Project
averaged GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
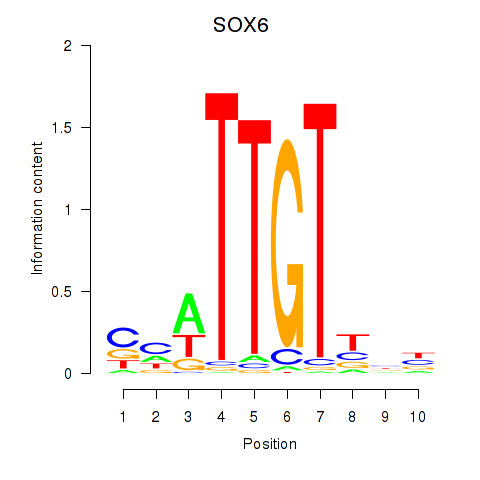
Results for SOX6
Z-value: 1.10
Transcription factors associated with SOX6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX6
|
ENSG00000110693.11 | SRY-box transcription factor 6 |
Activity profile of SOX6 motif
Sorted Z-values of SOX6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_37899323 | 15.74 |
ENST00000295324.3
ENST00000457889.1 |
CDC42EP3
|
CDC42 effector protein (Rho GTPase binding) 3 |
| chr2_+_232573222 | 15.58 |
ENST00000341369.7
ENST00000409683.1 |
PTMA
|
prothymosin, alpha |
| chr11_-_102323489 | 14.96 |
ENST00000361236.3
|
TMEM123
|
transmembrane protein 123 |
| chr12_+_69979446 | 14.35 |
ENST00000543146.2
|
CCT2
|
chaperonin containing TCP1, subunit 2 (beta) |
| chr11_-_102323740 | 14.14 |
ENST00000398136.2
|
TMEM123
|
transmembrane protein 123 |
| chr2_+_10262857 | 13.99 |
ENST00000304567.5
|
RRM2
|
ribonucleotide reductase M2 |
| chr2_+_232573208 | 13.11 |
ENST00000409115.3
|
PTMA
|
prothymosin, alpha |
| chr12_-_58146048 | 12.87 |
ENST00000547281.1
ENST00000546489.1 ENST00000552388.1 ENST00000540325.1 ENST00000312990.6 |
CDK4
|
cyclin-dependent kinase 4 |
| chr16_+_33605231 | 11.52 |
ENST00000570121.2
|
IGHV3OR16-12
|
immunoglobulin heavy variable 3/OR16-12 (non-functional) |
| chr16_-_29466285 | 11.23 |
ENST00000330978.3
|
BOLA2
|
bolA family member 2 |
| chr3_-_57583052 | 10.87 |
ENST00000496292.1
ENST00000489843.1 |
ARF4
|
ADP-ribosylation factor 4 |
| chr8_-_102218292 | 10.39 |
ENST00000518336.1
ENST00000520454.1 |
ZNF706
|
zinc finger protein 706 |
| chr16_+_14980632 | 10.07 |
ENST00000565655.1
|
NOMO1
|
NODAL modulator 1 |
| chr12_-_53320245 | 9.82 |
ENST00000552150.1
|
KRT8
|
keratin 8 |
| chr3_-_57583130 | 9.62 |
ENST00000303436.6
|
ARF4
|
ADP-ribosylation factor 4 |
| chr21_-_40720995 | 9.53 |
ENST00000380749.5
|
HMGN1
|
high mobility group nucleosome binding domain 1 |
| chr4_-_71705027 | 9.43 |
ENST00000545193.1
|
GRSF1
|
G-rich RNA sequence binding factor 1 |
| chr4_-_71705060 | 9.36 |
ENST00000514161.1
|
GRSF1
|
G-rich RNA sequence binding factor 1 |
| chr10_-_33247124 | 8.86 |
ENST00000414670.1
ENST00000302278.3 ENST00000374956.4 ENST00000488494.1 ENST00000417122.2 ENST00000474568.1 |
ITGB1
|
integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) |
| chr1_+_68150744 | 8.80 |
ENST00000370986.4
ENST00000370985.3 |
GADD45A
|
growth arrest and DNA-damage-inducible, alpha |
| chr12_-_58146128 | 8.76 |
ENST00000551800.1
ENST00000549606.1 ENST00000257904.6 |
CDK4
|
cyclin-dependent kinase 4 |
| chr19_+_5681153 | 8.60 |
ENST00000579559.1
ENST00000577222.1 |
HSD11B1L
RPL36
|
hydroxysteroid (11-beta) dehydrogenase 1-like ribosomal protein L36 |
| chr14_-_106725723 | 8.59 |
ENST00000390609.2
|
IGHV3-23
|
immunoglobulin heavy variable 3-23 |
| chr1_-_32801825 | 8.58 |
ENST00000329421.7
|
MARCKSL1
|
MARCKS-like 1 |
| chr1_-_26233423 | 8.41 |
ENST00000357865.2
|
STMN1
|
stathmin 1 |
| chr15_+_77224045 | 8.41 |
ENST00000320963.5
ENST00000394883.3 |
RCN2
|
reticulocalbin 2, EF-hand calcium binding domain |
| chr17_-_40288449 | 8.28 |
ENST00000552162.1
ENST00000550504.1 |
RAB5C
|
RAB5C, member RAS oncogene family |
| chr22_-_42336209 | 8.06 |
ENST00000472374.2
|
CENPM
|
centromere protein M |
| chr18_+_3449821 | 8.01 |
ENST00000407501.2
ENST00000405385.3 ENST00000546979.1 |
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr7_-_148725733 | 7.95 |
ENST00000286091.4
|
PDIA4
|
protein disulfide isomerase family A, member 4 |
| chr21_-_40720974 | 7.85 |
ENST00000380748.1
|
HMGN1
|
high mobility group nucleosome binding domain 1 |
| chr14_-_106926724 | 7.74 |
ENST00000434710.1
|
IGHV3-43
|
immunoglobulin heavy variable 3-43 |
| chr2_-_85641162 | 7.70 |
ENST00000447219.2
ENST00000409670.1 ENST00000409724.1 |
CAPG
|
capping protein (actin filament), gelsolin-like |
| chr18_+_3451646 | 7.56 |
ENST00000345133.5
ENST00000330513.5 ENST00000549546.1 |
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr12_+_104359576 | 7.50 |
ENST00000392872.3
ENST00000436021.2 |
TDG
|
thymine-DNA glycosylase |
| chrX_-_153599578 | 7.43 |
ENST00000360319.4
ENST00000344736.4 |
FLNA
|
filamin A, alpha |
| chr4_+_41614720 | 7.40 |
ENST00000509277.1
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr2_-_86790593 | 7.40 |
ENST00000263856.4
ENST00000409225.2 |
CHMP3
|
charged multivesicular body protein 3 |
| chr13_-_24007815 | 7.39 |
ENST00000382298.3
|
SACS
|
spastic ataxia of Charlevoix-Saguenay (sacsin) |
| chr17_+_75283973 | 7.24 |
ENST00000431235.2
ENST00000449803.2 |
SEPT9
|
septin 9 |
| chr12_+_69633372 | 7.02 |
ENST00000456847.3
ENST00000266679.8 |
CPSF6
|
cleavage and polyadenylation specific factor 6, 68kDa |
| chr3_+_52719936 | 6.93 |
ENST00000418458.1
ENST00000394799.2 |
GNL3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr15_+_77223960 | 6.91 |
ENST00000394885.3
|
RCN2
|
reticulocalbin 2, EF-hand calcium binding domain |
| chr12_+_104359641 | 6.90 |
ENST00000537100.1
|
TDG
|
thymine-DNA glycosylase |
| chr6_-_134639180 | 6.85 |
ENST00000367858.5
|
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr1_-_26232951 | 6.70 |
ENST00000426559.2
ENST00000455785.2 |
STMN1
|
stathmin 1 |
| chr8_+_126442563 | 6.60 |
ENST00000311922.3
|
TRIB1
|
tribbles pseudokinase 1 |
| chr13_-_41593425 | 6.58 |
ENST00000239882.3
|
ELF1
|
E74-like factor 1 (ets domain transcription factor) |
| chr15_+_52311398 | 6.46 |
ENST00000261845.5
|
MAPK6
|
mitogen-activated protein kinase 6 |
| chr14_-_106845789 | 6.42 |
ENST00000390617.2
|
IGHV3-35
|
immunoglobulin heavy variable 3-35 (non-functional) |
| chrX_+_12993336 | 6.38 |
ENST00000380635.1
|
TMSB4X
|
thymosin beta 4, X-linked |
| chr7_+_134464414 | 6.35 |
ENST00000361901.2
|
CALD1
|
caldesmon 1 |
| chr11_+_63754294 | 6.34 |
ENST00000543988.1
|
OTUB1
|
OTU domain, ubiquitin aldehyde binding 1 |
| chr11_-_64013663 | 6.33 |
ENST00000392210.2
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr7_+_99006232 | 6.23 |
ENST00000403633.2
|
BUD31
|
BUD31 homolog (S. cerevisiae) |
| chr14_-_71107921 | 6.13 |
ENST00000553982.1
ENST00000500016.1 |
CTD-2540L5.5
CTD-2540L5.6
|
CTD-2540L5.5 CTD-2540L5.6 |
| chr10_-_13390270 | 6.11 |
ENST00000378614.4
ENST00000545675.1 ENST00000327347.5 |
SEPHS1
|
selenophosphate synthetase 1 |
| chr1_-_212004090 | 6.07 |
ENST00000366997.4
|
LPGAT1
|
lysophosphatidylglycerol acyltransferase 1 |
| chr14_-_35344093 | 6.05 |
ENST00000382422.2
|
BAZ1A
|
bromodomain adjacent to zinc finger domain, 1A |
| chr13_+_100153665 | 6.00 |
ENST00000376387.4
|
TM9SF2
|
transmembrane 9 superfamily member 2 |
| chr3_+_151986709 | 5.96 |
ENST00000495875.2
ENST00000493459.1 ENST00000324210.5 ENST00000459747.1 |
MBNL1
|
muscleblind-like splicing regulator 1 |
| chr13_-_95131923 | 5.88 |
ENST00000377028.5
ENST00000446125.1 |
DCT
|
dopachrome tautomerase |
| chr12_+_11081828 | 5.81 |
ENST00000381847.3
ENST00000396400.3 |
PRH2
|
proline-rich protein HaeIII subfamily 2 |
| chr12_+_104359614 | 5.81 |
ENST00000266775.9
ENST00000544861.1 |
TDG
|
thymine-DNA glycosylase |
| chr17_+_75372165 | 5.76 |
ENST00000427674.2
|
SEPT9
|
septin 9 |
| chr4_+_41614909 | 5.68 |
ENST00000509454.1
ENST00000396595.3 ENST00000381753.4 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr4_-_143227088 | 5.66 |
ENST00000511838.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr12_-_11036844 | 5.60 |
ENST00000428168.2
|
PRH1
|
proline-rich protein HaeIII subfamily 1 |
| chr10_+_104535994 | 5.59 |
ENST00000369889.4
|
WBP1L
|
WW domain binding protein 1-like |
| chr19_+_47105309 | 5.57 |
ENST00000599839.1
ENST00000596362.1 |
CALM3
|
calmodulin 3 (phosphorylase kinase, delta) |
| chr17_-_1588101 | 5.57 |
ENST00000577001.1
ENST00000572621.1 ENST00000304992.6 |
PRPF8
|
pre-mRNA processing factor 8 |
| chr16_-_67970990 | 5.53 |
ENST00000358514.4
|
PSMB10
|
proteasome (prosome, macropain) subunit, beta type, 10 |
| chr16_+_56970567 | 5.49 |
ENST00000563911.1
|
HERPUD1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr9_-_86595105 | 5.45 |
ENST00000457156.1
ENST00000360384.5 ENST00000376263.3 |
HNRNPK
|
heterogeneous nuclear ribonucleoprotein K |
| chr14_+_23791159 | 5.15 |
ENST00000557702.1
|
PABPN1
|
poly(A) binding protein, nuclear 1 |
| chr12_-_49582593 | 5.11 |
ENST00000295766.5
|
TUBA1A
|
tubulin, alpha 1a |
| chr7_+_134464376 | 5.08 |
ENST00000454108.1
ENST00000361675.2 |
CALD1
|
caldesmon 1 |
| chr12_+_69633407 | 5.00 |
ENST00000551516.1
|
CPSF6
|
cleavage and polyadenylation specific factor 6, 68kDa |
| chr5_-_39425068 | 4.96 |
ENST00000515700.1
ENST00000339788.6 |
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chrX_+_70503433 | 4.94 |
ENST00000276079.8
ENST00000373856.3 ENST00000373841.1 ENST00000420903.1 |
NONO
|
non-POU domain containing, octamer-binding |
| chr17_+_52978156 | 4.85 |
ENST00000348161.4
|
TOM1L1
|
target of myb1 (chicken)-like 1 |
| chr3_-_195270162 | 4.80 |
ENST00000438848.1
ENST00000328432.3 |
PPP1R2
|
protein phosphatase 1, regulatory (inhibitor) subunit 2 |
| chr7_-_107643674 | 4.79 |
ENST00000222399.6
|
LAMB1
|
laminin, beta 1 |
| chr5_-_16936340 | 4.67 |
ENST00000507288.1
ENST00000513610.1 |
MYO10
|
myosin X |
| chr2_+_54683419 | 4.64 |
ENST00000356805.4
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1 |
| chr5_-_157002775 | 4.53 |
ENST00000257527.4
|
ADAM19
|
ADAM metallopeptidase domain 19 |
| chr7_-_99699538 | 4.53 |
ENST00000343023.6
ENST00000303887.5 |
MCM7
|
minichromosome maintenance complex component 7 |
| chr1_-_89357179 | 4.47 |
ENST00000448623.1
ENST00000418217.1 ENST00000370500.5 |
GTF2B
|
general transcription factor IIB |
| chr9_-_74979420 | 4.45 |
ENST00000343431.2
ENST00000376956.3 |
ZFAND5
|
zinc finger, AN1-type domain 5 |
| chr6_+_114178512 | 4.41 |
ENST00000368635.4
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate |
| chr12_-_49582978 | 4.29 |
ENST00000301071.7
|
TUBA1A
|
tubulin, alpha 1a |
| chr16_-_15736881 | 4.22 |
ENST00000540441.2
|
KIAA0430
|
KIAA0430 |
| chr7_+_69064300 | 4.19 |
ENST00000342771.4
|
AUTS2
|
autism susceptibility candidate 2 |
| chr4_+_70861647 | 4.19 |
ENST00000246895.4
ENST00000381060.2 |
STATH
|
statherin |
| chr8_+_11660120 | 4.19 |
ENST00000220584.4
|
FDFT1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr11_+_19798964 | 4.18 |
ENST00000527559.2
|
NAV2
|
neuron navigator 2 |
| chr11_+_64949899 | 4.18 |
ENST00000531068.1
ENST00000527699.1 ENST00000533909.1 ENST00000527323.1 |
CAPN1
|
calpain 1, (mu/I) large subunit |
| chr6_-_24877490 | 4.14 |
ENST00000540914.1
ENST00000378023.4 |
FAM65B
|
family with sequence similarity 65, member B |
| chr1_+_40505891 | 4.10 |
ENST00000372797.3
ENST00000372802.1 ENST00000449311.1 |
CAP1
|
CAP, adenylate cyclase-associated protein 1 (yeast) |
| chr4_+_70916119 | 4.05 |
ENST00000246896.3
ENST00000511674.1 |
HTN1
|
histatin 1 |
| chr18_+_7946941 | 3.98 |
ENST00000444013.1
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M |
| chr4_-_109090106 | 3.97 |
ENST00000379951.2
|
LEF1
|
lymphoid enhancer-binding factor 1 |
| chr17_-_56065484 | 3.92 |
ENST00000581208.1
|
VEZF1
|
vascular endothelial zinc finger 1 |
| chr11_+_19799327 | 3.90 |
ENST00000540292.1
|
NAV2
|
neuron navigator 2 |
| chr1_+_166808692 | 3.80 |
ENST00000367876.4
|
POGK
|
pogo transposable element with KRAB domain |
| chr10_+_22605374 | 3.79 |
ENST00000448361.1
|
COMMD3
|
COMM domain containing 3 |
| chr8_+_11660227 | 3.78 |
ENST00000443614.2
ENST00000525900.1 |
FDFT1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr5_-_39425222 | 3.73 |
ENST00000320816.6
|
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr7_+_23146271 | 3.72 |
ENST00000545771.1
|
KLHL7
|
kelch-like family member 7 |
| chr4_-_143226979 | 3.71 |
ENST00000514525.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr9_+_108463234 | 3.67 |
ENST00000374688.1
|
TMEM38B
|
transmembrane protein 38B |
| chr5_-_176900610 | 3.67 |
ENST00000477391.2
ENST00000393565.1 ENST00000309007.5 |
DBN1
|
drebrin 1 |
| chr5_+_134094461 | 3.63 |
ENST00000452510.2
ENST00000354283.4 |
DDX46
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 46 |
| chr2_-_55276320 | 3.62 |
ENST00000357376.3
|
RTN4
|
reticulon 4 |
| chr11_+_64949343 | 3.60 |
ENST00000279247.6
ENST00000532285.1 ENST00000534373.1 |
CAPN1
|
calpain 1, (mu/I) large subunit |
| chr5_-_39425290 | 3.56 |
ENST00000545653.1
|
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr15_+_64388166 | 3.53 |
ENST00000353874.4
ENST00000261889.5 ENST00000559844.1 ENST00000561026.1 ENST00000558040.1 |
SNX1
|
sorting nexin 1 |
| chr4_-_105416039 | 3.50 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chr12_+_22778009 | 3.47 |
ENST00000266517.4
ENST00000335148.3 |
ETNK1
|
ethanolamine kinase 1 |
| chr1_-_26232522 | 3.44 |
ENST00000399728.1
|
STMN1
|
stathmin 1 |
| chr13_-_110959478 | 3.40 |
ENST00000543140.1
ENST00000375820.4 |
COL4A1
|
collagen, type IV, alpha 1 |
| chr8_-_141810634 | 3.38 |
ENST00000521986.1
ENST00000523539.1 ENST00000538769.1 |
PTK2
|
protein tyrosine kinase 2 |
| chr21_-_16437126 | 3.36 |
ENST00000318948.4
|
NRIP1
|
nuclear receptor interacting protein 1 |
| chr8_-_30515693 | 3.27 |
ENST00000355904.4
|
GTF2E2
|
general transcription factor IIE, polypeptide 2, beta 34kDa |
| chr8_+_107738240 | 3.25 |
ENST00000449762.2
ENST00000297447.6 |
OXR1
|
oxidation resistance 1 |
| chr11_+_125496619 | 3.20 |
ENST00000532669.1
ENST00000278916.3 |
CHEK1
|
checkpoint kinase 1 |
| chr10_+_22605304 | 3.19 |
ENST00000475460.2
ENST00000602390.1 ENST00000489125.2 ENST00000456711.1 ENST00000444869.1 |
COMMD3-BMI1
COMMD3
|
COMMD3-BMI1 readthrough COMM domain containing 3 |
| chrX_+_12993202 | 3.11 |
ENST00000451311.2
ENST00000380636.1 |
TMSB4X
|
thymosin beta 4, X-linked |
| chr14_-_106518922 | 3.09 |
ENST00000390598.2
|
IGHV3-7
|
immunoglobulin heavy variable 3-7 |
| chr17_-_79479789 | 3.03 |
ENST00000571691.1
ENST00000571721.1 ENST00000573283.1 ENST00000575842.1 ENST00000575087.1 ENST00000570382.1 ENST00000331925.2 |
ACTG1
|
actin, gamma 1 |
| chr1_-_155880672 | 3.00 |
ENST00000609492.1
ENST00000368322.3 |
RIT1
|
Ras-like without CAAX 1 |
| chr14_+_50234827 | 2.99 |
ENST00000554589.1
ENST00000557247.1 |
KLHDC2
|
kelch domain containing 2 |
| chr19_+_50354462 | 2.86 |
ENST00000601675.1
|
PTOV1
|
prostate tumor overexpressed 1 |
| chr5_-_176836577 | 2.85 |
ENST00000253496.3
|
F12
|
coagulation factor XII (Hageman factor) |
| chr17_-_49124230 | 2.80 |
ENST00000510283.1
ENST00000510855.1 |
SPAG9
|
sperm associated antigen 9 |
| chr11_-_62414070 | 2.77 |
ENST00000540933.1
ENST00000346178.4 ENST00000356638.3 ENST00000534779.1 ENST00000525994.1 |
GANAB
|
glucosidase, alpha; neutral AB |
| chr6_-_132834184 | 2.76 |
ENST00000367941.2
ENST00000367937.4 |
STX7
|
syntaxin 7 |
| chr6_-_128841503 | 2.75 |
ENST00000368215.3
ENST00000532331.1 ENST00000368213.5 ENST00000368207.3 ENST00000525459.1 ENST00000368210.3 ENST00000368226.4 ENST00000368227.3 |
PTPRK
|
protein tyrosine phosphatase, receptor type, K |
| chr16_+_89989687 | 2.65 |
ENST00000315491.7
ENST00000555576.1 ENST00000554336.1 ENST00000553967.1 |
TUBB3
|
Tubulin beta-3 chain |
| chr6_-_109777128 | 2.55 |
ENST00000358807.3
ENST00000358577.3 |
MICAL1
|
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
| chr19_+_50354430 | 2.53 |
ENST00000599732.1
|
PTOV1
|
prostate tumor overexpressed 1 |
| chr2_+_120517174 | 2.52 |
ENST00000263708.2
|
PTPN4
|
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) |
| chrX_+_80457442 | 2.46 |
ENST00000373212.5
|
SH3BGRL
|
SH3 domain binding glutamic acid-rich protein like |
| chr1_+_172502336 | 2.45 |
ENST00000263688.3
|
SUCO
|
SUN domain containing ossification factor |
| chr13_+_111767650 | 2.42 |
ENST00000449979.1
ENST00000370623.3 |
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr12_-_8815215 | 2.39 |
ENST00000544889.1
ENST00000543369.1 |
MFAP5
|
microfibrillar associated protein 5 |
| chr17_-_53046058 | 2.36 |
ENST00000571584.1
ENST00000299335.3 |
COX11
|
cytochrome c oxidase assembly homolog 11 (yeast) |
| chr21_-_16437255 | 2.36 |
ENST00000400199.1
ENST00000400202.1 |
NRIP1
|
nuclear receptor interacting protein 1 |
| chr15_+_79166065 | 2.35 |
ENST00000559690.1
ENST00000559158.1 |
MORF4L1
|
mortality factor 4 like 1 |
| chr1_+_24286287 | 2.29 |
ENST00000334351.7
ENST00000374468.1 |
PNRC2
|
proline-rich nuclear receptor coactivator 2 |
| chr16_-_29478016 | 2.28 |
ENST00000549858.1
ENST00000551411.1 |
RP11-345J4.3
|
Uncharacterized protein |
| chr1_+_150039369 | 2.24 |
ENST00000369130.3
ENST00000369128.5 |
VPS45
|
vacuolar protein sorting 45 homolog (S. cerevisiae) |
| chr12_-_8815299 | 2.19 |
ENST00000535336.1
|
MFAP5
|
microfibrillar associated protein 5 |
| chr7_+_107224364 | 2.18 |
ENST00000491150.1
|
BCAP29
|
B-cell receptor-associated protein 29 |
| chr20_-_43150601 | 2.15 |
ENST00000541235.1
ENST00000255175.1 ENST00000342374.4 |
SERINC3
|
serine incorporator 3 |
| chr15_+_85923797 | 2.11 |
ENST00000559362.1
|
AKAP13
|
A kinase (PRKA) anchor protein 13 |
| chr11_+_46402583 | 2.09 |
ENST00000359803.3
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr20_-_39317868 | 1.93 |
ENST00000373313.2
|
MAFB
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog B |
| chr14_-_106791536 | 1.88 |
ENST00000390613.2
|
IGHV3-30
|
immunoglobulin heavy variable 3-30 |
| chr20_+_60718785 | 1.87 |
ENST00000421564.1
ENST00000450482.1 ENST00000331758.3 |
SS18L1
|
synovial sarcoma translocation gene on chromosome 18-like 1 |
| chr15_-_73076030 | 1.86 |
ENST00000311669.8
|
ADPGK
|
ADP-dependent glucokinase |
| chr1_-_21503337 | 1.83 |
ENST00000400422.1
ENST00000602326.1 ENST00000411888.1 ENST00000438975.1 |
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3 |
| chr11_+_63706444 | 1.74 |
ENST00000377793.4
ENST00000456907.2 ENST00000539656.1 |
NAA40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit |
| chr11_+_60691924 | 1.73 |
ENST00000544065.1
ENST00000453848.2 ENST00000005286.4 |
TMEM132A
|
transmembrane protein 132A |
| chr17_-_19648916 | 1.73 |
ENST00000444455.1
ENST00000439102.2 |
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr6_+_64282447 | 1.68 |
ENST00000370650.2
ENST00000578299.1 |
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr2_-_1748214 | 1.68 |
ENST00000433670.1
ENST00000425171.1 ENST00000252804.4 |
PXDN
|
peroxidasin homolog (Drosophila) |
| chr19_+_49468558 | 1.63 |
ENST00000331825.6
|
FTL
|
ferritin, light polypeptide |
| chr11_+_65339820 | 1.61 |
ENST00000316409.2
ENST00000449319.2 ENST00000530349.1 |
FAM89B
|
family with sequence similarity 89, member B |
| chr17_-_19648683 | 1.60 |
ENST00000573368.1
ENST00000457500.2 |
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr11_+_63753883 | 1.59 |
ENST00000538426.1
ENST00000543004.1 |
OTUB1
|
OTU domain, ubiquitin aldehyde binding 1 |
| chr1_-_93645818 | 1.59 |
ENST00000370280.1
ENST00000479918.1 |
TMED5
|
transmembrane emp24 protein transport domain containing 5 |
| chr3_-_18466026 | 1.55 |
ENST00000417717.2
|
SATB1
|
SATB homeobox 1 |
| chr4_+_26585686 | 1.48 |
ENST00000505206.1
ENST00000511789.1 |
TBC1D19
|
TBC1 domain family, member 19 |
| chr14_-_106866934 | 1.47 |
ENST00000390618.2
|
IGHV3-38
|
immunoglobulin heavy variable 3-38 (non-functional) |
| chr16_+_30205754 | 1.37 |
ENST00000354723.6
ENST00000355544.5 |
SULT1A3
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3 |
| chr14_-_55878538 | 1.37 |
ENST00000247178.5
|
ATG14
|
autophagy related 14 |
| chr2_-_39348137 | 1.36 |
ENST00000426016.1
|
SOS1
|
son of sevenless homolog 1 (Drosophila) |
| chr17_+_9066252 | 1.32 |
ENST00000436734.1
|
NTN1
|
netrin 1 |
| chr4_+_88896819 | 1.32 |
ENST00000237623.7
ENST00000395080.3 ENST00000508233.1 ENST00000360804.4 |
SPP1
|
secreted phosphoprotein 1 |
| chr1_+_182808474 | 1.31 |
ENST00000367549.3
|
DHX9
|
DEAH (Asp-Glu-Ala-His) box helicase 9 |
| chr16_-_22385901 | 1.28 |
ENST00000268383.2
|
CDR2
|
cerebellar degeneration-related protein 2, 62kDa |
| chr17_-_76719807 | 1.18 |
ENST00000589297.1
|
CYTH1
|
cytohesin 1 |
| chr8_+_23104130 | 1.18 |
ENST00000313219.7
ENST00000519984.1 |
CHMP7
|
charged multivesicular body protein 7 |
| chr15_+_89631381 | 1.17 |
ENST00000352732.5
|
ABHD2
|
abhydrolase domain containing 2 |
| chr12_+_25205666 | 1.17 |
ENST00000547044.1
|
LRMP
|
lymphoid-restricted membrane protein |
| chr7_+_73868439 | 1.11 |
ENST00000424337.2
|
GTF2IRD1
|
GTF2I repeat domain containing 1 |
| chr3_-_65583561 | 1.11 |
ENST00000460329.2
|
MAGI1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr15_+_63335899 | 1.11 |
ENST00000561266.1
|
TPM1
|
tropomyosin 1 (alpha) |
| chr13_+_49684445 | 1.09 |
ENST00000398316.3
|
FNDC3A
|
fibronectin type III domain containing 3A |
| chr8_+_42195972 | 1.09 |
ENST00000532157.1
ENST00000265421.4 ENST00000520008.1 |
POLB
|
polymerase (DNA directed), beta |
| chr11_+_46402744 | 1.06 |
ENST00000533952.1
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr16_+_3704822 | 1.06 |
ENST00000414110.2
|
DNASE1
|
deoxyribonuclease I |
| chr18_+_13611763 | 1.00 |
ENST00000585931.1
|
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
| chr2_-_231989808 | 1.00 |
ENST00000258400.3
|
HTR2B
|
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chr12_-_11463353 | 0.97 |
ENST00000279575.1
ENST00000535904.1 ENST00000445719.2 |
PRB4
|
proline-rich protein BstNI subfamily 4 |
| chr6_+_13272904 | 0.97 |
ENST00000379335.3
ENST00000379329.1 |
PHACTR1
|
phosphatase and actin regulator 1 |
| chr22_+_39052632 | 0.97 |
ENST00000411557.1
ENST00000396811.2 ENST00000216029.3 ENST00000416285.1 |
CBY1
|
chibby homolog 1 (Drosophila) |
| chr11_-_69633792 | 0.84 |
ENST00000334134.2
|
FGF3
|
fibroblast growth factor 3 |
| chr21_+_38445539 | 0.82 |
ENST00000418766.1
ENST00000450533.1 ENST00000438055.1 ENST00000355666.1 ENST00000540756.1 ENST00000399010.1 |
TTC3
|
tetratricopeptide repeat domain 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX6
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.1 | 20.2 | GO:1902544 | oxidative DNA demethylation(GO:0035511) regulation of DNA N-glycosylase activity(GO:1902544) |
| 4.8 | 14.4 | GO:0051086 | chaperone mediated protein folding independent of cofactor(GO:0051086) |
| 4.3 | 21.6 | GO:1904637 | response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 3.9 | 19.3 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 3.1 | 12.3 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 2.8 | 11.2 | GO:0044571 | [2Fe-2S] cluster assembly(GO:0044571) |
| 2.3 | 21.0 | GO:0021943 | formation of radial glial scaffolds(GO:0021943) |
| 2.2 | 6.6 | GO:0045659 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 2.1 | 29.1 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 2.0 | 6.1 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 2.0 | 8.1 | GO:0021564 | vagus nerve development(GO:0021564) |
| 1.9 | 20.5 | GO:0031584 | activation of phospholipase D activity(GO:0031584) apical protein localization(GO:0045176) |
| 1.7 | 5.2 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 1.6 | 8.0 | GO:0008204 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 1.4 | 5.4 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 1.4 | 9.5 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 1.2 | 4.8 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 1.2 | 4.6 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 1.1 | 7.9 | GO:1901315 | negative regulation of histone ubiquitination(GO:0033183) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) negative regulation of protein polyubiquitination(GO:1902915) |
| 1.1 | 13.7 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 1.0 | 4.2 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 1.0 | 10.4 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 1.0 | 6.2 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 1.0 | 6.9 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.9 | 2.8 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.9 | 7.4 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.9 | 2.6 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.9 | 3.4 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.8 | 5.9 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.8 | 4.9 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.7 | 4.5 | GO:1904798 | positive regulation of core promoter binding(GO:1904798) |
| 0.7 | 9.8 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.7 | 1.3 | GO:0035549 | positive regulation of interferon-beta secretion(GO:0035549) |
| 0.6 | 1.9 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.6 | 1.9 | GO:0021571 | rhombomere 5 development(GO:0021571) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.6 | 8.3 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.6 | 13.5 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.6 | 3.7 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.6 | 14.0 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.6 | 17.4 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.6 | 2.4 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.6 | 3.5 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.5 | 3.2 | GO:0010767 | regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010767) histone H3-T11 phosphorylation(GO:0035407) |
| 0.5 | 3.2 | GO:0030421 | defecation(GO:0030421) |
| 0.5 | 35.7 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.5 | 8.8 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.5 | 7.4 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.5 | 6.9 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) positive regulation of protein localization to chromosome, telomeric region(GO:1904816) |
| 0.5 | 4.5 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.5 | 6.8 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.4 | 1.3 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.4 | 1.3 | GO:0035937 | estrogen secretion(GO:0035937) estradiol secretion(GO:0035938) regulation of estrogen secretion(GO:2000861) regulation of estradiol secretion(GO:2000864) |
| 0.4 | 23.7 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.4 | 7.7 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.4 | 3.6 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.4 | 1.6 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.4 | 2.8 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.4 | 5.5 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.4 | 4.2 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.4 | 5.7 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.4 | 3.4 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.4 | 1.1 | GO:0006286 | base-excision repair, base-free sugar-phosphate removal(GO:0006286) |
| 0.4 | 6.1 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.3 | 5.6 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.3 | 4.9 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.3 | 2.2 | GO:0009597 | detection of virus(GO:0009597) |
| 0.3 | 2.8 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.3 | 2.3 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.3 | 1.1 | GO:0070997 | neuron death(GO:0070997) |
| 0.3 | 1.4 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.3 | 1.4 | GO:1904693 | midbrain morphogenesis(GO:1904693) |
| 0.3 | 2.2 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.3 | 3.3 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.3 | 8.1 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.3 | 4.4 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.3 | 3.5 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.2 | 6.6 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.2 | 1.6 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.2 | 1.1 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.2 | 1.0 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.2 | 1.4 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.2 | 2.8 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.2 | 2.4 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.2 | 0.8 | GO:0006581 | acetylcholine catabolic process in synaptic cleft(GO:0001507) acetylcholine catabolic process(GO:0006581) |
| 0.2 | 3.5 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.2 | 1.2 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.2 | 4.1 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 4.0 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 0.4 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 | 1.1 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) negative regulation of vascular associated smooth muscle cell migration(GO:1904753) |
| 0.1 | 2.5 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 0.5 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 4.4 | GO:0003016 | respiratory system process(GO:0003016) |
| 0.1 | 4.7 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.1 | 0.5 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 1.6 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 1.2 | GO:0039702 | viral budding via host ESCRT complex(GO:0039702) |
| 0.1 | 4.8 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.1 | 0.7 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.1 | 0.6 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.1 | 4.1 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.1 | 7.9 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.1 | 6.5 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 4.7 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.1 | 6.5 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.1 | 1.9 | GO:0050775 | positive regulation of dendrite morphogenesis(GO:0050775) |
| 0.1 | 1.6 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.1 | 4.1 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.1 | 0.4 | GO:1990834 | response to odorant(GO:1990834) |
| 0.1 | 2.5 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.1 | 1.6 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.8 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.1 | 8.6 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.1 | 3.6 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.1 | 0.4 | GO:0046549 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.1 | 3.0 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.1 | 1.1 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.1 | 6.0 | GO:0045445 | myoblast differentiation(GO:0045445) |
| 0.1 | 5.5 | GO:0002479 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent(GO:0002479) |
| 0.1 | 0.4 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 1.1 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.1 | 3.9 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.1 | 0.6 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 5.0 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 9.4 | GO:0010389 | regulation of G2/M transition of mitotic cell cycle(GO:0010389) |
| 0.0 | 6.2 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 2.2 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 8.0 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 3.3 | GO:0009301 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 1.9 | GO:0006735 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.7 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 2.6 | GO:0006081 | cellular aldehyde metabolic process(GO:0006081) |
| 0.0 | 1.7 | GO:0001960 | negative regulation of cytokine-mediated signaling pathway(GO:0001960) |
| 0.0 | 1.8 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.7 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 1.1 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.0 | 1.6 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.2 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.0 | 9.5 | GO:0042493 | response to drug(GO:0042493) |
| 0.0 | 0.1 | GO:0003105 | negative regulation of glomerular filtration(GO:0003105) filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) negative regulation of retinal ganglion cell axon guidance(GO:0090260) |
| 0.0 | 0.7 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.8 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 1.0 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 1.2 | GO:0006906 | vesicle fusion(GO:0006906) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 21.6 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 3.5 | 14.0 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 3.0 | 8.9 | GO:0034680 | integrin alpha1-beta1 complex(GO:0034665) integrin alpha3-beta1 complex(GO:0034667) integrin alpha8-beta1 complex(GO:0034678) integrin alpha10-beta1 complex(GO:0034680) |
| 2.5 | 7.4 | GO:0031523 | Myb complex(GO:0031523) |
| 2.1 | 17.0 | GO:0042382 | paraspeckles(GO:0042382) |
| 1.6 | 4.8 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 1.4 | 12.3 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 1.1 | 7.4 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.9 | 5.5 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.9 | 11.4 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.9 | 20.0 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.8 | 11.4 | GO:0030478 | actin cap(GO:0030478) |
| 0.7 | 8.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.7 | 35.7 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.7 | 5.5 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.7 | 4.6 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.7 | 8.6 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.6 | 3.5 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.6 | 6.1 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.6 | 7.7 | GO:0090543 | Flemming body(GO:0090543) |
| 0.5 | 1.4 | GO:1903349 | omegasome membrane(GO:1903349) |
| 0.4 | 4.7 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.4 | 3.4 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.4 | 1.6 | GO:0070288 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.4 | 4.5 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.3 | 4.5 | GO:0042555 | MCM complex(GO:0042555) |
| 0.3 | 5.2 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.3 | 18.8 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.3 | 5.9 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.3 | 2.4 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.3 | 2.1 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.3 | 9.8 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.3 | 4.8 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.3 | 4.4 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.2 | 4.7 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.2 | 3.0 | GO:0097433 | dense body(GO:0097433) |
| 0.2 | 4.7 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.2 | 5.6 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.2 | 21.8 | GO:0016605 | PML body(GO:0016605) |
| 0.2 | 0.8 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.1 | 21.6 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 5.6 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 9.4 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.1 | 7.2 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 0.4 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 2.3 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 9.9 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.1 | 27.1 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 0.5 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 0.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 6.0 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 2.5 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.1 | 14.8 | GO:1904813 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.1 | 1.1 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 1.6 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 6.9 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.1 | 9.5 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 9.5 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 8.6 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 4.8 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 3.3 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 3.7 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 4.1 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 1.6 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 4.4 | GO:0000922 | spindle pole(GO:0000922) |
| 0.1 | 0.8 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 5.4 | GO:0042641 | actomyosin(GO:0042641) |
| 0.1 | 3.2 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.1 | 13.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 3.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 15.6 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 4.0 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.6 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 13.9 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 2.6 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 4.7 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 2.8 | GO:0031256 | leading edge membrane(GO:0031256) |
| 0.0 | 8.9 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 0.3 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 1.3 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 1.8 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 2.3 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 1.9 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 9.2 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 1.7 | GO:0005819 | spindle(GO:0005819) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.7 | 20.2 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 3.5 | 14.0 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 2.0 | 6.1 | GO:0004756 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 2.0 | 8.0 | GO:0051996 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 2.0 | 6.0 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 1.9 | 9.4 | GO:0052828 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 1.9 | 7.4 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 1.9 | 5.6 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 1.8 | 8.9 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 1.4 | 4.2 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 1.0 | 5.9 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.9 | 15.7 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.9 | 22.1 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.8 | 3.2 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.8 | 7.9 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.7 | 3.5 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.7 | 35.7 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.7 | 3.3 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.6 | 4.8 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.6 | 12.3 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.5 | 15.6 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.5 | 21.6 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.5 | 4.7 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.4 | 3.4 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.4 | 7.4 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.4 | 6.9 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.4 | 4.7 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.4 | 6.6 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.4 | 14.4 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.4 | 3.5 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.4 | 9.6 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.4 | 7.9 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.4 | 7.5 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.4 | 10.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.4 | 17.4 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.3 | 1.7 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.3 | 2.8 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.3 | 1.7 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.3 | 1.3 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.3 | 3.4 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.3 | 27.8 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.3 | 1.1 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.3 | 4.1 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.2 | 1.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.2 | 5.3 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.2 | 1.4 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.2 | 0.7 | GO:0036134 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.2 | 9.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 1.9 | GO:0016773 | phosphotransferase activity, alcohol group as acceptor(GO:0016773) |
| 0.2 | 7.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.2 | 6.9 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.2 | 2.7 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.2 | 7.3 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.2 | 5.5 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.2 | 4.6 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.2 | 2.8 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.2 | 3.0 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 1.6 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.1 | 3.7 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 1.8 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.1 | 30.8 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.1 | 4.5 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.1 | 1.6 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 0.4 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.1 | 0.8 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.1 | 4.5 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.1 | 6.1 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 8.3 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 1.9 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 4.4 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 7.5 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.1 | 5.2 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.1 | 2.6 | GO:0071949 | FAD binding(GO:0071949) |
| 0.1 | 2.3 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 4.9 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.1 | 1.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 7.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 1.1 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.1 | 1.0 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.1 | 2.4 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 1.0 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 11.1 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.1 | 21.3 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 0.8 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 7.4 | GO:0004386 | helicase activity(GO:0004386) |
| 0.1 | 3.6 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 5.4 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 8.6 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 3.7 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 1.7 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 1.1 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.5 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.8 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 1.3 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 5.0 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 1.6 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.2 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.7 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 4.4 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 3.7 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 2.2 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 2.7 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 1.2 | GO:0008565 | protein transporter activity(GO:0008565) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 18.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.7 | 16.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.3 | 10.5 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.3 | 8.1 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.3 | 8.8 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.2 | 18.3 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.2 | 4.8 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.2 | 9.4 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.2 | 28.7 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.2 | 11.7 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.2 | 11.3 | PID FOXO PATHWAY | FoxO family signaling |
| 0.2 | 14.0 | PID E2F PATHWAY | E2F transcription factor network |
| 0.2 | 8.2 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.2 | 9.8 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.2 | 9.8 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.2 | 17.8 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.2 | 6.6 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 4.0 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 7.8 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 4.5 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 11.5 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.1 | 6.3 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 5.5 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.1 | 7.9 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.1 | 3.2 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 2.8 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 4.7 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 1.4 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 1.6 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 4.7 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 2.5 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.6 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.4 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 21.3 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.8 | 26.0 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.7 | 12.3 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.6 | 9.4 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.5 | 8.9 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.5 | 14.0 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.4 | 6.1 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.4 | 4.4 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.4 | 15.6 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.4 | 21.6 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.3 | 8.6 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.3 | 1.4 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.3 | 4.5 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.3 | 12.0 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.3 | 5.2 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.3 | 5.6 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.3 | 8.0 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.2 | 7.7 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.2 | 4.6 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.2 | 2.8 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.2 | 4.7 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.2 | 10.9 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.2 | 3.2 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.2 | 5.6 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.2 | 3.5 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.2 | 5.5 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.2 | 2.8 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.2 | 3.0 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.1 | 27.4 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.1 | 4.7 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 9.9 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 4.1 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 2.8 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 9.5 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 8.1 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 9.5 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 5.5 | REACTOME P53 INDEPENDENT G1 S DNA DAMAGE CHECKPOINT | Genes involved in p53-Independent G1/S DNA damage checkpoint |
| 0.1 | 1.0 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 1.4 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.8 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.0 | 1.9 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 2.1 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 1.6 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 1.6 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 2.2 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.7 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |