Project
averaged GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
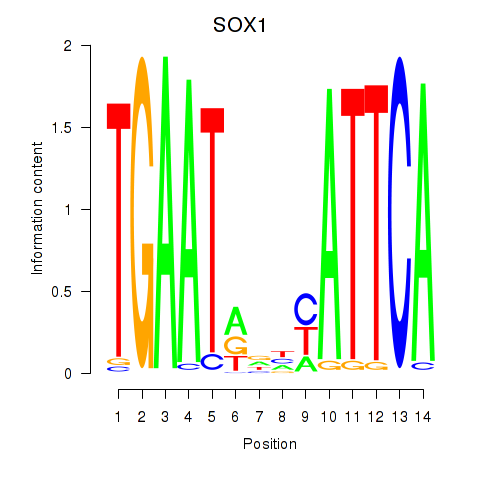
Results for SOX1
Z-value: 0.60
Transcription factors associated with SOX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX1
|
ENSG00000182968.3 | SRY-box transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX1 | hg19_v2_chr13_+_112721913_112721913 | -0.12 | 7.3e-02 | Click! |
Activity profile of SOX1 motif
Sorted Z-values of SOX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_74417096 | 5.95 |
ENST00000286544.3
|
FAM161B
|
family with sequence similarity 161, member B |
| chr5_-_95158644 | 5.74 |
ENST00000237858.6
|
GLRX
|
glutaredoxin (thioltransferase) |
| chr7_-_36634181 | 5.72 |
ENST00000538464.1
|
AOAH
|
acyloxyacyl hydrolase (neutrophil) |
| chr13_-_22178284 | 5.49 |
ENST00000468222.2
ENST00000382374.4 |
MICU2
|
mitochondrial calcium uptake 2 |
| chr4_+_154622652 | 5.28 |
ENST00000260010.6
|
TLR2
|
toll-like receptor 2 |
| chr16_+_12058961 | 5.12 |
ENST00000053243.1
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr16_+_12059050 | 4.99 |
ENST00000396495.3
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr19_-_10491234 | 4.86 |
ENST00000524462.1
ENST00000531836.1 ENST00000525621.1 |
TYK2
|
tyrosine kinase 2 |
| chr12_+_75874460 | 4.58 |
ENST00000266659.3
|
GLIPR1
|
GLI pathogenesis-related 1 |
| chr11_-_47399942 | 4.57 |
ENST00000227163.4
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr3_-_16524357 | 4.55 |
ENST00000432519.1
|
RFTN1
|
raftlin, lipid raft linker 1 |
| chr11_-_47400078 | 4.22 |
ENST00000378538.3
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr1_+_174769006 | 4.00 |
ENST00000489615.1
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr14_+_21249200 | 3.93 |
ENST00000304677.2
|
RNASE6
|
ribonuclease, RNase A family, k6 |
| chr16_+_12059091 | 3.90 |
ENST00000562385.1
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr12_+_9980113 | 3.74 |
ENST00000537723.1
|
KLRF1
|
killer cell lectin-like receptor subfamily F, member 1 |
| chr12_+_9980069 | 3.70 |
ENST00000354855.3
ENST00000324214.4 ENST00000279544.3 |
KLRF1
|
killer cell lectin-like receptor subfamily F, member 1 |
| chr11_-_47400062 | 3.29 |
ENST00000533030.1
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr16_-_11876408 | 3.24 |
ENST00000396516.2
|
ZC3H7A
|
zinc finger CCCH-type containing 7A |
| chr19_+_49588690 | 2.70 |
ENST00000221448.5
|
SNRNP70
|
small nuclear ribonucleoprotein 70kDa (U1) |
| chr3_-_189840223 | 2.60 |
ENST00000427335.2
|
LEPREL1
|
leprecan-like 1 |
| chr12_-_10607084 | 2.57 |
ENST00000408006.3
ENST00000544822.1 ENST00000536188.1 |
KLRC1
|
killer cell lectin-like receptor subfamily C, member 1 |
| chr14_-_25078864 | 2.57 |
ENST00000216338.4
ENST00000557220.2 ENST00000382548.4 |
GZMH
|
granzyme H (cathepsin G-like 2, protein h-CCPX) |
| chr19_+_49588677 | 2.53 |
ENST00000598984.1
ENST00000598441.1 |
SNRNP70
|
small nuclear ribonucleoprotein 70kDa (U1) |
| chr5_-_150460914 | 2.48 |
ENST00000389378.2
|
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr11_-_104827425 | 2.47 |
ENST00000393150.3
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr19_+_21324827 | 2.45 |
ENST00000600692.1
ENST00000599296.1 ENST00000594425.1 ENST00000311048.7 |
ZNF431
|
zinc finger protein 431 |
| chr17_+_75181292 | 2.26 |
ENST00000431431.2
|
SEC14L1
|
SEC14-like 1 (S. cerevisiae) |
| chr1_-_212004090 | 2.12 |
ENST00000366997.4
|
LPGAT1
|
lysophosphatidylglycerol acyltransferase 1 |
| chr3_-_185655795 | 1.90 |
ENST00000342294.4
ENST00000382191.4 ENST00000453386.2 |
TRA2B
|
transformer 2 beta homolog (Drosophila) |
| chr10_+_47894572 | 1.76 |
ENST00000355876.5
|
FAM21B
|
family with sequence similarity 21, member B |
| chr11_-_78052923 | 1.72 |
ENST00000340149.2
|
GAB2
|
GRB2-associated binding protein 2 |
| chr2_+_234296792 | 1.68 |
ENST00000409813.3
|
DGKD
|
diacylglycerol kinase, delta 130kDa |
| chr12_-_110434096 | 1.64 |
ENST00000320063.9
ENST00000457474.2 ENST00000547815.1 ENST00000361006.5 |
GIT2
|
G protein-coupled receptor kinase interacting ArfGAP 2 |
| chr12_-_110434183 | 1.59 |
ENST00000360185.4
ENST00000354574.4 ENST00000338373.5 ENST00000343646.5 ENST00000356259.4 ENST00000553118.1 |
GIT2
|
G protein-coupled receptor kinase interacting ArfGAP 2 |
| chr18_-_51751132 | 1.51 |
ENST00000256429.3
|
MBD2
|
methyl-CpG binding domain protein 2 |
| chr12_-_10605929 | 1.42 |
ENST00000347831.5
ENST00000359151.3 |
KLRC1
|
killer cell lectin-like receptor subfamily C, member 1 |
| chr1_-_200379129 | 1.41 |
ENST00000367353.1
|
ZNF281
|
zinc finger protein 281 |
| chr22_-_17302589 | 1.39 |
ENST00000331428.5
|
XKR3
|
XK, Kell blood group complex subunit-related family, member 3 |
| chr1_+_144811943 | 1.39 |
ENST00000281815.8
|
NBPF9
|
neuroblastoma breakpoint family, member 9 |
| chr12_-_65146636 | 1.36 |
ENST00000418919.2
|
GNS
|
glucosamine (N-acetyl)-6-sulfatase |
| chr1_+_148560843 | 1.30 |
ENST00000442702.2
ENST00000369187.3 |
NBPF15
|
neuroblastoma breakpoint family, member 15 |
| chr11_-_57177586 | 1.29 |
ENST00000529411.1
|
RP11-872D17.8
|
Uncharacterized protein |
| chr1_+_206516200 | 1.19 |
ENST00000295713.5
|
SRGAP2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr2_+_102615416 | 1.12 |
ENST00000393414.2
|
IL1R2
|
interleukin 1 receptor, type II |
| chr20_-_49575058 | 1.10 |
ENST00000371584.4
ENST00000371583.5 ENST00000413082.1 |
DPM1
|
dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit |
| chr3_-_58613323 | 1.07 |
ENST00000474531.1
ENST00000465970.1 |
FAM107A
|
family with sequence similarity 107, member A |
| chr7_+_142457315 | 1.07 |
ENST00000486171.1
ENST00000311737.7 |
PRSS1
|
protease, serine, 1 (trypsin 1) |
| chr17_-_29624343 | 1.02 |
ENST00000247271.4
|
OMG
|
oligodendrocyte myelin glycoprotein |
| chr1_-_200379104 | 0.97 |
ENST00000367352.3
|
ZNF281
|
zinc finger protein 281 |
| chr6_+_149539767 | 0.96 |
ENST00000606202.1
ENST00000536230.1 ENST00000445901.1 |
TAB2
RP1-111D6.3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 2 RP1-111D6.3 |
| chr3_-_58200398 | 0.89 |
ENST00000318316.3
ENST00000460422.1 ENST00000483681.1 |
DNASE1L3
|
deoxyribonuclease I-like 3 |
| chr11_-_5255861 | 0.88 |
ENST00000380299.3
|
HBD
|
hemoglobin, delta |
| chr1_-_200379180 | 0.86 |
ENST00000294740.3
|
ZNF281
|
zinc finger protein 281 |
| chr13_+_53602894 | 0.85 |
ENST00000219022.2
|
OLFM4
|
olfactomedin 4 |
| chr9_-_2844058 | 0.85 |
ENST00000397885.2
|
KIAA0020
|
KIAA0020 |
| chr12_-_120763739 | 0.75 |
ENST00000549767.1
|
PLA2G1B
|
phospholipase A2, group IB (pancreas) |
| chr12_+_57810198 | 0.73 |
ENST00000598001.1
|
AC126614.1
|
HCG1818482; Uncharacterized protein |
| chrX_+_129040122 | 0.70 |
ENST00000394422.3
ENST00000371051.5 |
UTP14A
|
UTP14, U3 small nucleolar ribonucleoprotein, homolog A (yeast) |
| chr14_+_22356029 | 0.69 |
ENST00000390437.2
|
TRAV12-2
|
T cell receptor alpha variable 12-2 |
| chrX_+_129040094 | 0.68 |
ENST00000425117.2
|
UTP14A
|
UTP14, U3 small nucleolar ribonucleoprotein, homolog A (yeast) |
| chr4_+_156680153 | 0.67 |
ENST00000502959.1
ENST00000505764.1 ENST00000507146.1 ENST00000264424.8 ENST00000503520.1 |
GUCY1B3
|
guanylate cyclase 1, soluble, beta 3 |
| chr3_+_138327417 | 0.57 |
ENST00000338446.4
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr14_-_20801427 | 0.57 |
ENST00000557665.1
ENST00000358932.4 ENST00000353689.4 |
CCNB1IP1
|
cyclin B1 interacting protein 1, E3 ubiquitin protein ligase |
| chr4_+_26585538 | 0.56 |
ENST00000264866.4
|
TBC1D19
|
TBC1 domain family, member 19 |
| chr12_+_100594557 | 0.52 |
ENST00000546902.1
ENST00000552376.1 ENST00000551617.1 |
ACTR6
|
ARP6 actin-related protein 6 homolog (yeast) |
| chr8_-_102181718 | 0.48 |
ENST00000565617.1
|
KB-1460A1.5
|
KB-1460A1.5 |
| chr11_-_113577014 | 0.43 |
ENST00000544634.1
ENST00000539732.1 ENST00000538770.1 ENST00000536856.1 ENST00000544476.1 |
TMPRSS5
|
transmembrane protease, serine 5 |
| chr8_-_86290333 | 0.43 |
ENST00000521846.1
ENST00000523022.1 ENST00000524324.1 ENST00000519991.1 ENST00000520663.1 ENST00000517590.1 ENST00000522579.1 ENST00000522814.1 ENST00000522662.1 ENST00000523858.1 ENST00000519129.1 |
CA1
|
carbonic anhydrase I |
| chr22_-_30662828 | 0.41 |
ENST00000403463.1
ENST00000215781.2 |
OSM
|
oncostatin M |
| chr3_+_138327542 | 0.38 |
ENST00000360570.3
ENST00000393035.2 |
FAIM
|
Fas apoptotic inhibitory molecule |
| chr11_-_5255696 | 0.37 |
ENST00000292901.3
ENST00000417377.1 |
HBD
|
hemoglobin, delta |
| chr20_-_49575081 | 0.23 |
ENST00000371588.5
ENST00000371582.4 |
DPM1
|
dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit |
| chr10_-_92681033 | 0.23 |
ENST00000371697.3
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle) |
| chr8_+_110551925 | 0.21 |
ENST00000395785.2
|
EBAG9
|
estrogen receptor binding site associated, antigen, 9 |
| chrX_-_62974941 | 0.18 |
ENST00000374872.1
ENST00000253401.6 ENST00000374870.4 |
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9 |
| chr10_+_15085895 | 0.18 |
ENST00000378228.3
|
OLAH
|
oleoyl-ACP hydrolase |
| chr17_-_39646116 | 0.17 |
ENST00000328119.6
|
KRT36
|
keratin 36 |
| chr13_+_109248500 | 0.15 |
ENST00000356711.2
|
MYO16
|
myosin XVI |
| chr11_-_113577052 | 0.14 |
ENST00000540540.1
ENST00000545579.1 ENST00000538955.1 ENST00000299882.5 |
TMPRSS5
|
transmembrane protease, serine 5 |
| chr11_-_18062872 | 0.09 |
ENST00000250018.2
|
TPH1
|
tryptophan hydroxylase 1 |
| chr19_+_17830051 | 0.07 |
ENST00000594625.1
ENST00000324096.4 ENST00000600186.1 ENST00000597735.1 |
MAP1S
|
microtubule-associated protein 1S |
| chr11_+_102980126 | 0.07 |
ENST00000375735.2
|
DYNC2H1
|
dynein, cytoplasmic 2, heavy chain 1 |
| chr9_-_13165457 | 0.07 |
ENST00000542239.1
ENST00000538841.1 ENST00000433359.2 |
MPDZ
|
multiple PDZ domain protein |
| chr1_+_70876926 | 0.06 |
ENST00000370938.3
ENST00000346806.2 |
CTH
|
cystathionase (cystathionine gamma-lyase) |
| chr9_-_95244781 | 0.06 |
ENST00000375544.3
ENST00000375543.1 ENST00000395538.3 ENST00000450139.2 |
ASPN
|
asporin |
| chr11_+_62009653 | 0.03 |
ENST00000244926.3
|
SCGB1D2
|
secretoglobin, family 1D, member 2 |
| chr19_+_55385682 | 0.02 |
ENST00000391726.3
|
FCAR
|
Fc fragment of IgA, receptor for |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 12.1 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 1.8 | 5.3 | GO:0032289 | central nervous system myelin formation(GO:0032289) detection of bacterial lipoprotein(GO:0042494) detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) |
| 0.9 | 5.5 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.9 | 5.2 | GO:0061083 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) |
| 0.8 | 2.5 | GO:0085032 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.7 | 4.6 | GO:0032596 | protein transport within lipid bilayer(GO:0032594) protein transport into membrane raft(GO:0032596) |
| 0.5 | 1.9 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.3 | 5.7 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.3 | 1.1 | GO:0050712 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.3 | 1.3 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.2 | 2.5 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.2 | 0.7 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.2 | 2.3 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.2 | 0.8 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.2 | 14.0 | GO:0002260 | lymphocyte homeostasis(GO:0002260) |
| 0.1 | 3.2 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.1 | 2.1 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.1 | 0.5 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.1 | 0.9 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) |
| 0.1 | 1.2 | GO:0021815 | modulation of microtubule cytoskeleton involved in cerebral cortex radial glia guided migration(GO:0021815) |
| 0.1 | 0.4 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.1 | 1.5 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.1 | 1.7 | GO:0033008 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.1 | 4.9 | GO:0060338 | regulation of type I interferon-mediated signaling pathway(GO:0060338) |
| 0.1 | 1.2 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 3.9 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.1 | 1.7 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.1 | 0.9 | GO:0010623 | programmed cell death involved in cell development(GO:0010623) |
| 0.1 | 0.6 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 4.0 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 2.6 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 1.1 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.1 | GO:0042427 | serotonin biosynthetic process(GO:0042427) |
| 0.0 | 1.4 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 1.0 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.0 | 0.1 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.0 | 3.2 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 0.1 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 2.5 | GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity(GO:0043433) |
| 0.0 | 0.1 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) positive regulation of vascular smooth muscle cell differentiation(GO:1905065) |
| 0.0 | 0.2 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 1.0 | GO:0050771 | negative regulation of axonogenesis(GO:0050771) |
| 0.0 | 0.6 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 4.6 | GO:0019216 | regulation of lipid metabolic process(GO:0019216) |
| 0.0 | 10.7 | GO:0050776 | regulation of immune response(GO:0050776) |
| 0.0 | 4.7 | GO:0006954 | inflammatory response(GO:0006954) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.3 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 1.4 | 5.5 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.4 | 1.3 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.4 | 2.5 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.4 | 5.2 | GO:0000243 | commitment complex(GO:0000243) |
| 0.1 | 1.2 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.1 | 1.2 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 4.6 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 1.4 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 13.6 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 1.9 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.7 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 4.9 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 2.2 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.6 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 4.6 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 4.0 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 3.7 | GO:0005769 | early endosome(GO:0005769) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.7 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 1.1 | 12.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.8 | 4.0 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.6 | 5.3 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.6 | 5.7 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.6 | 7.4 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.5 | 5.2 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.4 | 1.3 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.4 | 4.9 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.4 | 2.3 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.3 | 1.4 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.3 | 2.1 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.2 | 1.1 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.2 | 2.5 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.2 | 1.5 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.1 | 1.2 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 1.7 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) diacylglycerol binding(GO:0019992) |
| 0.1 | 1.7 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 0.2 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 0.8 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 3.4 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.1 | 0.2 | GO:0016296 | oleoyl-[acyl-carrier-protein] hydrolase activity(GO:0004320) myristoyl-[acyl-carrier-protein] hydrolase activity(GO:0016295) palmitoyl-[acyl-carrier-protein] hydrolase activity(GO:0016296) acyl-[acyl-carrier-protein] hydrolase activity(GO:0016297) |
| 0.1 | 0.4 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.1 | 1.9 | GO:0070717 | poly-purine tract binding(GO:0070717) |
| 0.0 | 4.2 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 4.8 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.0 | 0.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.7 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 4.0 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 5.7 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 1.2 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 4.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.1 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 0.2 | GO:0016505 | peptidase activator activity involved in apoptotic process(GO:0016505) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.9 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.2 | 5.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 12.1 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 2.1 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 2.1 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 3.2 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 2.5 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.5 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 1.0 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 1.0 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.8 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 5.3 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.3 | 4.9 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.2 | 2.9 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.2 | 5.7 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 1.0 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.1 | 1.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 2.5 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.1 | 1.7 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 4.0 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 2.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.7 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 4.8 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 1.4 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 1.1 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 4.7 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.4 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 1.1 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.0 | 1.2 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 1.2 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |