Project
averaged GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
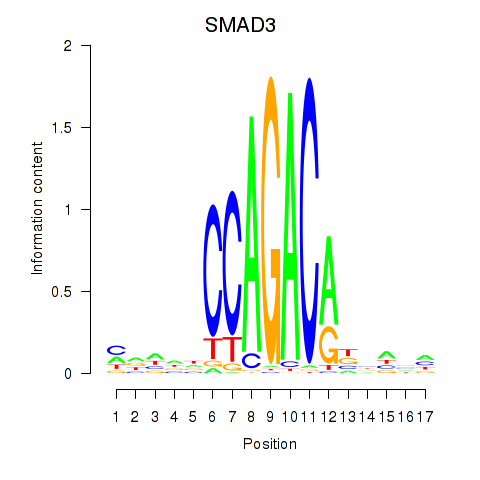
Results for SMAD3
Z-value: 0.96
Transcription factors associated with SMAD3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SMAD3
|
ENSG00000166949.11 | SMAD family member 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SMAD3 | hg19_v2_chr15_+_67358163_67358192 | -0.30 | 8.3e-06 | Click! |
Activity profile of SMAD3 motif
Sorted Z-values of SMAD3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_27114861 | 9.23 |
ENST00000377459.1
|
HIST1H2AH
|
histone cluster 1, H2ah |
| chr2_-_220110111 | 8.06 |
ENST00000428427.1
ENST00000356283.3 ENST00000432839.1 ENST00000424620.1 |
GLB1L
|
galactosidase, beta 1-like |
| chr2_-_220110187 | 7.79 |
ENST00000295759.7
ENST00000392089.2 |
GLB1L
|
galactosidase, beta 1-like |
| chr10_-_90712520 | 6.80 |
ENST00000224784.6
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta |
| chr19_-_11688447 | 6.58 |
ENST00000590420.1
|
ACP5
|
acid phosphatase 5, tartrate resistant |
| chr22_-_36013368 | 6.25 |
ENST00000442617.1
ENST00000397326.2 ENST00000397328.1 ENST00000451685.1 |
MB
|
myoglobin |
| chr15_-_83316254 | 6.12 |
ENST00000567678.1
ENST00000450751.2 |
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr6_-_27782548 | 6.08 |
ENST00000333151.3
|
HIST1H2AJ
|
histone cluster 1, H2aj |
| chr3_+_12838161 | 6.02 |
ENST00000456430.2
|
CAND2
|
cullin-associated and neddylation-dissociated 2 (putative) |
| chr3_+_125687987 | 5.93 |
ENST00000514116.1
ENST00000251776.4 ENST00000504401.1 ENST00000513830.1 ENST00000508088.1 |
ROPN1B
|
rhophilin associated tail protein 1B |
| chr14_+_93389425 | 5.92 |
ENST00000216492.5
ENST00000334654.4 |
CHGA
|
chromogranin A (parathyroid secretory protein 1) |
| chr6_+_27775899 | 5.88 |
ENST00000358739.3
|
HIST1H2AI
|
histone cluster 1, H2ai |
| chr3_-_123710893 | 5.45 |
ENST00000467907.1
ENST00000459660.1 ENST00000495093.1 ENST00000460743.1 ENST00000405845.3 ENST00000484329.1 ENST00000479867.1 ENST00000496145.1 |
ROPN1
|
rhophilin associated tail protein 1 |
| chr19_-_11688500 | 5.05 |
ENST00000433365.2
|
ACP5
|
acid phosphatase 5, tartrate resistant |
| chr1_-_9129735 | 4.92 |
ENST00000377424.4
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr15_-_83316711 | 4.88 |
ENST00000568128.1
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr4_+_55524085 | 4.88 |
ENST00000412167.2
ENST00000288135.5 |
KIT
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog |
| chr6_-_27100529 | 4.78 |
ENST00000607124.1
ENST00000339812.2 ENST00000541790.1 |
HIST1H2BJ
|
histone cluster 1, H2bj |
| chr6_-_27775694 | 4.73 |
ENST00000377401.2
|
HIST1H2BL
|
histone cluster 1, H2bl |
| chr17_+_30813576 | 4.69 |
ENST00000313401.3
|
CDK5R1
|
cyclin-dependent kinase 5, regulatory subunit 1 (p35) |
| chr7_+_139529040 | 4.68 |
ENST00000455353.1
ENST00000458722.1 ENST00000411653.1 |
TBXAS1
|
thromboxane A synthase 1 (platelet) |
| chr14_+_91580732 | 4.64 |
ENST00000519019.1
ENST00000523816.1 ENST00000517518.1 |
C14orf159
|
chromosome 14 open reading frame 159 |
| chr7_+_139528952 | 4.61 |
ENST00000416849.2
ENST00000436047.2 ENST00000414508.2 ENST00000448866.1 |
TBXAS1
|
thromboxane A synthase 1 (platelet) |
| chr14_+_91580777 | 4.59 |
ENST00000525393.2
ENST00000428926.2 ENST00000517362.1 |
C14orf159
|
chromosome 14 open reading frame 159 |
| chr19_+_51728316 | 4.58 |
ENST00000436584.2
ENST00000421133.2 ENST00000391796.3 ENST00000262262.4 |
CD33
|
CD33 molecule |
| chr6_-_11807277 | 4.44 |
ENST00000379415.2
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr15_-_83316087 | 4.32 |
ENST00000568757.1
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr14_+_91580708 | 4.26 |
ENST00000518868.1
|
C14orf159
|
chromosome 14 open reading frame 159 |
| chr12_-_110939870 | 4.16 |
ENST00000447578.2
ENST00000546588.1 ENST00000360579.7 ENST00000549970.1 ENST00000549578.1 |
VPS29
|
vacuolar protein sorting 29 homolog (S. cerevisiae) |
| chr6_+_27100811 | 4.16 |
ENST00000359193.2
|
HIST1H2AG
|
histone cluster 1, H2ag |
| chr6_+_292051 | 4.15 |
ENST00000344450.5
|
DUSP22
|
dual specificity phosphatase 22 |
| chr14_+_91580357 | 4.04 |
ENST00000298858.4
ENST00000521081.1 ENST00000520328.1 ENST00000256324.10 ENST00000524232.1 ENST00000522170.1 ENST00000519950.1 ENST00000523879.1 ENST00000521077.2 ENST00000518665.2 |
C14orf159
|
chromosome 14 open reading frame 159 |
| chr6_-_27114577 | 3.98 |
ENST00000356950.1
ENST00000396891.4 |
HIST1H2BK
|
histone cluster 1, H2bk |
| chr11_-_72145426 | 3.91 |
ENST00000535990.1
ENST00000437826.2 ENST00000340729.5 |
CLPB
|
ClpB caseinolytic peptidase B homolog (E. coli) |
| chr14_+_91581011 | 3.90 |
ENST00000523894.1
ENST00000522322.1 ENST00000523771.1 |
C14orf159
|
chromosome 14 open reading frame 159 |
| chr16_+_30483962 | 3.87 |
ENST00000356798.6
|
ITGAL
|
integrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide) |
| chr1_-_156399184 | 3.65 |
ENST00000368243.1
ENST00000357975.4 ENST00000310027.5 ENST00000400991.2 |
C1orf61
|
chromosome 1 open reading frame 61 |
| chr19_+_42724423 | 3.56 |
ENST00000301215.3
ENST00000597945.1 |
ZNF526
|
zinc finger protein 526 |
| chr6_-_26216872 | 3.43 |
ENST00000244601.3
|
HIST1H2BG
|
histone cluster 1, H2bg |
| chr6_-_26189304 | 3.42 |
ENST00000340756.2
|
HIST1H4D
|
histone cluster 1, H4d |
| chr14_-_24551137 | 3.37 |
ENST00000396995.1
|
NRL
|
neural retina leucine zipper |
| chr15_+_81591757 | 3.36 |
ENST00000558332.1
|
IL16
|
interleukin 16 |
| chr6_+_27806319 | 3.35 |
ENST00000606613.1
ENST00000396980.3 |
HIST1H2BN
|
histone cluster 1, H2bn |
| chr18_+_29598335 | 3.33 |
ENST00000217740.3
|
RNF125
|
ring finger protein 125, E3 ubiquitin protein ligase |
| chr6_+_27782788 | 3.19 |
ENST00000359465.4
|
HIST1H2BM
|
histone cluster 1, H2bm |
| chr6_+_26204825 | 3.15 |
ENST00000360441.4
|
HIST1H4E
|
histone cluster 1, H4e |
| chr16_+_30484021 | 3.14 |
ENST00000358164.5
|
ITGAL
|
integrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide) |
| chr1_-_9129598 | 3.06 |
ENST00000535586.1
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr11_+_113258495 | 3.04 |
ENST00000303941.3
|
ANKK1
|
ankyrin repeat and kinase domain containing 1 |
| chr1_-_160832642 | 3.04 |
ENST00000368034.4
|
CD244
|
CD244 molecule, natural killer cell receptor 2B4 |
| chr6_+_27791862 | 3.04 |
ENST00000355057.1
|
HIST1H4J
|
histone cluster 1, H4j |
| chr22_+_41956767 | 2.98 |
ENST00000306149.7
|
CSDC2
|
cold shock domain containing C2, RNA binding |
| chr13_-_114898016 | 2.94 |
ENST00000542651.1
ENST00000334062.7 |
RASA3
|
RAS p21 protein activator 3 |
| chr10_+_94451574 | 2.94 |
ENST00000492654.2
|
HHEX
|
hematopoietically expressed homeobox |
| chr2_+_204732666 | 2.92 |
ENST00000295854.6
ENST00000472206.1 |
CTLA4
|
cytotoxic T-lymphocyte-associated protein 4 |
| chr1_+_66797687 | 2.90 |
ENST00000371045.5
ENST00000531025.1 ENST00000526197.1 |
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr1_-_8877692 | 2.88 |
ENST00000400908.2
|
RERE
|
arginine-glutamic acid dipeptide (RE) repeats |
| chr12_-_9913489 | 2.86 |
ENST00000228434.3
ENST00000536709.1 |
CD69
|
CD69 molecule |
| chr3_+_51741072 | 2.76 |
ENST00000395052.3
|
GRM2
|
glutamate receptor, metabotropic 2 |
| chr14_-_24551195 | 2.70 |
ENST00000560550.1
|
NRL
|
neural retina leucine zipper |
| chr18_+_13612613 | 2.61 |
ENST00000586765.1
|
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
| chr12_+_26111823 | 2.59 |
ENST00000381352.3
ENST00000535907.1 ENST00000405154.2 |
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr6_+_26124373 | 2.53 |
ENST00000377791.2
ENST00000602637.1 |
HIST1H2AC
|
histone cluster 1, H2ac |
| chr20_+_24449821 | 2.53 |
ENST00000376862.3
|
SYNDIG1
|
synapse differentiation inducing 1 |
| chr19_-_36304201 | 2.51 |
ENST00000301175.3
|
PRODH2
|
proline dehydrogenase (oxidase) 2 |
| chr6_+_26217159 | 2.47 |
ENST00000303910.2
|
HIST1H2AE
|
histone cluster 1, H2ae |
| chr7_+_50348268 | 2.42 |
ENST00000438033.1
ENST00000439701.1 |
IKZF1
|
IKAROS family zinc finger 1 (Ikaros) |
| chr2_+_16080659 | 2.36 |
ENST00000281043.3
|
MYCN
|
v-myc avian myelocytomatosis viral oncogene neuroblastoma derived homolog |
| chr20_-_43280325 | 2.32 |
ENST00000537820.1
|
ADA
|
adenosine deaminase |
| chr20_-_43280361 | 2.25 |
ENST00000372874.4
|
ADA
|
adenosine deaminase |
| chr20_-_23030296 | 2.23 |
ENST00000377103.2
|
THBD
|
thrombomodulin |
| chr1_-_226129083 | 2.20 |
ENST00000420304.2
|
LEFTY2
|
left-right determination factor 2 |
| chr16_-_84220633 | 2.17 |
ENST00000566732.1
ENST00000561955.1 ENST00000564454.1 ENST00000341690.6 ENST00000541676.1 ENST00000570117.1 ENST00000564345.1 |
TAF1C
|
TATA box binding protein (TBP)-associated factor, RNA polymerase I, C, 110kDa |
| chrX_-_153523462 | 2.15 |
ENST00000361930.3
ENST00000369926.1 |
TEX28
|
testis expressed 28 |
| chr4_-_39033963 | 2.14 |
ENST00000381938.3
|
TMEM156
|
transmembrane protein 156 |
| chr17_-_39093672 | 2.13 |
ENST00000209718.3
ENST00000436344.3 ENST00000485751.1 |
KRT23
|
keratin 23 (histone deacetylase inducible) |
| chr5_-_180242534 | 2.06 |
ENST00000333055.3
ENST00000513431.1 |
MGAT1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr4_-_186696425 | 2.06 |
ENST00000430503.1
ENST00000319454.6 ENST00000450341.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr2_+_204732487 | 2.06 |
ENST00000302823.3
|
CTLA4
|
cytotoxic T-lymphocyte-associated protein 4 |
| chr6_+_146920097 | 2.05 |
ENST00000397944.3
ENST00000522242.1 |
ADGB
|
androglobin |
| chr16_-_20681177 | 2.03 |
ENST00000524149.1
|
ACSM1
|
acyl-CoA synthetase medium-chain family member 1 |
| chr15_+_71184931 | 2.02 |
ENST00000560369.1
ENST00000260382.5 |
LRRC49
|
leucine rich repeat containing 49 |
| chr6_-_27860956 | 2.02 |
ENST00000359611.2
|
HIST1H2AM
|
histone cluster 1, H2am |
| chr12_+_65563329 | 1.99 |
ENST00000308330.2
|
LEMD3
|
LEM domain containing 3 |
| chrX_-_110513703 | 1.98 |
ENST00000324068.1
|
CAPN6
|
calpain 6 |
| chr9_+_21967137 | 1.95 |
ENST00000441769.2
|
C9orf53
|
chromosome 9 open reading frame 53 |
| chr19_-_55691472 | 1.95 |
ENST00000537500.1
|
SYT5
|
synaptotagmin V |
| chr11_+_66406149 | 1.94 |
ENST00000578778.1
ENST00000483858.1 ENST00000398692.4 ENST00000510173.2 ENST00000506523.2 ENST00000530235.1 ENST00000532968.1 |
RBM4
|
RNA binding motif protein 4 |
| chr11_+_116700614 | 1.93 |
ENST00000375345.1
|
APOC3
|
apolipoprotein C-III |
| chr8_+_38585704 | 1.91 |
ENST00000519416.1
ENST00000520615.1 |
TACC1
|
transforming, acidic coiled-coil containing protein 1 |
| chr4_+_2814011 | 1.88 |
ENST00000502260.1
ENST00000435136.2 |
SH3BP2
|
SH3-domain binding protein 2 |
| chr1_+_149804218 | 1.86 |
ENST00000610125.1
|
HIST2H4A
|
histone cluster 2, H4a |
| chr11_-_133402410 | 1.85 |
ENST00000524381.1
|
OPCML
|
opioid binding protein/cell adhesion molecule-like |
| chr17_-_3195876 | 1.83 |
ENST00000323404.1
|
OR3A1
|
olfactory receptor, family 3, subfamily A, member 1 |
| chr19_-_55691614 | 1.81 |
ENST00000592470.1
ENST00000354308.3 |
SYT5
|
synaptotagmin V |
| chr16_-_84220604 | 1.77 |
ENST00000567759.1
|
TAF1C
|
TATA box binding protein (TBP)-associated factor, RNA polymerase I, C, 110kDa |
| chr11_+_116700600 | 1.76 |
ENST00000227667.3
|
APOC3
|
apolipoprotein C-III |
| chr2_+_220110177 | 1.72 |
ENST00000409638.3
ENST00000396738.2 ENST00000409516.3 |
STK16
|
serine/threonine kinase 16 |
| chr6_+_146920116 | 1.65 |
ENST00000367493.3
|
ADGB
|
androglobin |
| chr6_-_27806117 | 1.65 |
ENST00000330180.2
|
HIST1H2AK
|
histone cluster 1, H2ak |
| chr2_-_136594740 | 1.61 |
ENST00000264162.2
|
LCT
|
lactase |
| chr6_-_41888843 | 1.60 |
ENST00000434077.1
ENST00000409312.1 |
MED20
|
mediator complex subunit 20 |
| chr11_+_60869867 | 1.56 |
ENST00000347785.3
|
CD5
|
CD5 molecule |
| chr9_-_34691201 | 1.56 |
ENST00000378800.3
ENST00000311925.2 |
CCL19
|
chemokine (C-C motif) ligand 19 |
| chr1_-_226129189 | 1.53 |
ENST00000366820.5
|
LEFTY2
|
left-right determination factor 2 |
| chr2_-_208031943 | 1.51 |
ENST00000421199.1
ENST00000457962.1 |
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr4_-_90759440 | 1.49 |
ENST00000336904.3
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr14_+_102786063 | 1.43 |
ENST00000442396.2
ENST00000262236.5 |
ZNF839
|
zinc finger protein 839 |
| chr6_-_26027480 | 1.43 |
ENST00000377364.3
|
HIST1H4B
|
histone cluster 1, H4b |
| chr1_+_25757376 | 1.36 |
ENST00000399766.3
ENST00000399763.3 ENST00000374343.4 |
TMEM57
|
transmembrane protein 57 |
| chr19_-_46148820 | 1.35 |
ENST00000587152.1
|
EML2
|
echinoderm microtubule associated protein like 2 |
| chr7_+_39125365 | 1.34 |
ENST00000559001.1
ENST00000464276.2 |
POU6F2
|
POU class 6 homeobox 2 |
| chr9_+_5890802 | 1.33 |
ENST00000381477.3
ENST00000381476.1 ENST00000381471.1 |
MLANA
|
melan-A |
| chr11_+_62538775 | 1.32 |
ENST00000294168.3
ENST00000526261.1 |
TAF6L
|
TAF6-like RNA polymerase II, p300/CBP-associated factor (PCAF)-associated factor, 65kDa |
| chr18_+_43753974 | 1.27 |
ENST00000282059.6
ENST00000321319.6 |
C18orf25
|
chromosome 18 open reading frame 25 |
| chr11_+_72983246 | 1.24 |
ENST00000393590.2
|
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr5_-_127674883 | 1.23 |
ENST00000507835.1
|
FBN2
|
fibrillin 2 |
| chr4_+_2813946 | 1.22 |
ENST00000442312.2
|
SH3BP2
|
SH3-domain binding protein 2 |
| chr6_-_26033796 | 1.20 |
ENST00000259791.2
|
HIST1H2AB
|
histone cluster 1, H2ab |
| chr6_-_46922659 | 1.19 |
ENST00000265417.7
|
GPR116
|
G protein-coupled receptor 116 |
| chr19_+_41594377 | 1.17 |
ENST00000330436.3
|
CYP2A13
|
cytochrome P450, family 2, subfamily A, polypeptide 13 |
| chr6_+_27861190 | 1.15 |
ENST00000303806.4
|
HIST1H2BO
|
histone cluster 1, H2bo |
| chr19_-_55691377 | 1.14 |
ENST00000589172.1
|
SYT5
|
synaptotagmin V |
| chr16_-_3306587 | 1.14 |
ENST00000541159.1
ENST00000536379.1 ENST00000219596.1 ENST00000339854.4 |
MEFV
|
Mediterranean fever |
| chr6_+_26273144 | 1.08 |
ENST00000377733.2
|
HIST1H2BI
|
histone cluster 1, H2bi |
| chr8_+_10530155 | 1.02 |
ENST00000521818.1
|
C8orf74
|
chromosome 8 open reading frame 74 |
| chr16_-_15180257 | 1.02 |
ENST00000540462.1
|
RRN3
|
RRN3 RNA polymerase I transcription factor homolog (S. cerevisiae) |
| chr4_-_38666430 | 1.00 |
ENST00000436901.1
|
AC021860.1
|
Uncharacterized protein |
| chr14_+_96858454 | 0.96 |
ENST00000555570.1
|
AK7
|
adenylate kinase 7 |
| chr18_+_77623668 | 0.93 |
ENST00000316249.3
|
KCNG2
|
potassium voltage-gated channel, subfamily G, member 2 |
| chr11_+_124824000 | 0.92 |
ENST00000529051.1
ENST00000344762.5 |
CCDC15
|
coiled-coil domain containing 15 |
| chr18_+_13611763 | 0.89 |
ENST00000585931.1
|
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
| chr3_+_190105909 | 0.82 |
ENST00000456423.1
|
CLDN16
|
claudin 16 |
| chr5_+_79950463 | 0.80 |
ENST00000265081.6
|
MSH3
|
mutS homolog 3 |
| chr6_-_41888814 | 0.78 |
ENST00000409060.1
ENST00000265350.4 |
MED20
|
mediator complex subunit 20 |
| chr19_+_36132631 | 0.77 |
ENST00000379026.2
ENST00000379023.4 ENST00000402764.2 ENST00000479824.1 |
ETV2
|
ets variant 2 |
| chr20_-_60795316 | 0.75 |
ENST00000317393.6
|
HRH3
|
histamine receptor H3 |
| chr1_-_9189229 | 0.75 |
ENST00000377411.4
|
GPR157
|
G protein-coupled receptor 157 |
| chr5_-_68628543 | 0.72 |
ENST00000396496.2
ENST00000511257.1 ENST00000383374.2 |
CCDC125
|
coiled-coil domain containing 125 |
| chr9_+_136399929 | 0.69 |
ENST00000393060.1
|
ADAMTSL2
|
ADAMTS-like 2 |
| chr1_+_52682052 | 0.69 |
ENST00000371591.1
|
ZFYVE9
|
zinc finger, FYVE domain containing 9 |
| chr1_-_9129631 | 0.68 |
ENST00000377414.3
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr1_-_160040038 | 0.62 |
ENST00000368089.3
|
KCNJ10
|
potassium inwardly-rectifying channel, subfamily J, member 10 |
| chrX_-_2847366 | 0.60 |
ENST00000381154.1
|
ARSD
|
arylsulfatase D |
| chrX_-_46618490 | 0.56 |
ENST00000328306.4
|
SLC9A7
|
solute carrier family 9, subfamily A (NHE7, cation proton antiporter 7), member 7 |
| chrX_-_48931648 | 0.56 |
ENST00000376386.3
ENST00000376390.4 |
PRAF2
|
PRA1 domain family, member 2 |
| chr6_-_26124138 | 0.53 |
ENST00000314332.5
ENST00000396984.1 |
HIST1H2BC
|
histone cluster 1, H2bc |
| chr11_+_46383121 | 0.53 |
ENST00000454345.1
|
DGKZ
|
diacylglycerol kinase, zeta |
| chr4_+_40337340 | 0.47 |
ENST00000310169.2
|
CHRNA9
|
cholinergic receptor, nicotinic, alpha 9 (neuronal) |
| chr15_-_26874230 | 0.46 |
ENST00000400188.3
|
GABRB3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr12_+_14927270 | 0.45 |
ENST00000544848.1
|
H2AFJ
|
H2A histone family, member J |
| chr2_-_49381646 | 0.45 |
ENST00000346173.3
ENST00000406846.2 |
FSHR
|
follicle stimulating hormone receptor |
| chr4_-_26492076 | 0.40 |
ENST00000295589.3
|
CCKAR
|
cholecystokinin A receptor |
| chr16_-_425205 | 0.38 |
ENST00000448854.1
|
TMEM8A
|
transmembrane protein 8A |
| chr2_-_49381572 | 0.36 |
ENST00000454032.1
ENST00000304421.4 |
FSHR
|
follicle stimulating hormone receptor |
| chrX_+_49832231 | 0.36 |
ENST00000376108.3
|
CLCN5
|
chloride channel, voltage-sensitive 5 |
| chr7_-_142247606 | 0.33 |
ENST00000390361.3
|
TRBV7-3
|
T cell receptor beta variable 7-3 |
| chr1_+_59762642 | 0.33 |
ENST00000371218.4
ENST00000303721.7 |
FGGY
|
FGGY carbohydrate kinase domain containing |
| chr11_+_117070037 | 0.30 |
ENST00000392951.4
ENST00000525531.1 ENST00000278968.6 |
TAGLN
|
transgelin |
| chr19_-_51192661 | 0.30 |
ENST00000391813.1
|
SHANK1
|
SH3 and multiple ankyrin repeat domains 1 |
| chr4_+_186990298 | 0.25 |
ENST00000296795.3
ENST00000513189.1 |
TLR3
|
toll-like receptor 3 |
| chr7_-_14942944 | 0.24 |
ENST00000403951.2
|
DGKB
|
diacylglycerol kinase, beta 90kDa |
| chr2_-_128784846 | 0.16 |
ENST00000259235.3
ENST00000357702.5 ENST00000424298.1 |
SAP130
|
Sin3A-associated protein, 130kDa |
| chr5_+_140625147 | 0.10 |
ENST00000231173.3
|
PCDHB15
|
protocadherin beta 15 |
| chr20_-_60795296 | 0.10 |
ENST00000340177.5
|
HRH3
|
histamine receptor H3 |
| chr15_-_22473353 | 0.09 |
ENST00000557788.2
|
IGHV4OR15-8
|
immunoglobulin heavy variable 4/OR15-8 (non-functional) |
| chr5_+_140529630 | 0.05 |
ENST00000543635.1
|
PCDHB6
|
protocadherin beta 6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SMAD3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 11.6 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 2.0 | 5.9 | GO:2000705 | dense core granule biogenesis(GO:0061110) positive regulation of relaxation of cardiac muscle(GO:1901899) regulation of dense core granule biogenesis(GO:2000705) |
| 1.9 | 15.3 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 1.6 | 4.9 | GO:0048170 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) erythropoietin-mediated signaling pathway(GO:0038162) positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 1.6 | 4.7 | GO:0021718 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 1.5 | 4.6 | GO:0046102 | hypoxanthine salvage(GO:0043103) inosine metabolic process(GO:0046102) |
| 1.5 | 6.1 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 1.2 | 3.7 | GO:2000910 | regulation of cholesterol import(GO:0060620) negative regulation of cholesterol import(GO:0060621) regulation of sterol import(GO:2000909) negative regulation of sterol import(GO:2000910) |
| 1.1 | 6.8 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 1.0 | 3.0 | GO:2000566 | positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 1.0 | 5.0 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 1.0 | 2.9 | GO:0010621 | negative regulation of transcription by transcription factor localization(GO:0010621) gall bladder development(GO:0061010) |
| 0.9 | 6.0 | GO:1990539 | fructose transport(GO:0015755) fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.8 | 6.3 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.7 | 2.9 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.7 | 2.0 | GO:0018874 | benzoate metabolic process(GO:0018874) butyrate metabolic process(GO:0019605) |
| 0.5 | 23.9 | GO:0006536 | glutamate metabolic process(GO:0006536) |
| 0.5 | 1.6 | GO:2000547 | dendritic cell dendrite assembly(GO:0097026) regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.5 | 2.6 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.5 | 9.3 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.5 | 2.1 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.5 | 7.0 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.5 | 2.0 | GO:0044821 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.5 | 9.3 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.4 | 1.3 | GO:0048867 | stem cell fate determination(GO:0048867) |
| 0.4 | 4.4 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.4 | 1.1 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 0.3 | 5.9 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.3 | 3.3 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.3 | 1.5 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) regulation of peroxidase activity(GO:2000468) positive regulation of peroxidase activity(GO:2000470) |
| 0.3 | 3.5 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.3 | 2.5 | GO:0097091 | synaptic vesicle clustering(GO:0097091) |
| 0.3 | 23.6 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.2 | 4.1 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.2 | 2.9 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.2 | 1.6 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.2 | 0.9 | GO:0014063 | negative regulation of serotonin secretion(GO:0014063) |
| 0.2 | 2.2 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.2 | 0.8 | GO:0001545 | primary ovarian follicle growth(GO:0001545) |
| 0.2 | 1.2 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 0.2 | 2.8 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.2 | 4.2 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.2 | 2.4 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.2 | 0.8 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.2 | 3.4 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.1 | 1.2 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.1 | 1.4 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.1 | 0.4 | GO:0038188 | cholecystokinin signaling pathway(GO:0038188) |
| 0.1 | 4.9 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.1 | 5.0 | GO:0006356 | regulation of transcription from RNA polymerase I promoter(GO:0006356) |
| 0.1 | 0.6 | GO:0051935 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.1 | 12.4 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 0.3 | GO:0050894 | determination of affect(GO:0050894) |
| 0.1 | 1.2 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.1 | 0.8 | GO:0043570 | maintenance of DNA repeat elements(GO:0043570) |
| 0.1 | 0.7 | GO:0060481 | lobar bronchus epithelium development(GO:0060481) |
| 0.1 | 0.5 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.1 | 0.5 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.1 | 0.3 | GO:0034344 | microglial cell activation involved in immune response(GO:0002282) type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.1 | 3.7 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.1 | 2.1 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 0.8 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.1 | 0.7 | GO:0051270 | regulation of cellular component movement(GO:0051270) |
| 0.1 | 0.7 | GO:0060019 | radial glial cell differentiation(GO:0060019) |
| 0.1 | 0.7 | GO:0007183 | SMAD protein complex assembly(GO:0007183) |
| 0.0 | 2.9 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) |
| 0.0 | 1.8 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.0 | 1.8 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.6 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 2.9 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.0 | 2.0 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 3.2 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.3 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.0 | 0.5 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 0.5 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.6 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.0 | 2.4 | GO:0030218 | erythrocyte differentiation(GO:0030218) |
| 0.0 | 1.3 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 1.0 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 | 2.1 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 1.9 | GO:0021987 | cerebral cortex development(GO:0021987) |
| 0.0 | 1.6 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.0 | 0.6 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 3.0 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 0.2 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 1.5 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.0 | 0.4 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 6.8 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 1.4 | 4.2 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 1.2 | 4.7 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 1.2 | 7.0 | GO:0034687 | integrin alphaL-beta2 complex(GO:0034687) |
| 0.8 | 4.9 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.8 | 15.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.8 | 45.3 | GO:0000786 | nucleosome(GO:0000786) |
| 0.6 | 3.9 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.5 | 3.7 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.3 | 5.9 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.3 | 0.8 | GO:0032302 | MutSbeta complex(GO:0032302) |
| 0.2 | 3.3 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.2 | 2.4 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.2 | 2.9 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 4.9 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 2.9 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 5.0 | GO:0098636 | protein complex involved in cell adhesion(GO:0098636) |
| 0.1 | 2.0 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.1 | 10.6 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.1 | 1.3 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 1.5 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 2.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 2.4 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 1.2 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 2.5 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 5.9 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 12.0 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 2.0 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 2.3 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 2.8 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.5 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 2.3 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 2.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 2.8 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 1.7 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.0 | 0.3 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.0 | 26.0 | GO:0005773 | vacuole(GO:0005773) |
| 0.0 | 1.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 2.0 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 1.3 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 1.9 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 2.1 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.5 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.7 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.9 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.8 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.9 | GO:0044306 | neuron projection terminus(GO:0044306) |
| 0.0 | 1.4 | GO:0005875 | microtubule associated complex(GO:0005875) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.3 | 21.4 | GO:0047820 | D-glutamate cyclase activity(GO:0047820) |
| 3.1 | 9.3 | GO:0036134 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 2.0 | 15.8 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 1.6 | 4.7 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 1.0 | 7.0 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 1.0 | 2.9 | GO:0008267 | poly-glutamine tract binding(GO:0008267) |
| 1.0 | 15.3 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.9 | 3.7 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.9 | 11.6 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.9 | 6.0 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.6 | 2.5 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.6 | 4.6 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.6 | 2.8 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.5 | 2.1 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.5 | 6.3 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.4 | 1.6 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.4 | 1.6 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.3 | 1.0 | GO:0001181 | transcription factor activity, core RNA polymerase I binding(GO:0001181) |
| 0.3 | 6.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.3 | 1.5 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.3 | 4.9 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.3 | 0.8 | GO:0000404 | heteroduplex DNA loop binding(GO:0000404) double-strand/single-strand DNA junction binding(GO:0000406) dinucleotide repeat insertion binding(GO:0032181) |
| 0.2 | 2.0 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.2 | 3.9 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.2 | 1.2 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.2 | 0.8 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 0.2 | 2.9 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.2 | 6.2 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.2 | 0.5 | GO:0097158 | pre-mRNA intronic pyrimidine-rich binding(GO:0097158) |
| 0.2 | 1.2 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.2 | 3.4 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 5.7 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 0.7 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.1 | 4.8 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.1 | 0.4 | GO:0004951 | cholecystokinin receptor activity(GO:0004951) |
| 0.1 | 2.9 | GO:0015278 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.1 | 4.9 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 0.6 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 2.9 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 0.9 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.1 | 2.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 4.1 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 3.0 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.1 | 2.9 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 1.0 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 3.7 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 3.5 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 40.7 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.1 | 1.2 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 3.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 0.8 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 0.6 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.8 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 2.5 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.0 | 0.6 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.7 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 3.0 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.9 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 1.8 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.5 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 1.6 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 1.7 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 2.0 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.5 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.3 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 1.5 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 3.9 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.3 | GO:0017025 | TBP-class protein binding(GO:0017025) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 17.2 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.2 | 7.0 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.1 | 4.7 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 7.4 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 9.5 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 3.1 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 1.5 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 7.1 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 1.0 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.6 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 6.0 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 48.9 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.3 | 6.0 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.2 | 4.6 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.2 | 4.9 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.2 | 3.7 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.2 | 4.7 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.2 | 10.5 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.2 | 5.0 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.2 | 2.8 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 1.6 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 2.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 3.7 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 5.0 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.1 | 6.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 6.3 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 7.0 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 2.9 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 1.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 3.3 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 1.1 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 0.8 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 1.8 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 2.1 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.6 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 1.5 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 3.4 | REACTOME REGULATION OF INSULIN SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.0 | 0.3 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.5 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.7 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 2.4 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.8 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.9 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 2.0 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.5 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |