Project
averaged GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
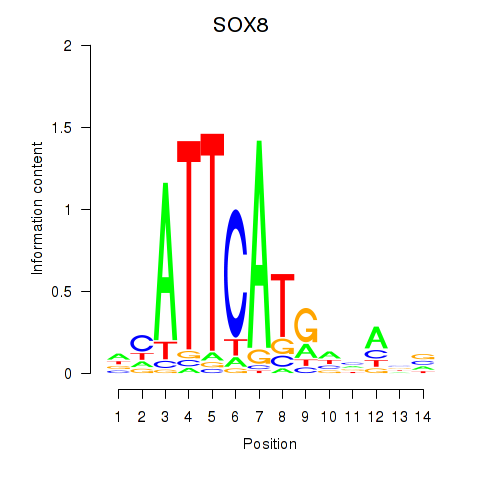
Results for SOX8
Z-value: 1.00
Transcription factors associated with SOX8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX8
|
ENSG00000005513.9 | SRY-box transcription factor 8 |
Activity profile of SOX8 motif
Sorted Z-values of SOX8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_153330322 | 15.51 |
ENST00000368738.3
|
S100A9
|
S100 calcium binding protein A9 |
| chr2_+_233527443 | 12.14 |
ENST00000410095.1
|
EFHD1
|
EF-hand domain family, member D1 |
| chr8_-_82359662 | 10.49 |
ENST00000519260.1
ENST00000256103.2 |
PMP2
|
peripheral myelin protein 2 |
| chr12_-_15104040 | 9.86 |
ENST00000541644.1
ENST00000545895.1 |
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr2_-_175711133 | 9.68 |
ENST00000409597.1
ENST00000413882.1 |
CHN1
|
chimerin 1 |
| chr6_-_32498046 | 8.56 |
ENST00000374975.3
|
HLA-DRB5
|
major histocompatibility complex, class II, DR beta 5 |
| chr14_-_106642049 | 8.40 |
ENST00000390605.2
|
IGHV1-18
|
immunoglobulin heavy variable 1-18 |
| chr14_+_75745477 | 8.32 |
ENST00000303562.4
ENST00000554617.1 ENST00000554212.1 ENST00000535987.1 ENST00000555242.1 |
FOS
|
FBJ murine osteosarcoma viral oncogene homolog |
| chr16_+_31271274 | 8.31 |
ENST00000287497.8
ENST00000544665.3 |
ITGAM
|
integrin, alpha M (complement component 3 receptor 3 subunit) |
| chr6_+_32605195 | 7.80 |
ENST00000374949.2
|
HLA-DQA1
|
major histocompatibility complex, class II, DQ alpha 1 |
| chr12_-_15103621 | 7.79 |
ENST00000536592.1
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr12_-_11548496 | 7.08 |
ENST00000389362.4
ENST00000565533.1 ENST00000546254.1 |
PRB2
PRB1
|
proline-rich protein BstNI subfamily 2 proline-rich protein BstNI subfamily 1 |
| chr1_+_111415757 | 6.76 |
ENST00000429072.2
ENST00000271324.5 |
CD53
|
CD53 molecule |
| chr3_-_39322728 | 6.74 |
ENST00000541347.1
ENST00000412814.1 |
CX3CR1
|
chemokine (C-X3-C motif) receptor 1 |
| chr22_-_50523760 | 6.72 |
ENST00000395876.2
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr12_-_10282836 | 6.65 |
ENST00000304084.8
ENST00000353231.5 ENST00000525605.1 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr8_-_120605194 | 6.16 |
ENST00000522167.1
|
ENPP2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr5_-_179499108 | 5.96 |
ENST00000521389.1
|
RNF130
|
ring finger protein 130 |
| chr7_+_80275752 | 5.92 |
ENST00000419819.2
|
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr11_-_125366089 | 5.53 |
ENST00000366139.3
ENST00000278919.3 |
FEZ1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr5_-_179499086 | 5.53 |
ENST00000261947.4
|
RNF130
|
ring finger protein 130 |
| chr5_-_160279207 | 5.51 |
ENST00000327245.5
|
ATP10B
|
ATPase, class V, type 10B |
| chr2_+_219646462 | 5.31 |
ENST00000258415.4
|
CYP27A1
|
cytochrome P450, family 27, subfamily A, polypeptide 1 |
| chr1_+_20959943 | 5.28 |
ENST00000321556.4
|
PINK1
|
PTEN induced putative kinase 1 |
| chr6_+_32605134 | 5.14 |
ENST00000343139.5
ENST00000395363.1 ENST00000496318.1 |
HLA-DQA1
|
major histocompatibility complex, class II, DQ alpha 1 |
| chr14_+_95078714 | 5.04 |
ENST00000393078.3
ENST00000393080.4 ENST00000467132.1 |
SERPINA3
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 |
| chr8_+_21915368 | 4.86 |
ENST00000265800.5
ENST00000517418.1 |
DMTN
|
dematin actin binding protein |
| chr15_+_25101698 | 4.73 |
ENST00000400097.1
|
SNRPN
|
small nuclear ribonucleoprotein polypeptide N |
| chr11_+_128563652 | 4.69 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr1_-_38512450 | 4.53 |
ENST00000373012.2
|
POU3F1
|
POU class 3 homeobox 1 |
| chr4_+_155484155 | 4.38 |
ENST00000509493.1
|
FGB
|
fibrinogen beta chain |
| chr2_+_89986318 | 4.36 |
ENST00000491977.1
|
IGKV2D-29
|
immunoglobulin kappa variable 2D-29 |
| chr1_-_153029980 | 4.27 |
ENST00000392653.2
|
SPRR2A
|
small proline-rich protein 2A |
| chr7_+_80275621 | 4.23 |
ENST00000426978.1
ENST00000432207.1 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr12_-_10007448 | 4.20 |
ENST00000538152.1
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr14_-_106967788 | 4.18 |
ENST00000390622.2
|
IGHV1-46
|
immunoglobulin heavy variable 1-46 |
| chr1_-_200992827 | 4.07 |
ENST00000332129.2
ENST00000422435.2 |
KIF21B
|
kinesin family member 21B |
| chr8_-_86253888 | 4.07 |
ENST00000522389.1
ENST00000432364.2 ENST00000517618.1 |
CA1
|
carbonic anhydrase I |
| chr1_+_161475208 | 4.06 |
ENST00000367972.4
ENST00000271450.6 |
FCGR2A
|
Fc fragment of IgG, low affinity IIa, receptor (CD32) |
| chr7_+_139478030 | 4.04 |
ENST00000425687.1
ENST00000263552.6 ENST00000438104.1 ENST00000336425.5 |
TBXAS1
|
thromboxane A synthase 1 (platelet) |
| chr9_-_113761720 | 4.02 |
ENST00000541779.1
ENST00000374430.2 |
LPAR1
|
lysophosphatidic acid receptor 1 |
| chr19_+_1269324 | 3.91 |
ENST00000589710.1
ENST00000588230.1 ENST00000413636.2 ENST00000586472.1 ENST00000589686.1 ENST00000444172.2 ENST00000587323.1 ENST00000320936.5 ENST00000587896.1 ENST00000589235.1 ENST00000591659.1 |
CIRBP
|
cold inducible RNA binding protein |
| chr18_-_77711625 | 3.89 |
ENST00000357575.4
ENST00000590381.1 ENST00000397778.2 |
PQLC1
|
PQ loop repeat containing 1 |
| chr10_+_7745232 | 3.89 |
ENST00000358415.4
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr7_+_79998864 | 3.77 |
ENST00000435819.1
|
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr2_+_102615416 | 3.73 |
ENST00000393414.2
|
IL1R2
|
interleukin 1 receptor, type II |
| chr5_+_140186647 | 3.71 |
ENST00000512229.2
ENST00000356878.4 ENST00000530339.1 |
PCDHA4
|
protocadherin alpha 4 |
| chr11_+_128563948 | 3.70 |
ENST00000534087.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chrX_+_15767971 | 3.67 |
ENST00000479740.1
ENST00000454127.2 |
CA5B
|
carbonic anhydrase VB, mitochondrial |
| chr16_+_30075783 | 3.58 |
ENST00000412304.2
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr19_+_19144384 | 3.57 |
ENST00000392335.2
ENST00000535612.1 ENST00000537263.1 ENST00000540707.1 ENST00000541725.1 ENST00000269932.6 ENST00000546344.1 ENST00000540792.1 ENST00000536098.1 ENST00000541898.1 ENST00000543877.1 |
ARMC6
|
armadillo repeat containing 6 |
| chr15_+_80733570 | 3.57 |
ENST00000533983.1
ENST00000527771.1 ENST00000525103.1 |
ARNT2
|
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr4_+_40192656 | 3.55 |
ENST00000505618.1
|
RHOH
|
ras homolog family member H |
| chr12_-_11508520 | 3.51 |
ENST00000545626.1
ENST00000500254.2 |
PRB1
|
proline-rich protein BstNI subfamily 1 |
| chr14_-_107283278 | 3.43 |
ENST00000390639.2
|
IGHV7-81
|
immunoglobulin heavy variable 7-81 (non-functional) |
| chr22_-_42466782 | 3.31 |
ENST00000396398.3
ENST00000403363.1 ENST00000402937.1 |
NAGA
|
N-acetylgalactosaminidase, alpha- |
| chr19_-_45681482 | 3.28 |
ENST00000592647.1
ENST00000006275.4 ENST00000588062.1 ENST00000585934.1 |
TRAPPC6A
|
trafficking protein particle complex 6A |
| chr19_+_51728316 | 3.21 |
ENST00000436584.2
ENST00000421133.2 ENST00000391796.3 ENST00000262262.4 |
CD33
|
CD33 molecule |
| chr15_+_43985084 | 3.12 |
ENST00000434505.1
ENST00000411750.1 |
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr6_-_32160622 | 3.12 |
ENST00000487761.1
ENST00000375040.3 |
GPSM3
|
G-protein signaling modulator 3 |
| chr19_+_18794470 | 3.09 |
ENST00000321949.8
ENST00000338797.6 |
CRTC1
|
CREB regulated transcription coactivator 1 |
| chr7_-_80551671 | 3.06 |
ENST00000419255.2
ENST00000544525.1 |
SEMA3C
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr1_-_203274418 | 2.94 |
ENST00000457348.1
|
RP11-134P9.1
|
long intergenic non-protein coding RNA 1136 |
| chr18_+_61442629 | 2.94 |
ENST00000398019.2
ENST00000540675.1 |
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr17_-_34207295 | 2.92 |
ENST00000463941.1
ENST00000293272.3 |
CCL5
|
chemokine (C-C motif) ligand 5 |
| chr12_-_123201337 | 2.87 |
ENST00000528880.2
|
HCAR3
|
hydroxycarboxylic acid receptor 3 |
| chr15_-_89755034 | 2.84 |
ENST00000563254.1
|
RLBP1
|
retinaldehyde binding protein 1 |
| chr9_-_116837249 | 2.81 |
ENST00000466610.2
|
AMBP
|
alpha-1-microglobulin/bikunin precursor |
| chr2_-_89521942 | 2.79 |
ENST00000482769.1
|
IGKV2-28
|
immunoglobulin kappa variable 2-28 |
| chr7_-_36634181 | 2.78 |
ENST00000538464.1
|
AOAH
|
acyloxyacyl hydrolase (neutrophil) |
| chr4_+_155484103 | 2.64 |
ENST00000302068.4
|
FGB
|
fibrinogen beta chain |
| chr3_-_49459865 | 2.64 |
ENST00000427987.1
|
AMT
|
aminomethyltransferase |
| chr3_+_137906154 | 2.63 |
ENST00000466749.1
ENST00000358441.2 ENST00000489213.1 |
ARMC8
|
armadillo repeat containing 8 |
| chr1_+_152956549 | 2.59 |
ENST00000307122.2
|
SPRR1A
|
small proline-rich protein 1A |
| chr3_-_49459878 | 2.57 |
ENST00000546031.1
ENST00000458307.2 ENST00000430521.1 |
AMT
|
aminomethyltransferase |
| chr1_+_13516066 | 2.55 |
ENST00000332192.6
|
PRAMEF21
|
PRAME family member 21 |
| chr15_+_43885252 | 2.53 |
ENST00000453782.1
ENST00000300283.6 ENST00000437924.1 ENST00000450086.2 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr22_+_24236191 | 2.52 |
ENST00000215754.7
|
MIF
|
macrophage migration inhibitory factor (glycosylation-inhibiting factor) |
| chr14_-_107049312 | 2.49 |
ENST00000390627.2
|
IGHV3-53
|
immunoglobulin heavy variable 3-53 |
| chr4_+_156680153 | 2.46 |
ENST00000502959.1
ENST00000505764.1 ENST00000507146.1 ENST00000264424.8 ENST00000503520.1 |
GUCY1B3
|
guanylate cyclase 1, soluble, beta 3 |
| chr6_+_37012607 | 2.44 |
ENST00000423336.1
|
COX6A1P2
|
cytochrome c oxidase subunit VIa polypeptide 1 pseudogene 2 |
| chr14_-_106733624 | 2.40 |
ENST00000390610.2
|
IGHV1-24
|
immunoglobulin heavy variable 1-24 |
| chr7_-_38289173 | 2.36 |
ENST00000436911.2
|
TRGC2
|
T cell receptor gamma constant 2 |
| chr5_+_140227048 | 2.32 |
ENST00000532602.1
|
PCDHA9
|
protocadherin alpha 9 |
| chr16_+_30075595 | 2.30 |
ENST00000563060.2
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr17_+_2240916 | 2.28 |
ENST00000574563.1
|
SGSM2
|
small G protein signaling modulator 2 |
| chr11_+_125774362 | 2.28 |
ENST00000530414.1
ENST00000530129.2 |
DDX25
|
DEAD (Asp-Glu-Ala-Asp) box helicase 25 |
| chr1_-_9811600 | 2.26 |
ENST00000435891.1
|
CLSTN1
|
calsyntenin 1 |
| chr17_-_8021710 | 2.23 |
ENST00000380149.1
ENST00000448843.2 |
ALOXE3
|
arachidonate lipoxygenase 3 |
| chr16_+_57769635 | 2.23 |
ENST00000379661.3
ENST00000562592.1 ENST00000566726.1 |
KATNB1
|
katanin p80 (WD repeat containing) subunit B 1 |
| chr22_-_36903069 | 2.22 |
ENST00000216187.6
ENST00000423980.1 |
FOXRED2
|
FAD-dependent oxidoreductase domain containing 2 |
| chr8_+_1993152 | 2.09 |
ENST00000262113.4
|
MYOM2
|
myomesin 2 |
| chr16_+_87985029 | 2.08 |
ENST00000439677.1
ENST00000286122.7 ENST00000355163.5 ENST00000454563.1 ENST00000479780.2 ENST00000393208.2 ENST00000412691.1 ENST00000355022.4 |
BANP
|
BTG3 associated nuclear protein |
| chr8_-_91095099 | 2.08 |
ENST00000265431.3
|
CALB1
|
calbindin 1, 28kDa |
| chr6_+_31707725 | 2.06 |
ENST00000375755.3
ENST00000375742.3 ENST00000375750.3 ENST00000425703.1 ENST00000534153.4 ENST00000375703.3 ENST00000375740.3 |
MSH5
|
mutS homolog 5 |
| chr1_+_192605252 | 2.05 |
ENST00000391995.2
ENST00000543215.1 |
RGS13
|
regulator of G-protein signaling 13 |
| chr6_-_133079022 | 2.05 |
ENST00000525289.1
ENST00000326499.6 |
VNN2
|
vanin 2 |
| chr16_+_30075463 | 2.03 |
ENST00000562168.1
ENST00000569545.1 |
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr15_-_20170354 | 2.03 |
ENST00000338912.5
|
IGHV1OR15-9
|
immunoglobulin heavy variable 1/OR15-9 (non-functional) |
| chr13_-_47012325 | 2.02 |
ENST00000409879.2
|
KIAA0226L
|
KIAA0226-like |
| chr5_-_94417339 | 1.99 |
ENST00000429576.2
ENST00000508509.1 ENST00000510732.1 |
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr15_+_34261089 | 1.96 |
ENST00000383263.5
|
CHRM5
|
cholinergic receptor, muscarinic 5 |
| chr2_+_113816215 | 1.89 |
ENST00000346807.3
|
IL36RN
|
interleukin 36 receptor antagonist |
| chr18_+_71815743 | 1.89 |
ENST00000169551.6
ENST00000580087.1 |
TIMM21
|
translocase of inner mitochondrial membrane 21 homolog (yeast) |
| chr12_-_123187890 | 1.89 |
ENST00000328880.5
|
HCAR2
|
hydroxycarboxylic acid receptor 2 |
| chr5_+_140792614 | 1.88 |
ENST00000398610.2
|
PCDHGA10
|
protocadherin gamma subfamily A, 10 |
| chr1_+_158901329 | 1.87 |
ENST00000368140.1
ENST00000368138.3 ENST00000392254.2 ENST00000392252.3 ENST00000368135.4 |
PYHIN1
|
pyrin and HIN domain family, member 1 |
| chr18_+_11857439 | 1.81 |
ENST00000602628.1
|
GNAL
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr9_-_139891165 | 1.81 |
ENST00000494426.1
|
CLIC3
|
chloride intracellular channel 3 |
| chr3_+_137906353 | 1.79 |
ENST00000461822.1
ENST00000485396.1 ENST00000471453.1 ENST00000470821.1 ENST00000471709.1 ENST00000538260.1 ENST00000393058.3 ENST00000463485.1 |
ARMC8
|
armadillo repeat containing 8 |
| chr4_+_156680143 | 1.77 |
ENST00000505154.1
|
GUCY1B3
|
guanylate cyclase 1, soluble, beta 3 |
| chrX_+_100474906 | 1.76 |
ENST00000541709.1
|
DRP2
|
dystrophin related protein 2 |
| chr18_-_3845321 | 1.73 |
ENST00000539435.1
ENST00000400147.2 |
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1 |
| chr8_+_107738240 | 1.72 |
ENST00000449762.2
ENST00000297447.6 |
OXR1
|
oxidation resistance 1 |
| chr8_+_1993173 | 1.70 |
ENST00000523438.1
|
MYOM2
|
myomesin 2 |
| chr10_-_1779663 | 1.69 |
ENST00000381312.1
|
ADARB2
|
adenosine deaminase, RNA-specific, B2 (non-functional) |
| chr6_-_136788001 | 1.69 |
ENST00000544465.1
|
MAP7
|
microtubule-associated protein 7 |
| chr4_-_47983519 | 1.65 |
ENST00000358519.4
ENST00000544810.1 ENST00000402813.3 |
CNGA1
|
cyclic nucleotide gated channel alpha 1 |
| chr10_-_74114714 | 1.63 |
ENST00000338820.3
ENST00000394903.2 ENST00000444643.2 |
DNAJB12
|
DnaJ (Hsp40) homolog, subfamily B, member 12 |
| chr3_+_137906109 | 1.57 |
ENST00000481646.1
ENST00000469044.1 ENST00000491704.1 ENST00000461600.1 |
ARMC8
|
armadillo repeat containing 8 |
| chr12_+_29376673 | 1.51 |
ENST00000547116.1
|
FAR2
|
fatty acyl CoA reductase 2 |
| chr1_-_13673511 | 1.48 |
ENST00000344998.3
ENST00000334600.6 |
PRAMEF14
|
PRAME family member 14 |
| chr3_+_38347427 | 1.48 |
ENST00000273173.4
|
SLC22A14
|
solute carrier family 22, member 14 |
| chr1_-_12958101 | 1.47 |
ENST00000235347.4
|
PRAMEF10
|
PRAME family member 10 |
| chr12_-_9760482 | 1.45 |
ENST00000229402.3
|
KLRB1
|
killer cell lectin-like receptor subfamily B, member 1 |
| chr17_-_66951474 | 1.44 |
ENST00000269080.2
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr17_-_73663245 | 1.42 |
ENST00000584999.1
ENST00000317905.5 ENST00000420326.2 ENST00000340830.5 |
RECQL5
|
RecQ protein-like 5 |
| chr11_-_57177586 | 1.42 |
ENST00000529411.1
|
RP11-872D17.8
|
Uncharacterized protein |
| chr3_-_186288097 | 1.40 |
ENST00000446782.1
|
TBCCD1
|
TBCC domain containing 1 |
| chr22_-_29784519 | 1.35 |
ENST00000357586.2
ENST00000356015.2 ENST00000432560.2 ENST00000317368.7 |
AP1B1
|
adaptor-related protein complex 1, beta 1 subunit |
| chr18_-_53253323 | 1.33 |
ENST00000540999.1
ENST00000563888.2 |
TCF4
|
transcription factor 4 |
| chr4_-_144940477 | 1.33 |
ENST00000513128.1
ENST00000429670.2 ENST00000502664.1 |
GYPB
|
glycophorin B (MNS blood group) |
| chr4_+_95972822 | 1.32 |
ENST00000509540.1
ENST00000440890.2 |
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr12_-_10282742 | 1.31 |
ENST00000298523.5
ENST00000396484.2 ENST00000310002.4 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr3_-_21792838 | 1.29 |
ENST00000281523.2
|
ZNF385D
|
zinc finger protein 385D |
| chr18_-_3845293 | 1.27 |
ENST00000400145.2
|
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1 |
| chr15_+_62853562 | 1.26 |
ENST00000561311.1
|
TLN2
|
talin 2 |
| chr2_-_228497888 | 1.25 |
ENST00000264387.4
ENST00000409066.1 |
C2orf83
|
chromosome 2 open reading frame 83 |
| chr5_+_140800638 | 1.24 |
ENST00000398587.2
ENST00000518882.1 |
PCDHGA11
|
protocadherin gamma subfamily A, 11 |
| chr6_-_138539627 | 1.24 |
ENST00000527246.2
|
PBOV1
|
prostate and breast cancer overexpressed 1 |
| chr3_+_113465866 | 1.23 |
ENST00000273398.3
ENST00000538620.1 ENST00000496747.1 ENST00000475322.1 |
ATP6V1A
|
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A |
| chr11_-_129062093 | 1.23 |
ENST00000310343.9
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr6_-_49712147 | 1.23 |
ENST00000433368.2
ENST00000354620.4 |
CRISP3
|
cysteine-rich secretory protein 3 |
| chr4_+_71248795 | 1.20 |
ENST00000304915.3
|
SMR3B
|
submaxillary gland androgen regulated protein 3B |
| chr18_+_32173276 | 1.19 |
ENST00000591816.1
ENST00000588125.1 ENST00000598334.1 ENST00000588684.1 ENST00000554864.3 ENST00000399121.5 ENST00000595022.1 ENST00000269190.7 ENST00000399097.3 |
DTNA
|
dystrobrevin, alpha |
| chr14_-_106471723 | 1.18 |
ENST00000390595.2
|
IGHV1-3
|
immunoglobulin heavy variable 1-3 |
| chrX_-_9734004 | 1.14 |
ENST00000467482.1
ENST00000380929.2 |
GPR143
|
G protein-coupled receptor 143 |
| chr18_-_5396271 | 1.14 |
ENST00000579951.1
|
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3 |
| chr10_-_101825151 | 1.13 |
ENST00000441382.1
|
CPN1
|
carboxypeptidase N, polypeptide 1 |
| chr12_+_29376592 | 1.12 |
ENST00000182377.4
|
FAR2
|
fatty acyl CoA reductase 2 |
| chr20_+_32150140 | 1.12 |
ENST00000344201.3
ENST00000346541.3 ENST00000397800.1 ENST00000397798.2 ENST00000492345.1 |
CBFA2T2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2 |
| chr4_-_38858428 | 1.12 |
ENST00000436693.2
ENST00000508254.1 ENST00000514655.1 ENST00000506146.1 |
TLR6
TLR1
|
toll-like receptor 6 toll-like receptor 1 |
| chr1_+_13742808 | 1.11 |
ENST00000602960.1
|
PRAMEF20
|
PRAME family member 20 |
| chr15_-_58306295 | 1.11 |
ENST00000559517.1
|
ALDH1A2
|
aldehyde dehydrogenase 1 family, member A2 |
| chr16_+_30387141 | 1.11 |
ENST00000566955.1
|
MYLPF
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chrX_-_132887729 | 1.08 |
ENST00000406757.2
|
GPC3
|
glypican 3 |
| chr3_-_54962100 | 1.04 |
ENST00000273286.5
|
LRTM1
|
leucine-rich repeats and transmembrane domains 1 |
| chr17_+_1646130 | 1.04 |
ENST00000453066.1
ENST00000324015.3 ENST00000450523.2 ENST00000453723.1 ENST00000382061.4 |
SERPINF2
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2 |
| chr14_+_88471468 | 1.03 |
ENST00000267549.3
|
GPR65
|
G protein-coupled receptor 65 |
| chr6_-_22297730 | 1.03 |
ENST00000306482.1
|
PRL
|
prolactin |
| chr12_-_10151773 | 1.03 |
ENST00000298527.6
ENST00000348658.4 |
CLEC1B
|
C-type lectin domain family 1, member B |
| chr5_-_160973649 | 1.02 |
ENST00000393959.1
ENST00000517547.1 |
GABRB2
|
gamma-aminobutyric acid (GABA) A receptor, beta 2 |
| chr17_-_34313685 | 1.00 |
ENST00000435911.2
ENST00000586216.1 ENST00000394509.4 |
CCL14
|
chemokine (C-C motif) ligand 14 |
| chr3_-_58196939 | 0.98 |
ENST00000394549.2
ENST00000461914.3 |
DNASE1L3
|
deoxyribonuclease I-like 3 |
| chr5_+_140739537 | 0.98 |
ENST00000522605.1
|
PCDHGB2
|
protocadherin gamma subfamily B, 2 |
| chr18_-_53253112 | 0.98 |
ENST00000568673.1
ENST00000562847.1 ENST00000568147.1 |
TCF4
|
transcription factor 4 |
| chr6_+_63921351 | 0.97 |
ENST00000370659.1
|
FKBP1C
|
FK506 binding protein 1C |
| chr7_-_14942283 | 0.96 |
ENST00000402815.1
|
DGKB
|
diacylglycerol kinase, beta 90kDa |
| chr16_-_30122717 | 0.95 |
ENST00000566613.1
|
GDPD3
|
glycerophosphodiester phosphodiesterase domain containing 3 |
| chr14_+_22337014 | 0.95 |
ENST00000390436.2
|
TRAV13-1
|
T cell receptor alpha variable 13-1 |
| chr19_+_22235279 | 0.95 |
ENST00000594363.1
ENST00000597927.1 ENST00000594947.1 |
ZNF257
|
zinc finger protein 257 |
| chr4_-_69434245 | 0.94 |
ENST00000317746.2
|
UGT2B17
|
UDP glucuronosyltransferase 2 family, polypeptide B17 |
| chr7_+_151771377 | 0.94 |
ENST00000434507.1
|
GALNT11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) |
| chr1_+_70876926 | 0.94 |
ENST00000370938.3
ENST00000346806.2 |
CTH
|
cystathionase (cystathionine gamma-lyase) |
| chr5_-_138534071 | 0.93 |
ENST00000394817.2
|
SIL1
|
SIL1 nucleotide exchange factor |
| chr9_+_2157655 | 0.92 |
ENST00000452193.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr22_-_50708781 | 0.90 |
ENST00000449719.2
ENST00000330651.6 |
MAPK11
|
mitogen-activated protein kinase 11 |
| chr4_-_100356551 | 0.89 |
ENST00000209665.4
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr1_+_87012753 | 0.88 |
ENST00000370563.3
|
CLCA4
|
chloride channel accessory 4 |
| chr12_+_56325812 | 0.87 |
ENST00000394147.1
ENST00000551156.1 ENST00000553783.1 ENST00000557080.1 ENST00000432422.3 ENST00000556001.1 |
DGKA
|
diacylglycerol kinase, alpha 80kDa |
| chr3_-_49851313 | 0.86 |
ENST00000333486.3
|
UBA7
|
ubiquitin-like modifier activating enzyme 7 |
| chr16_-_90096309 | 0.86 |
ENST00000408886.2
|
C16orf3
|
chromosome 16 open reading frame 3 |
| chr5_+_140753444 | 0.86 |
ENST00000517434.1
|
PCDHGA6
|
protocadherin gamma subfamily A, 6 |
| chr18_-_11148587 | 0.86 |
ENST00000302079.6
ENST00000580640.1 ENST00000503781.3 |
PIEZO2
|
piezo-type mechanosensitive ion channel component 2 |
| chr4_-_145061788 | 0.85 |
ENST00000512064.1
ENST00000512789.1 ENST00000504786.1 ENST00000503627.1 ENST00000535709.1 ENST00000324022.10 ENST00000360771.4 ENST00000283126.7 |
GYPA
GYPB
|
glycophorin A (MNS blood group) glycophorin B (MNS blood group) |
| chr16_+_87636474 | 0.83 |
ENST00000284262.2
|
JPH3
|
junctophilin 3 |
| chr5_-_132948216 | 0.83 |
ENST00000265342.7
|
FSTL4
|
follistatin-like 4 |
| chr6_-_49712123 | 0.82 |
ENST00000263045.4
|
CRISP3
|
cysteine-rich secretory protein 3 |
| chr6_+_27100811 | 0.81 |
ENST00000359193.2
|
HIST1H2AG
|
histone cluster 1, H2ag |
| chr12_+_113354341 | 0.81 |
ENST00000553152.1
|
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr3_-_52090461 | 0.79 |
ENST00000296483.6
ENST00000495880.1 |
DUSP7
|
dual specificity phosphatase 7 |
| chr11_+_74862140 | 0.78 |
ENST00000525650.1
ENST00000454962.2 |
SLCO2B1
|
solute carrier organic anion transporter family, member 2B1 |
| chr3_-_58196688 | 0.78 |
ENST00000486455.1
|
DNASE1L3
|
deoxyribonuclease I-like 3 |
| chr8_+_38758737 | 0.78 |
ENST00000521746.1
ENST00000420274.1 |
PLEKHA2
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 2 |
| chr2_-_207024233 | 0.77 |
ENST00000423725.1
ENST00000233190.6 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr18_-_51750948 | 0.72 |
ENST00000583046.1
ENST00000398398.2 |
MBD2
|
methyl-CpG binding domain protein 2 |
| chr1_-_109935819 | 0.71 |
ENST00000538502.1
|
SORT1
|
sortilin 1 |
| chr2_+_166326157 | 0.70 |
ENST00000421875.1
ENST00000314499.7 ENST00000409664.1 |
CSRNP3
|
cysteine-serine-rich nuclear protein 3 |
| chr11_+_60995849 | 0.69 |
ENST00000537932.1
|
PGA4
|
pepsinogen 4, group I (pepsinogen A) |
| chr1_-_67142710 | 0.67 |
ENST00000502413.2
|
AL139147.1
|
Uncharacterized protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX8
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.2 | 15.5 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 4.4 | 17.6 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 2.2 | 6.7 | GO:0002881 | negative regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002881) |
| 2.0 | 13.9 | GO:2000334 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 1.8 | 10.8 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 1.4 | 4.2 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 1.0 | 5.2 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 1.0 | 8.3 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 1.0 | 3.1 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 1.0 | 4.0 | GO:1904566 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.9 | 3.7 | GO:0050712 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.9 | 2.6 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.8 | 4.9 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.8 | 3.8 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.7 | 2.9 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.7 | 2.9 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.7 | 7.0 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.6 | 8.3 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.6 | 2.2 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.5 | 6.7 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.5 | 3.1 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.5 | 2.5 | GO:0002905 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.5 | 1.4 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.5 | 3.3 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.5 | 1.4 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.5 | 0.9 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.4 | 7.9 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.4 | 1.3 | GO:0015888 | thiamine transport(GO:0015888) thiamine transmembrane transport(GO:0071934) |
| 0.4 | 3.3 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.4 | 6.2 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.4 | 25.4 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.4 | 2.0 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.4 | 8.5 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.4 | 1.9 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.4 | 1.1 | GO:0070340 | detection of bacterial lipoprotein(GO:0042494) detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) |
| 0.4 | 2.2 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.3 | 3.9 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.3 | 1.9 | GO:1902714 | negative regulation of interferon-gamma secretion(GO:1902714) |
| 0.3 | 0.9 | GO:0099543 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.3 | 0.9 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.3 | 6.8 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.3 | 1.1 | GO:0043324 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) regulation of melanosome transport(GO:1902908) |
| 0.3 | 3.1 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.3 | 0.8 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.3 | 9.7 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.3 | 1.1 | GO:0072180 | mesonephric duct morphogenesis(GO:0072180) |
| 0.3 | 1.3 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.3 | 3.1 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.3 | 2.1 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 0.3 | 5.0 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.2 | 4.0 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.2 | 0.9 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.2 | 3.9 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.2 | 2.1 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.2 | 1.1 | GO:0030070 | insulin processing(GO:0030070) |
| 0.2 | 1.0 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.2 | 0.6 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.2 | 2.0 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.2 | 5.5 | GO:0034204 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.2 | 0.9 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.2 | 2.8 | GO:0018298 | porphyrin-containing compound catabolic process(GO:0006787) protein-chromophore linkage(GO:0018298) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.2 | 19.6 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.2 | 1.1 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.2 | 1.4 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.2 | 0.5 | GO:0001172 | transcription, RNA-templated(GO:0001172) |
| 0.2 | 4.5 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.1 | 1.0 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.1 | 1.9 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.1 | 0.6 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.1 | 1.7 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.1 | 0.4 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.1 | 7.7 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 1.6 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 0.7 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.1 | 0.9 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 9.5 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.1 | 1.8 | GO:0010623 | programmed cell death involved in cell development(GO:0010623) |
| 0.1 | 0.3 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.1 | 1.3 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.1 | 1.8 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.1 | 0.5 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) positive regulation of t-circle formation(GO:1904431) |
| 0.1 | 4.6 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 2.0 | GO:0007163 | establishment or maintenance of cell polarity(GO:0007163) |
| 0.1 | 4.2 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 1.2 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.1 | 0.4 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.1 | 3.5 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.1 | 2.3 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 0.5 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.1 | 0.9 | GO:0050974 | detection of mechanical stimulus involved in sensory perception(GO:0050974) |
| 0.1 | 8.6 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 1.0 | GO:0030220 | platelet formation(GO:0030220) |
| 0.1 | 1.0 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.1 | 1.1 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.1 | 1.5 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 1.0 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 2.1 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 | 0.5 | GO:0060371 | regulation of atrial cardiac muscle cell membrane depolarization(GO:0060371) |
| 0.0 | 0.7 | GO:0006346 | chromatin silencing at rDNA(GO:0000183) methylation-dependent chromatin silencing(GO:0006346) |
| 0.0 | 1.0 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 1.9 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 0.2 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.0 | 1.4 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 2.3 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 4.1 | GO:0038096 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 0.0 | 0.1 | GO:0001694 | histamine biosynthetic process(GO:0001694) |
| 0.0 | 0.5 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.3 | GO:0070836 | plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 0.8 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.0 | 4.4 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.1 | GO:0036337 | Fas signaling pathway(GO:0036337) regulation of Fas signaling pathway(GO:1902044) |
| 0.0 | 2.1 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.4 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 2.6 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 2.2 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.8 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.4 | GO:0010804 | negative regulation of tumor necrosis factor-mediated signaling pathway(GO:0010804) |
| 0.0 | 0.1 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 3.2 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.0 | 0.1 | GO:0035947 | regulation of gluconeogenesis by regulation of transcription from RNA polymerase II promoter(GO:0035947) positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.0 | 0.4 | GO:0044705 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.0 | 0.6 | GO:0007183 | SMAD protein complex assembly(GO:0007183) |
| 0.0 | 0.1 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) Sertoli cell proliferation(GO:0060011) |
| 0.0 | 1.0 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.2 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.3 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.2 | GO:0045408 | regulation of interleukin-6 biosynthetic process(GO:0045408) |
| 0.0 | 1.7 | GO:0007268 | chemical synaptic transmission(GO:0007268) anterograde trans-synaptic signaling(GO:0098916) trans-synaptic signaling(GO:0099537) |
| 0.0 | 0.9 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.4 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.2 | GO:0090331 | negative regulation of platelet aggregation(GO:0090331) |
| 0.0 | 0.9 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.2 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.8 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 | 1.3 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.5 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.0 | 0.3 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.8 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 2.6 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 0.2 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.9 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 1.8 | GO:0050808 | synapse organization(GO:0050808) |
| 0.0 | 0.7 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 8.3 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 1.0 | 8.3 | GO:0035976 | AP1 complex(GO:0035976) |
| 1.0 | 21.5 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.8 | 5.3 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.6 | 8.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.4 | 13.9 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.4 | 1.1 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.3 | 17.4 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.3 | 4.9 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.3 | 4.7 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.3 | 3.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.2 | 6.9 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.2 | 11.7 | GO:0031430 | M band(GO:0031430) |
| 0.2 | 0.8 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.2 | 2.3 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.2 | 4.2 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.2 | 1.9 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.2 | 0.8 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.1 | 2.1 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.1 | 7.0 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 1.6 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 2.6 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 1.3 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 1.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 4.6 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 4.9 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 1.4 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 4.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 12.2 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 1.1 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.1 | 0.5 | GO:0070522 | nucleotide-excision repair factor 1 complex(GO:0000110) ERCC4-ERCC1 complex(GO:0070522) |
| 0.1 | 0.7 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 0.1 | 7.6 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 4.8 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.1 | 0.5 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 1.1 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 0.5 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 16.5 | GO:0034774 | secretory granule lumen(GO:0034774) |
| 0.0 | 1.1 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 21.1 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 2.6 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 2.8 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.9 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 2.4 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 2.3 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 1.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 3.2 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 8.9 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.5 | GO:1904813 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.0 | 3.4 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 1.0 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 0.5 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.7 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 9.7 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 3.7 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 1.5 | GO:0097060 | synaptic membrane(GO:0097060) |
| 0.0 | 4.5 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 2.8 | GO:0044297 | cell body(GO:0044297) |
| 0.0 | 0.7 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 1.0 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 2.3 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 3.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 11.0 | GO:0031410 | cytoplasmic vesicle(GO:0031410) |
| 0.0 | 0.0 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.0 | 1.4 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.0 | 0.5 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 1.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.9 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 16.4 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.6 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 22.8 | GO:0016021 | integral component of membrane(GO:0016021) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.2 | 15.5 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 2.3 | 13.9 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 2.2 | 6.7 | GO:0016495 | C-X3-C chemokine receptor activity(GO:0016495) |
| 1.8 | 17.6 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 1.5 | 6.2 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 1.3 | 4.0 | GO:0036134 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 1.3 | 5.2 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.9 | 2.8 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.9 | 12.9 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.9 | 2.6 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.8 | 8.3 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.7 | 3.7 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.7 | 7.9 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.7 | 2.8 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.7 | 2.8 | GO:0019862 | IgA binding(GO:0019862) |
| 0.6 | 1.1 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.6 | 2.2 | GO:0051120 | hepoxilin A3 synthase activity(GO:0051120) |
| 0.5 | 4.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.5 | 3.9 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.5 | 5.3 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.4 | 4.0 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.4 | 2.5 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.4 | 1.3 | GO:0015403 | thiamine transmembrane transporter activity(GO:0015234) thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.4 | 2.9 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.4 | 2.0 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.4 | 2.0 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.4 | 1.9 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.4 | 8.0 | GO:0038187 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.4 | 2.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.3 | 2.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.3 | 1.0 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.3 | 3.3 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.3 | 17.4 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.3 | 8.6 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.3 | 1.1 | GO:0060422 | peptidyl-dipeptidase inhibitor activity(GO:0060422) |
| 0.3 | 3.7 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.3 | 3.6 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.3 | 4.1 | GO:0019864 | IgG binding(GO:0019864) |
| 0.2 | 1.4 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.2 | 1.4 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.2 | 0.9 | GO:0035276 | aldehyde oxidase activity(GO:0004031) ethanol binding(GO:0035276) |
| 0.2 | 5.5 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.2 | 3.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.2 | 1.7 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.2 | 3.1 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.2 | 3.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.2 | 5.5 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.2 | 9.8 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.2 | 9.7 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.2 | 2.3 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.2 | 1.4 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.2 | 4.2 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.2 | 3.8 | GO:0097493 | muscle alpha-actinin binding(GO:0051371) structural molecule activity conferring elasticity(GO:0097493) |
| 0.2 | 8.3 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.2 | 3.5 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.2 | 5.3 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.2 | 0.5 | GO:0003968 | RNA-directed RNA polymerase activity(GO:0003968) |
| 0.1 | 1.6 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.1 | 0.4 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.1 | 1.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 1.0 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.1 | 0.9 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 0.7 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.1 | 0.9 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 0.9 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 0.8 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.1 | 0.4 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.1 | 2.1 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 1.8 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 1.1 | GO:0016918 | retinal binding(GO:0016918) |
| 0.1 | 4.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 1.5 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.1 | 13.8 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 1.1 | GO:0035240 | dopamine binding(GO:0035240) |
| 0.1 | 5.5 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 8.9 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.1 | 0.8 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.1 | 0.9 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 1.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 0.5 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.1 | 8.1 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.1 | 7.0 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.1 | 0.9 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 3.3 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 1.6 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 6.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.9 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 2.6 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 4.1 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 2.4 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 0.8 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 1.2 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.4 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 3.4 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.9 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 1.0 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.8 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.8 | GO:0099604 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 0.6 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.7 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.2 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 1.8 | GO:0004520 | endodeoxyribonuclease activity(GO:0004520) |
| 0.0 | 1.5 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.6 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 2.3 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 1.0 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.6 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.4 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.5 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 0.8 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.3 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.5 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 2.2 | GO:0050660 | flavin adenine dinucleotide binding(GO:0050660) |
| 0.0 | 0.2 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.5 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.4 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.4 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 5.6 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.0 | 0.3 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 1.1 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.5 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 16.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.4 | 10.6 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.4 | 8.3 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.4 | 19.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.2 | 17.6 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.2 | 9.7 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 1.8 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 8.1 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 3.7 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 11.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 4.0 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.1 | 15.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 2.9 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 1.6 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 5.2 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.9 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 3.1 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 1.7 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.3 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.6 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.8 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.3 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.7 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.7 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 21.5 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.5 | 5.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.5 | 7.7 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.5 | 9.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.4 | 6.7 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.2 | 30.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.2 | 0.8 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.2 | 7.9 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.2 | 1.8 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 8.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 13.9 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.1 | 0.9 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 1.4 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 0.7 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 5.5 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 2.0 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 0.6 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.1 | 4.4 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.1 | 2.2 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 8.3 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.1 | 4.0 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 9.2 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 1.0 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 1.2 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 1.1 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 1.4 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.9 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 1.0 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 1.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.5 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 1.0 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 2.5 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 1.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.8 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.9 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.9 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 1.0 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 0.4 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.6 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.8 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.8 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.4 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 1.9 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.7 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |