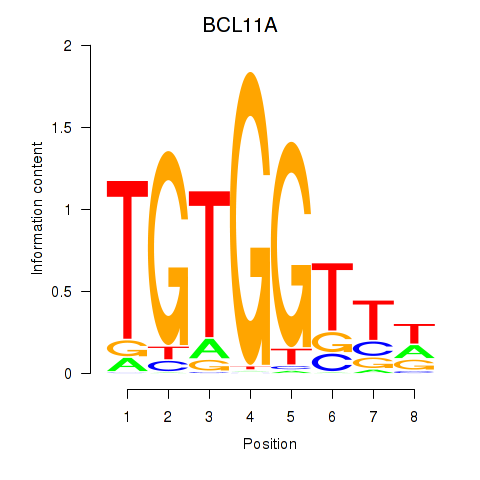
BCL11A
|
ENSG00000119866.16 | BAF chromatin remodeling complex subunit BCL11A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RUNX3 | hg19_v2_chr1_-_25291475_25291511 | 0.62 | 1.4e-24 | Click! |
| BCL11A | hg19_v2_chr2_-_60780536_60780541 | -0.14 | 4.6e-02 | Click! |
Activity profile of RUNX3_BCL11A motif
Sorted Z-values of RUNX3_BCL11A motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_25291475 | 34.47 |
ENST00000338888.3
ENST00000399916.1 |
RUNX3
|
runt-related transcription factor 3 |
| chr10_+_17272608 | 33.26 |
ENST00000421459.2
|
VIM
|
vimentin |
| chr2_-_158300556 | 31.12 |
ENST00000264192.3
|
CYTIP
|
cytohesin 1 interacting protein |
| chr11_-_58345569 | 30.52 |
ENST00000528954.1
ENST00000528489.1 |
LPXN
|
leupaxin |
| chr4_-_84035905 | 26.00 |
ENST00000311507.4
|
PLAC8
|
placenta-specific 8 |
| chr4_-_84035868 | 24.80 |
ENST00000426923.2
ENST00000509973.1 |
PLAC8
|
placenta-specific 8 |
| chr5_+_35856951 | 23.64 |
ENST00000303115.3
ENST00000343305.4 ENST00000506850.1 ENST00000511982.1 |
IL7R
|
interleukin 7 receptor |
| chr5_+_54398463 | 22.83 |
ENST00000274306.6
|
GZMA
|
granzyme A (granzyme 1, cytotoxic T-lymphocyte-associated serine esterase 3) |
| chr9_+_75766652 | 22.06 |
ENST00000257497.6
|
ANXA1
|
annexin A1 |
| chr5_+_156607829 | 20.99 |
ENST00000422843.3
|
ITK
|
IL2-inducible T-cell kinase |
| chrX_-_106960285 | 20.57 |
ENST00000503515.1
ENST00000372397.2 |
TSC22D3
|
TSC22 domain family, member 3 |
| chr17_+_7239821 | 19.23 |
ENST00000158762.3
ENST00000570457.2 |
ACAP1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chr10_+_17270214 | 19.13 |
ENST00000544301.1
|
VIM
|
vimentin |
| chr19_-_36233332 | 18.99 |
ENST00000592537.1
ENST00000246532.1 ENST00000344990.3 ENST00000588992.1 |
IGFLR1
|
IGF-like family receptor 1 |
| chr3_-_150920979 | 18.57 |
ENST00000309180.5
ENST00000480322.1 |
GPR171
|
G protein-coupled receptor 171 |
| chr12_-_15114492 | 18.48 |
ENST00000541546.1
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr1_+_198608146 | 18.17 |
ENST00000367376.2
ENST00000352140.3 ENST00000594404.1 ENST00000598951.1 ENST00000530727.1 ENST00000442510.2 ENST00000367367.4 ENST00000348564.6 ENST00000367364.1 ENST00000413409.2 |
PTPRC
|
protein tyrosine phosphatase, receptor type, C |
| chr13_-_46756351 | 17.64 |
ENST00000323076.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr1_+_209929377 | 17.62 |
ENST00000400959.3
ENST00000367025.3 |
TRAF3IP3
|
TRAF3 interacting protein 3 |
| chr12_-_51717875 | 17.60 |
ENST00000604560.1
|
BIN2
|
bridging integrator 2 |
| chrX_+_48542168 | 17.24 |
ENST00000376701.4
|
WAS
|
Wiskott-Aldrich syndrome |
| chr1_-_114414316 | 16.95 |
ENST00000528414.1
ENST00000538253.1 ENST00000460620.1 ENST00000420377.2 ENST00000525799.1 ENST00000359785.5 |
PTPN22
|
protein tyrosine phosphatase, non-receptor type 22 (lymphoid) |
| chr12_-_51717948 | 16.89 |
ENST00000267012.4
|
BIN2
|
bridging integrator 2 |
| chr12_-_51717922 | 16.59 |
ENST00000452142.2
|
BIN2
|
bridging integrator 2 |
| chr12_-_53594227 | 15.50 |
ENST00000550743.2
|
ITGB7
|
integrin, beta 7 |
| chr22_+_40297079 | 15.18 |
ENST00000344138.4
ENST00000543252.1 |
GRAP2
|
GRB2-related adaptor protein 2 |
| chr12_-_10542617 | 14.53 |
ENST00000240618.6
|
KLRK1
|
killer cell lectin-like receptor subfamily K, member 1 |
| chr11_-_2323290 | 13.87 |
ENST00000381153.3
|
C11orf21
|
chromosome 11 open reading frame 21 |
| chr5_+_133451254 | 13.85 |
ENST00000517851.1
ENST00000521639.1 ENST00000522375.1 ENST00000378560.4 ENST00000432532.2 ENST00000520958.1 ENST00000518915.1 ENST00000395023.1 |
TCF7
|
transcription factor 7 (T-cell specific, HMG-box) |
| chr1_+_32716840 | 13.42 |
ENST00000336890.5
|
LCK
|
lymphocyte-specific protein tyrosine kinase |
| chr1_+_209929494 | 13.20 |
ENST00000367026.3
|
TRAF3IP3
|
TRAF3 interacting protein 3 |
| chr8_-_134072593 | 12.91 |
ENST00000427060.2
|
SLA
|
Src-like-adaptor |
| chr22_+_40297105 | 12.85 |
ENST00000540310.1
|
GRAP2
|
GRB2-related adaptor protein 2 |
| chr14_-_25103388 | 12.64 |
ENST00000526004.1
ENST00000415355.3 |
GZMB
|
granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) |
| chr1_-_153517473 | 12.18 |
ENST00000368715.1
|
S100A4
|
S100 calcium binding protein A4 |
| chr1_+_32716857 | 12.12 |
ENST00000482949.1
ENST00000495610.2 |
LCK
|
lymphocyte-specific protein tyrosine kinase |
| chr14_+_64680854 | 12.09 |
ENST00000458046.2
|
SYNE2
|
spectrin repeat containing, nuclear envelope 2 |
| chr13_-_99910673 | 11.95 |
ENST00000397473.2
ENST00000397470.2 |
GPR18
|
G protein-coupled receptor 18 |
| chr14_-_25103472 | 11.79 |
ENST00000216341.4
ENST00000382542.1 ENST00000382540.1 |
GZMB
|
granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) |
| chr19_-_51875894 | 11.78 |
ENST00000600427.1
ENST00000595217.1 ENST00000221978.5 |
NKG7
|
natural killer cell group 7 sequence |
| chr1_+_113161778 | 11.72 |
ENST00000263168.3
|
CAPZA1
|
capping protein (actin filament) muscle Z-line, alpha 1 |
| chr5_+_75699149 | 11.45 |
ENST00000379730.3
|
IQGAP2
|
IQ motif containing GTPase activating protein 2 |
| chr5_+_75699040 | 10.78 |
ENST00000274364.6
|
IQGAP2
|
IQ motif containing GTPase activating protein 2 |
| chr17_-_34417479 | 10.72 |
ENST00000225245.5
|
CCL3
|
chemokine (C-C motif) ligand 3 |
| chr11_+_2323349 | 10.43 |
ENST00000381121.3
|
TSPAN32
|
tetraspanin 32 |
| chr6_+_151042224 | 10.40 |
ENST00000358517.2
|
PLEKHG1
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 1 |
| chr11_+_35198243 | 10.38 |
ENST00000528455.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr4_-_54930790 | 10.11 |
ENST00000263921.3
|
CHIC2
|
cysteine-rich hydrophobic domain 2 |
| chr11_-_128457446 | 9.78 |
ENST00000392668.4
|
ETS1
|
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
| chr12_-_54694807 | 9.59 |
ENST00000435572.2
|
NFE2
|
nuclear factor, erythroid 2 |
| chr12_-_4754339 | 9.25 |
ENST00000228850.1
|
AKAP3
|
A kinase (PRKA) anchor protein 3 |
| chr16_+_84801852 | 9.21 |
ENST00000569925.1
ENST00000567526.1 |
USP10
|
ubiquitin specific peptidase 10 |
| chr19_+_10197463 | 9.19 |
ENST00000590378.1
ENST00000397881.3 |
C19orf66
|
chromosome 19 open reading frame 66 |
| chr12_+_25205568 | 9.03 |
ENST00000548766.1
ENST00000556887.1 |
LRMP
|
lymphoid-restricted membrane protein |
| chr5_-_39203093 | 8.91 |
ENST00000515010.1
|
FYB
|
FYN binding protein |
| chr16_+_85942594 | 8.85 |
ENST00000566369.1
|
IRF8
|
interferon regulatory factor 8 |
| chr1_-_168513229 | 8.60 |
ENST00000367819.2
|
XCL2
|
chemokine (C motif) ligand 2 |
| chr8_-_21771182 | 8.60 |
ENST00000523932.1
ENST00000544659.1 |
DOK2
|
docking protein 2, 56kDa |
| chr11_+_35198118 | 8.52 |
ENST00000525211.1
ENST00000526000.1 ENST00000279452.6 ENST00000527889.1 |
CD44
|
CD44 molecule (Indian blood group) |
| chr12_-_54694758 | 8.32 |
ENST00000553070.1
|
NFE2
|
nuclear factor, erythroid 2 |
| chr17_-_29648761 | 8.31 |
ENST00000247270.3
ENST00000462804.2 |
EVI2A
|
ecotropic viral integration site 2A |
| chr2_-_198299726 | 8.30 |
ENST00000409915.4
ENST00000487698.1 ENST00000414963.2 ENST00000335508.6 |
SF3B1
|
splicing factor 3b, subunit 1, 155kDa |
| chr8_-_21771214 | 8.12 |
ENST00000276420.4
|
DOK2
|
docking protein 2, 56kDa |
| chr8_-_101719159 | 7.96 |
ENST00000520868.1
ENST00000522658.1 |
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr2_-_111291587 | 7.93 |
ENST00000437167.1
|
RGPD6
|
RANBP2-like and GRIP domain containing 6 |
| chr11_+_60739115 | 7.88 |
ENST00000344028.5
ENST00000346437.4 |
CD6
|
CD6 molecule |
| chr15_-_55563072 | 7.83 |
ENST00000567380.1
ENST00000565972.1 ENST00000569493.1 |
RAB27A
|
RAB27A, member RAS oncogene family |
| chr12_+_25205446 | 7.82 |
ENST00000557489.1
ENST00000354454.3 ENST00000536173.1 |
LRMP
|
lymphoid-restricted membrane protein |
| chr11_+_60739140 | 7.77 |
ENST00000313421.7
|
CD6
|
CD6 molecule |
| chr17_-_56082455 | 7.62 |
ENST00000578794.1
|
RP11-159D12.5
|
Uncharacterized protein |
| chr1_-_207095324 | 7.62 |
ENST00000530505.1
ENST00000367091.3 ENST00000442471.2 |
FAIM3
|
Fas apoptotic inhibitory molecule 3 |
| chr1_+_168545711 | 7.59 |
ENST00000367818.3
|
XCL1
|
chemokine (C motif) ligand 1 |
| chr8_-_101718991 | 7.43 |
ENST00000517990.1
|
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr3_+_50126341 | 7.42 |
ENST00000347869.3
ENST00000469838.1 ENST00000404526.2 ENST00000441305.1 |
RBM5
|
RNA binding motif protein 5 |
| chr4_-_109087872 | 7.22 |
ENST00000510624.1
|
LEF1
|
lymphoid enhancer-binding factor 1 |
| chr2_+_136499287 | 7.10 |
ENST00000415164.1
|
UBXN4
|
UBX domain protein 4 |
| chr2_+_103035102 | 7.04 |
ENST00000264260.2
|
IL18RAP
|
interleukin 18 receptor accessory protein |
| chr2_-_175462934 | 6.73 |
ENST00000392546.2
ENST00000436221.1 |
WIPF1
|
WAS/WASL interacting protein family, member 1 |
| chr1_-_145715565 | 6.68 |
ENST00000369288.2
ENST00000369290.1 ENST00000401557.3 |
CD160
|
CD160 molecule |
| chr15_-_55562582 | 6.67 |
ENST00000396307.2
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr5_-_39270725 | 6.60 |
ENST00000512138.1
ENST00000512982.1 ENST00000540520.1 |
FYB
|
FYN binding protein |
| chr20_+_36405665 | 6.56 |
ENST00000373469.1
|
CTNNBL1
|
catenin, beta like 1 |
| chr2_-_175462456 | 6.45 |
ENST00000409891.1
ENST00000410117.1 |
WIPF1
|
WAS/WASL interacting protein family, member 1 |
| chr15_-_55562479 | 6.34 |
ENST00000564609.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr3_+_49977440 | 6.33 |
ENST00000442092.1
ENST00000266022.4 ENST00000443081.1 |
RBM6
|
RNA binding motif protein 6 |
| chr2_+_136499179 | 6.25 |
ENST00000272638.9
|
UBXN4
|
UBX domain protein 4 |
| chr3_+_111260954 | 6.23 |
ENST00000283285.5
|
CD96
|
CD96 molecule |
| chr11_+_2323236 | 6.21 |
ENST00000182290.4
|
TSPAN32
|
tetraspanin 32 |
| chr17_-_73781567 | 6.20 |
ENST00000586607.1
|
H3F3B
|
H3 histone, family 3B (H3.3B) |
| chrX_+_37639264 | 6.13 |
ENST00000378588.4
|
CYBB
|
cytochrome b-245, beta polypeptide |
| chr1_-_92951607 | 6.05 |
ENST00000427103.1
|
GFI1
|
growth factor independent 1 transcription repressor |
| chr6_+_42584847 | 5.93 |
ENST00000372883.3
|
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2 |
| chr17_+_45810594 | 5.88 |
ENST00000177694.1
|
TBX21
|
T-box 21 |
| chr12_-_24103954 | 5.75 |
ENST00000441133.2
ENST00000545921.1 |
SOX5
|
SRY (sex determining region Y)-box 5 |
| chr5_+_148206156 | 5.68 |
ENST00000305988.4
|
ADRB2
|
adrenoceptor beta 2, surface |
| chr6_+_144904334 | 5.65 |
ENST00000367526.4
|
UTRN
|
utrophin |
| chr3_+_49977523 | 5.57 |
ENST00000422955.1
|
RBM6
|
RNA binding motif protein 6 |
| chr12_-_109025849 | 5.52 |
ENST00000228463.6
|
SELPLG
|
selectin P ligand |
| chr11_+_114310102 | 5.50 |
ENST00000265881.5
|
REXO2
|
RNA exonuclease 2 |
| chr11_+_114310164 | 5.46 |
ENST00000544196.1
ENST00000539754.1 ENST00000539275.1 |
REXO2
|
RNA exonuclease 2 |
| chr3_+_49977490 | 5.43 |
ENST00000539992.1
|
RBM6
|
RNA binding motif protein 6 |
| chr3_+_111260856 | 5.35 |
ENST00000352690.4
|
CD96
|
CD96 molecule |
| chr3_+_113251143 | 5.23 |
ENST00000264852.4
ENST00000393830.3 |
SIDT1
|
SID1 transmembrane family, member 1 |
| chr11_+_114310237 | 5.21 |
ENST00000539119.1
|
REXO2
|
RNA exonuclease 2 |
| chr15_-_72668805 | 5.17 |
ENST00000268097.5
|
HEXA
|
hexosaminidase A (alpha polypeptide) |
| chrX_+_47441712 | 5.12 |
ENST00000218388.4
ENST00000377018.2 ENST00000456754.2 ENST00000377017.1 ENST00000441738.1 |
TIMP1
|
TIMP metallopeptidase inhibitor 1 |
| chr11_+_108093559 | 5.11 |
ENST00000278616.4
|
ATM
|
ataxia telangiectasia mutated |
| chr4_-_71532339 | 5.10 |
ENST00000254801.4
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr1_+_89246647 | 5.04 |
ENST00000544045.1
|
PKN2
|
protein kinase N2 |
| chr22_+_40440804 | 4.97 |
ENST00000441751.1
ENST00000301923.9 |
TNRC6B
|
trinucleotide repeat containing 6B |
| chr20_+_44637526 | 4.95 |
ENST00000372330.3
|
MMP9
|
matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) |
| chr6_-_31508304 | 4.94 |
ENST00000376177.2
|
DDX39B
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39B |
| chr2_+_90077680 | 4.93 |
ENST00000390270.2
|
IGKV3D-20
|
immunoglobulin kappa variable 3D-20 |
| chr11_-_62457371 | 4.86 |
ENST00000317449.4
|
LRRN4CL
|
LRRN4 C-terminal like |
| chr4_-_109087906 | 4.83 |
ENST00000515500.1
|
LEF1
|
lymphoid enhancer-binding factor 1 |
| chr17_-_61777090 | 4.81 |
ENST00000578061.1
|
LIMD2
|
LIM domain containing 2 |
| chr1_+_221051699 | 4.76 |
ENST00000366903.6
|
HLX
|
H2.0-like homeobox |
| chr1_-_207095212 | 4.72 |
ENST00000420007.2
|
FAIM3
|
Fas apoptotic inhibitory molecule 3 |
| chr9_-_34048873 | 4.69 |
ENST00000449054.1
ENST00000379239.4 ENST00000539807.1 ENST00000379238.1 ENST00000418786.2 ENST00000360802.1 ENST00000412543.1 |
UBAP2
|
ubiquitin associated protein 2 |
| chr16_+_2802623 | 4.68 |
ENST00000576924.1
ENST00000575009.1 ENST00000576415.1 ENST00000571378.1 |
SRRM2
|
serine/arginine repetitive matrix 2 |
| chr3_-_135914615 | 4.67 |
ENST00000309993.2
|
MSL2
|
male-specific lethal 2 homolog (Drosophila) |
| chr19_-_42636543 | 4.57 |
ENST00000528894.4
ENST00000560804.2 ENST00000560558.1 ENST00000560398.1 ENST00000526816.2 |
POU2F2
|
POU class 2 homeobox 2 |
| chr14_+_75988851 | 4.47 |
ENST00000555504.1
|
BATF
|
basic leucine zipper transcription factor, ATF-like |
| chr9_+_34652164 | 4.39 |
ENST00000441545.2
ENST00000553620.1 |
IL11RA
|
interleukin 11 receptor, alpha |
| chr2_+_33359687 | 4.37 |
ENST00000402934.1
ENST00000404525.1 ENST00000407925.1 |
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr1_+_12524965 | 4.32 |
ENST00000471923.1
|
VPS13D
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr11_+_121447469 | 4.06 |
ENST00000532694.1
ENST00000534286.1 |
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing |
| chr2_+_33359646 | 4.05 |
ENST00000390003.4
ENST00000418533.2 |
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr3_+_51575596 | 3.93 |
ENST00000409535.2
|
RAD54L2
|
RAD54-like 2 (S. cerevisiae) |
| chr14_+_97263641 | 3.93 |
ENST00000216639.3
|
VRK1
|
vaccinia related kinase 1 |
| chr16_+_28505955 | 3.92 |
ENST00000564831.1
ENST00000328423.5 ENST00000431282.1 |
APOBR
|
apolipoprotein B receptor |
| chr19_-_42636617 | 3.90 |
ENST00000529067.1
ENST00000529952.1 ENST00000533720.1 ENST00000389341.5 ENST00000342301.4 |
POU2F2
|
POU class 2 homeobox 2 |
| chr7_-_148580563 | 3.81 |
ENST00000476773.1
|
EZH2
|
enhancer of zeste homolog 2 (Drosophila) |
| chr15_+_58430368 | 3.77 |
ENST00000558772.1
ENST00000219919.4 |
AQP9
|
aquaporin 9 |
| chr2_+_97203082 | 3.77 |
ENST00000454558.2
|
ARID5A
|
AT rich interactive domain 5A (MRF1-like) |
| chr3_-_11610255 | 3.76 |
ENST00000424529.2
|
VGLL4
|
vestigial like 4 (Drosophila) |
| chr16_+_31366455 | 3.76 |
ENST00000268296.4
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit) |
| chr7_-_115670804 | 3.75 |
ENST00000320239.7
|
TFEC
|
transcription factor EC |
| chr7_-_115670792 | 3.74 |
ENST00000265440.7
ENST00000393485.1 |
TFEC
|
transcription factor EC |
| chr16_-_425205 | 3.74 |
ENST00000448854.1
|
TMEM8A
|
transmembrane protein 8A |
| chr6_-_150067632 | 3.73 |
ENST00000460354.2
ENST00000367404.4 ENST00000543637.1 |
NUP43
|
nucleoporin 43kDa |
| chr16_+_69221028 | 3.61 |
ENST00000336278.4
|
SNTB2
|
syntrophin, beta 2 (dystrophin-associated protein A1, 59kDa, basic component 2) |
| chr1_+_62902308 | 3.56 |
ENST00000339950.4
|
USP1
|
ubiquitin specific peptidase 1 |
| chr16_+_22308717 | 3.53 |
ENST00000299853.5
ENST00000564209.1 ENST00000565358.1 ENST00000418581.2 ENST00000564883.1 ENST00000359210.4 ENST00000563024.1 |
POLR3E
|
polymerase (RNA) III (DNA directed) polypeptide E (80kD) |
| chr1_-_153348067 | 3.47 |
ENST00000368737.3
|
S100A12
|
S100 calcium binding protein A12 |
| chr10_-_65028938 | 3.40 |
ENST00000402544.1
|
JMJD1C
|
jumonji domain containing 1C |
| chr1_+_111772314 | 3.39 |
ENST00000466741.1
ENST00000477185.2 |
CHI3L2
|
chitinase 3-like 2 |
| chr19_-_49496557 | 3.39 |
ENST00000323798.3
ENST00000541188.1 ENST00000544287.1 ENST00000540532.1 ENST00000263276.6 |
GYS1
|
glycogen synthase 1 (muscle) |
| chr17_-_34524157 | 3.34 |
ENST00000378354.4
ENST00000394484.1 |
CCL3L3
|
chemokine (C-C motif) ligand 3-like 3 |
| chr9_-_35103105 | 3.34 |
ENST00000452248.2
ENST00000356493.5 |
STOML2
|
stomatin (EPB72)-like 2 |
| chr17_-_34625719 | 3.34 |
ENST00000422211.2
ENST00000542124.1 |
CCL3L1
|
chemokine (C-C motif) ligand 3-like 1 |
| chr10_-_65028817 | 3.34 |
ENST00000542921.1
|
JMJD1C
|
jumonji domain containing 1C |
| chr8_-_28747717 | 3.32 |
ENST00000416984.2
|
INTS9
|
integrator complex subunit 9 |
| chr15_+_58430567 | 3.30 |
ENST00000536493.1
|
AQP9
|
aquaporin 9 |
| chr1_-_1711508 | 3.29 |
ENST00000378625.1
|
NADK
|
NAD kinase |
| chr15_+_49715293 | 3.21 |
ENST00000267843.4
ENST00000560270.1 |
FGF7
|
fibroblast growth factor 7 |
| chr12_+_108079664 | 3.20 |
ENST00000541166.1
|
PWP1
|
PWP1 homolog (S. cerevisiae) |
| chr17_-_39661849 | 3.14 |
ENST00000246635.3
ENST00000336861.3 ENST00000587544.1 ENST00000587435.1 |
KRT13
|
keratin 13 |
| chr12_-_49245936 | 3.14 |
ENST00000308025.3
|
DDX23
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 23 |
| chr8_-_60031762 | 3.12 |
ENST00000361421.1
|
TOX
|
thymocyte selection-associated high mobility group box |
| chr19_-_51220176 | 3.05 |
ENST00000359082.3
ENST00000293441.1 |
SHANK1
|
SH3 and multiple ankyrin repeat domains 1 |
| chr8_-_28747424 | 3.04 |
ENST00000523436.1
ENST00000397363.4 ENST00000521777.1 ENST00000520184.1 ENST00000521022.1 |
INTS9
|
integrator complex subunit 9 |
| chr3_+_50284321 | 3.02 |
ENST00000451956.1
|
GNAI2
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2 |
| chr3_-_151047327 | 3.00 |
ENST00000325602.5
|
P2RY13
|
purinergic receptor P2Y, G-protein coupled, 13 |
| chr17_+_27047570 | 2.99 |
ENST00000472628.1
ENST00000578181.1 |
RPL23A
|
ribosomal protein L23a |
| chrX_-_70838306 | 2.99 |
ENST00000373691.4
ENST00000373693.3 |
CXCR3
|
chemokine (C-X-C motif) receptor 3 |
| chr11_+_117049910 | 2.98 |
ENST00000431081.2
ENST00000524842.1 |
SIDT2
|
SID1 transmembrane family, member 2 |
| chr17_-_39674668 | 2.97 |
ENST00000393981.3
|
KRT15
|
keratin 15 |
| chr1_+_111772435 | 2.93 |
ENST00000524472.1
|
CHI3L2
|
chitinase 3-like 2 |
| chr1_+_62901968 | 2.92 |
ENST00000452143.1
ENST00000442679.1 ENST00000371146.1 |
USP1
|
ubiquitin specific peptidase 1 |
| chrX_-_119005735 | 2.87 |
ENST00000371442.2
|
RNF113A
|
ring finger protein 113A |
| chr3_+_189507460 | 2.84 |
ENST00000434928.1
|
TP63
|
tumor protein p63 |
| chr12_+_75874984 | 2.79 |
ENST00000550491.1
|
GLIPR1
|
GLI pathogenesis-related 1 |
| chr14_+_35761580 | 2.77 |
ENST00000553809.1
ENST00000555764.1 ENST00000556506.1 |
PSMA6
|
proteasome (prosome, macropain) subunit, alpha type, 6 |
| chr2_+_217277466 | 2.67 |
ENST00000358207.5
ENST00000434435.1 |
SMARCAL1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a-like 1 |
| chr2_+_173292301 | 2.66 |
ENST00000264106.6
ENST00000375221.2 ENST00000343713.4 |
ITGA6
|
integrin, alpha 6 |
| chr7_+_120628731 | 2.64 |
ENST00000310396.5
|
CPED1
|
cadherin-like and PC-esterase domain containing 1 |
| chr2_+_74757050 | 2.64 |
ENST00000352222.3
ENST00000437202.1 |
HTRA2
|
HtrA serine peptidase 2 |
| chr9_-_20622478 | 2.63 |
ENST00000355930.6
ENST00000380338.4 |
MLLT3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr16_-_12009833 | 2.62 |
ENST00000420576.2
|
GSPT1
|
G1 to S phase transition 1 |
| chr22_-_19419205 | 2.61 |
ENST00000340170.4
ENST00000263208.5 |
HIRA
|
histone cell cycle regulator |
| chr19_-_14785698 | 2.59 |
ENST00000344373.4
ENST00000595472.1 |
EMR3
|
egf-like module containing, mucin-like, hormone receptor-like 3 |
| chr2_+_173292280 | 2.58 |
ENST00000264107.7
|
ITGA6
|
integrin, alpha 6 |
| chr8_+_27182862 | 2.57 |
ENST00000521164.1
ENST00000346049.5 |
PTK2B
|
protein tyrosine kinase 2 beta |
| chr2_+_97202480 | 2.53 |
ENST00000357485.3
|
ARID5A
|
AT rich interactive domain 5A (MRF1-like) |
| chr11_+_60739249 | 2.50 |
ENST00000542157.1
ENST00000433107.2 ENST00000452451.2 ENST00000352009.5 |
CD6
|
CD6 molecule |
| chr8_+_27183033 | 2.45 |
ENST00000420218.2
|
PTK2B
|
protein tyrosine kinase 2 beta |
| chrX_+_37639302 | 2.43 |
ENST00000545017.1
ENST00000536160.1 |
CYBB
|
cytochrome b-245, beta polypeptide |
| chr2_+_173292390 | 2.40 |
ENST00000442250.1
ENST00000458358.1 ENST00000409080.1 |
ITGA6
|
integrin, alpha 6 |
| chr11_-_45940343 | 2.37 |
ENST00000532681.1
|
PEX16
|
peroxisomal biogenesis factor 16 |
| chr22_-_29196030 | 2.35 |
ENST00000405219.3
|
XBP1
|
X-box binding protein 1 |
| chr1_+_158223923 | 2.33 |
ENST00000289429.5
|
CD1A
|
CD1a molecule |
| chr19_+_41882598 | 2.32 |
ENST00000447302.2
ENST00000544232.1 ENST00000542945.1 ENST00000540732.1 |
TMEM91
CTC-435M10.3
|
transmembrane protein 91 2-oxoisovalerate dehydrogenase subunit alpha, mitochondrial; Uncharacterized protein |
| chr11_+_118958689 | 2.30 |
ENST00000535253.1
ENST00000392841.1 |
HMBS
|
hydroxymethylbilane synthase |
| chr19_+_13135386 | 2.27 |
ENST00000360105.4
ENST00000588228.1 ENST00000591028.1 |
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr6_+_31543334 | 2.27 |
ENST00000449264.2
|
TNF
|
tumor necrosis factor |
| chr19_+_41257084 | 2.25 |
ENST00000601393.1
|
SNRPA
|
small nuclear ribonucleoprotein polypeptide A |
| chr4_+_152020736 | 2.21 |
ENST00000509736.1
ENST00000505243.1 ENST00000514682.1 ENST00000322686.6 ENST00000503002.1 |
RPS3A
|
ribosomal protein S3A |
| chr2_+_217277137 | 2.20 |
ENST00000430374.1
ENST00000357276.4 ENST00000444508.1 |
SMARCAL1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a-like 1 |
| chr14_-_75643296 | 2.17 |
ENST00000303575.4
|
TMED10
|
transmembrane emp24-like trafficking protein 10 (yeast) |
Network of associatons between targets according to the STRING database.
First level regulatory network of RUNX3_BCL11A
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.4 | 22.1 | GO:1900138 | negative regulation of phospholipase A2 activity(GO:1900138) |
| 5.7 | 17.2 | GO:0002625 | regulation of T cell antigen processing and presentation(GO:0002625) |
| 5.7 | 17.2 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 5.7 | 17.0 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 4.8 | 14.5 | GO:1901624 | negative regulation of lymphocyte chemotaxis(GO:1901624) |
| 4.8 | 52.4 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 4.6 | 18.5 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 3.9 | 15.5 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 3.9 | 11.6 | GO:0002370 | natural killer cell cytokine production(GO:0002370) regulation of natural killer cell cytokine production(GO:0002727) |
| 3.8 | 30.5 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 3.8 | 18.8 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 3.5 | 14.0 | GO:2000538 | regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 3.5 | 24.4 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 3.5 | 20.8 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 3.1 | 56.5 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 2.8 | 13.8 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 2.6 | 20.6 | GO:0070236 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 2.4 | 51.1 | GO:0097320 | membrane tubulation(GO:0097320) |
| 2.4 | 12.0 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 2.4 | 18.9 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 2.3 | 22.8 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 2.3 | 22.7 | GO:0033089 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 2.1 | 21.0 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 2.0 | 6.0 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 2.0 | 12.1 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 2.0 | 27.4 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 1.8 | 5.5 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 1.8 | 10.7 | GO:2000501 | regulation of natural killer cell chemotaxis(GO:2000501) |
| 1.7 | 8.6 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 1.7 | 15.4 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 1.7 | 5.1 | GO:0072425 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 1.4 | 7.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) pyrimidine nucleobase transport(GO:0015855) urea transmembrane transport(GO:0071918) purine nucleobase transmembrane transport(GO:1904823) |
| 1.4 | 8.2 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
| 1.4 | 4.1 | GO:1902771 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 1.3 | 12.1 | GO:1904996 | positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 1.3 | 22.4 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 1.3 | 5.1 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 1.3 | 3.8 | GO:0036333 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 1.2 | 3.7 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 1.2 | 4.9 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 1.2 | 12.0 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |
| 1.2 | 5.9 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 1.2 | 17.3 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 1.1 | 3.4 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 1.0 | 3.1 | GO:0000354 | cis assembly of pre-catalytic spliceosome(GO:0000354) |
| 1.0 | 3.1 | GO:0050894 | determination of affect(GO:0050894) |
| 1.0 | 22.2 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 1.0 | 15.9 | GO:0051639 | actin filament network formation(GO:0051639) |
| 1.0 | 5.8 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 1.0 | 4.8 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.9 | 8.0 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.9 | 2.6 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.8 | 3.3 | GO:0090296 | regulation of mitochondrial DNA replication(GO:0090296) |
| 0.8 | 4.2 | GO:0032571 | response to vitamin K(GO:0032571) |
| 0.8 | 2.4 | GO:0032581 | ER-dependent peroxisome organization(GO:0032581) |
| 0.8 | 2.4 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) regulation of lactation(GO:1903487) |
| 0.7 | 8.2 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.7 | 17.9 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.7 | 2.1 | GO:0065001 | specification of axis polarity(GO:0065001) negative regulation of tooth mineralization(GO:0070171) |
| 0.6 | 6.4 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.6 | 2.5 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.6 | 1.7 | GO:0038178 | complement component C5a signaling pathway(GO:0038178) |
| 0.6 | 6.3 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.6 | 5.6 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.6 | 3.9 | GO:0043987 | histone H3-S10 phosphorylation(GO:0043987) |
| 0.5 | 4.9 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.5 | 3.3 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.5 | 7.6 | GO:0010668 | ectodermal cell differentiation(GO:0010668) nail development(GO:0035878) |
| 0.5 | 2.2 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.5 | 6.5 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.5 | 12.7 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.5 | 1.1 | GO:1901072 | glucosamine-containing compound catabolic process(GO:1901072) |
| 0.5 | 2.0 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.5 | 5.9 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.5 | 4.9 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.5 | 8.5 | GO:0002335 | mature B cell differentiation(GO:0002335) |
| 0.5 | 44.0 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.5 | 2.8 | GO:0071104 | response to interleukin-9(GO:0071104) |
| 0.5 | 5.0 | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.4 | 15.5 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.4 | 16.2 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.4 | 1.6 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.4 | 2.9 | GO:0043144 | snoRNA processing(GO:0043144) |
| 0.4 | 16.0 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.4 | 2.1 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.4 | 0.7 | GO:0002329 | pre-B cell differentiation(GO:0002329) pre-B cell allelic exclusion(GO:0002331) |
| 0.4 | 2.1 | GO:1904550 | chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.3 | 11.7 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.3 | 6.7 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.3 | 1.3 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.3 | 1.5 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.3 | 3.0 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.3 | 5.5 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.3 | 1.3 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.3 | 9.3 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.2 | 1.7 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.2 | 2.6 | GO:0007379 | segment specification(GO:0007379) positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.2 | 2.3 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.2 | 3.9 | GO:0034447 | very-low-density lipoprotein particle clearance(GO:0034447) |
| 0.2 | 0.9 | GO:0034343 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.2 | 2.0 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.2 | 10.1 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.2 | 15.7 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.2 | 1.0 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.2 | 1.0 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.2 | 1.0 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.2 | 0.4 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.2 | 9.2 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.2 | 1.9 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.2 | 15.9 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.2 | 2.3 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.2 | 17.1 | GO:0007338 | single fertilization(GO:0007338) |
| 0.2 | 27.5 | GO:0050680 | negative regulation of epithelial cell proliferation(GO:0050680) |
| 0.2 | 1.6 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.2 | 0.9 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.2 | 3.0 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.1 | 1.5 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 | 4.7 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.1 | 7.2 | GO:0016445 | somatic diversification of immunoglobulins(GO:0016445) |
| 0.1 | 2.6 | GO:0031935 | regulation of chromatin silencing(GO:0031935) |
| 0.1 | 0.4 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
| 0.1 | 19.7 | GO:0002433 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 0.1 | 5.0 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.1 | 5.7 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.1 | 2.3 | GO:1900363 | regulation of mRNA polyadenylation(GO:1900363) |
| 0.1 | 3.5 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.1 | 0.2 | GO:1901899 | positive regulation of relaxation of cardiac muscle(GO:1901899) |
| 0.1 | 0.5 | GO:0098501 | polynucleotide dephosphorylation(GO:0098501) |
| 0.1 | 2.1 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.1 | 0.5 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 7.4 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.1 | 3.7 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.1 | 3.0 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 0.9 | GO:1902166 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902166) |
| 0.1 | 1.3 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.1 | 1.4 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.1 | 1.4 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 7.5 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.1 | 0.7 | GO:0010666 | positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.1 | 1.3 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.1 | 9.5 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.1 | 13.3 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.1 | 0.5 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 0.3 | GO:0046086 | adenosine biosynthetic process(GO:0046086) |
| 0.1 | 3.3 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.1 | 1.4 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.1 | 1.2 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.1 | 3.5 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.1 | 3.4 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.1 | 1.6 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.0 | 0.2 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 1.0 | GO:0070911 | global genome nucleotide-excision repair(GO:0070911) |
| 0.0 | 3.3 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 2.8 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.4 | GO:0060850 | regulation of transcription involved in cell fate commitment(GO:0060850) |
| 0.0 | 0.8 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.6 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 2.1 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.4 | GO:0032354 | response to follicle-stimulating hormone(GO:0032354) |
| 0.0 | 2.3 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.9 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 2.2 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.3 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.0 | 1.2 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.9 | GO:0060021 | palate development(GO:0060021) |
| 0.0 | 1.5 | GO:0051591 | response to cAMP(GO:0051591) |
| 0.0 | 0.4 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.1 | GO:0048484 | enteric nervous system development(GO:0048484) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.7 | 93.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 3.9 | 15.5 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 2.7 | 18.9 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 2.5 | 7.6 | GO:0034676 | integrin alpha6-beta4 complex(GO:0034676) |
| 1.7 | 20.1 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 1.7 | 11.7 | GO:0071203 | WASH complex(GO:0071203) |
| 1.7 | 94.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 1.1 | 3.4 | GO:0018444 | translation release factor complex(GO:0018444) |
| 1.0 | 10.8 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.9 | 12.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.8 | 9.3 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.8 | 15.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.7 | 6.4 | GO:0032039 | integrator complex(GO:0032039) |
| 0.7 | 4.7 | GO:0072487 | MSL complex(GO:0072487) |
| 0.6 | 9.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.6 | 5.6 | GO:0070938 | contractile ring(GO:0070938) |
| 0.6 | 1.8 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) CNTFR-CLCF1 complex(GO:0097059) |
| 0.6 | 27.8 | GO:0002102 | podosome(GO:0002102) |
| 0.5 | 8.6 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.5 | 20.4 | GO:0008305 | integrin complex(GO:0008305) |
| 0.5 | 50.8 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.5 | 56.6 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.4 | 4.9 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.4 | 9.6 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.4 | 20.1 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.4 | 15.6 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.4 | 5.5 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.4 | 6.6 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.4 | 8.0 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.3 | 4.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.3 | 54.8 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.3 | 4.9 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.3 | 25.5 | GO:0005884 | actin filament(GO:0005884) |
| 0.3 | 2.2 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.3 | 3.0 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.3 | 25.5 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.3 | 1.4 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.3 | 19.2 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.2 | 3.7 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.2 | 2.8 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.2 | 1.6 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.2 | 19.8 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.2 | 1.3 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.2 | 3.5 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.2 | 3.1 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.2 | 3.1 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.2 | 1.5 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.2 | 21.8 | GO:0016605 | PML body(GO:0016605) |
| 0.2 | 3.9 | GO:0042627 | low-density lipoprotein particle(GO:0034362) chylomicron(GO:0042627) |
| 0.2 | 3.8 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.2 | 2.1 | GO:0031105 | septin complex(GO:0031105) |
| 0.2 | 46.5 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 1.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 3.5 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 3.6 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 2.9 | GO:0005684 | U2-type spliceosomal complex(GO:0005684) |
| 0.1 | 36.8 | GO:0005769 | early endosome(GO:0005769) |
| 0.1 | 14.2 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.1 | 5.0 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 5.1 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.1 | 2.1 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 1.7 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 1.2 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 1.0 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 2.0 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 6.0 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 5.2 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 1.5 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.1 | 0.5 | GO:0031968 | mitochondrial outer membrane(GO:0005741) outer membrane(GO:0019867) organelle outer membrane(GO:0031968) |
| 0.1 | 0.4 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.1 | 0.3 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.1 | 2.1 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.1 | 0.4 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.1 | 2.6 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 0.5 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.9 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 3.4 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 0.4 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 1.0 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 17.6 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 2.2 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 1.8 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.0 | 1.8 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 3.5 | GO:0034774 | secretory granule lumen(GO:0034774) |
| 0.0 | 0.2 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 1.9 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 2.4 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 1.2 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 1.0 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.2 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 2.5 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 1.2 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.4 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.7 | 52.4 | GO:1990254 | keratin filament binding(GO:1990254) |
| 7.9 | 23.6 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 4.3 | 25.5 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 3.7 | 22.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 2.8 | 22.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 2.6 | 18.1 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 2.3 | 20.6 | GO:0043426 | MRF binding(GO:0043426) |
| 2.1 | 10.7 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 2.1 | 6.4 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 1.9 | 5.7 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 1.8 | 18.5 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 1.8 | 7.1 | GO:0015254 | glycerol channel activity(GO:0015254) urea channel activity(GO:0015265) |
| 1.7 | 8.4 | GO:0050436 | microfibril binding(GO:0050436) |
| 1.4 | 8.2 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 1.2 | 5.9 | GO:0070728 | leucine binding(GO:0070728) |
| 1.1 | 22.4 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 1.0 | 3.1 | GO:0031877 | somatostatin receptor binding(GO:0031877) |
| 1.0 | 3.9 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.9 | 15.4 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.9 | 16.7 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.9 | 5.1 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.8 | 20.8 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.8 | 15.7 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.8 | 64.9 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.7 | 17.2 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.7 | 13.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.7 | 5.8 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.7 | 2.2 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.7 | 12.0 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.7 | 4.9 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.6 | 18.9 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.6 | 7.6 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.6 | 1.7 | GO:0004878 | complement component C5a receptor activity(GO:0004878) |
| 0.6 | 6.3 | GO:0008061 | chitinase activity(GO:0004568) chitin binding(GO:0008061) |
| 0.6 | 3.9 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.5 | 4.9 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.5 | 22.3 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.5 | 6.7 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.5 | 20.6 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.5 | 13.5 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.5 | 1.4 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.4 | 16.2 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.4 | 8.6 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.4 | 3.8 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.4 | 1.2 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.4 | 6.7 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.4 | 3.3 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.4 | 1.8 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.4 | 1.8 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.3 | 5.0 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.3 | 3.0 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.3 | 3.4 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.3 | 3.0 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.3 | 4.1 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.3 | 2.1 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.3 | 1.3 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.3 | 0.5 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.2 | 8.9 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.2 | 5.1 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.2 | 2.3 | GO:0030884 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.2 | 5.6 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.2 | 2.3 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.2 | 1.3 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.2 | 6.4 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.2 | 20.7 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.2 | 1.0 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.2 | 5.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 16.2 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.2 | 1.1 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.2 | 48.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.2 | 0.5 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.2 | 1.7 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.2 | 1.3 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.2 | 9.9 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.2 | 3.5 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.2 | 0.8 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.1 | 1.5 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.1 | 2.5 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.1 | 1.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 2.9 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 12.2 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 18.6 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 3.3 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.1 | 1.2 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.1 | 0.9 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.1 | 3.2 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 0.4 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.1 | 1.5 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 9.5 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 2.8 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 11.6 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 2.0 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 3.0 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 1.0 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 0.4 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 1.6 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 2.0 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.1 | 3.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 1.6 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 2.2 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 25.9 | GO:0005543 | phospholipid binding(GO:0005543) |
| 0.1 | 3.7 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 0.3 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 2.4 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.1 | 12.8 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 1.6 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 2.0 | GO:0016887 | ATPase activity(GO:0016887) |
| 0.1 | 16.6 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 9.7 | GO:0004386 | helicase activity(GO:0004386) |
| 0.1 | 1.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 1.0 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 2.6 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.1 | 0.2 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 3.0 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 5.8 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 16.5 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 1.7 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 1.5 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.4 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 2.1 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.6 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 3.8 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.4 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 1.1 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 2.2 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 2.9 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 3.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 5.4 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 7.5 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.2 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 3.8 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 1.3 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.3 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.5 | GO:0005548 | phospholipid transporter activity(GO:0005548) |
| 0.0 | 1.5 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 3.4 | GO:0005525 | GTP binding(GO:0005525) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 28.0 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 1.0 | 97.4 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.9 | 94.4 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.8 | 15.5 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.7 | 26.9 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.6 | 49.7 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.5 | 23.7 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.5 | 23.1 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.4 | 16.8 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.3 | 10.9 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.3 | 3.0 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.2 | 5.1 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.2 | 6.9 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.2 | 25.3 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.2 | 9.2 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.2 | 3.9 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.2 | 1.3 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.2 | 2.1 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.1 | 3.0 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.1 | 5.0 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 5.1 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 3.4 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 14.4 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.1 | 4.1 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 4.9 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 17.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 5.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 9.6 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 2.0 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 1.7 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 1.7 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 1.6 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 3.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.1 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 0.3 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 2.3 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 1.1 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.9 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 2.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 3.2 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.8 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 1.5 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.3 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.4 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 4.3 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.9 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 18.2 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 2.0 | 107.3 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 1.4 | 49.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 1.3 | 24.7 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 1.1 | 23.6 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.8 | 20.8 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.8 | 10.3 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.6 | 37.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.6 | 16.7 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.5 | 48.3 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.5 | 24.4 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.5 | 7.1 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.5 | 15.2 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.4 | 15.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.4 | 2.1 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.3 | 3.5 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.3 | 5.0 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.3 | 6.4 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.3 | 9.7 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.2 | 12.7 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.2 | 33.2 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.2 | 3.8 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.2 | 3.0 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.2 | 3.5 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.2 | 22.9 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.2 | 2.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.2 | 9.2 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 16.7 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.1 | 12.1 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 5.4 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.1 | 3.4 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 3.0 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 4.0 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 2.8 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.1 | 11.3 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.1 | 17.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 1.7 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.1 | 0.9 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.1 | 8.1 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 1.5 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 1.8 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 2.2 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.5 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 0.3 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 1.1 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.9 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 4.1 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.6 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 2.4 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.7 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 0.6 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |