Project
averaged GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
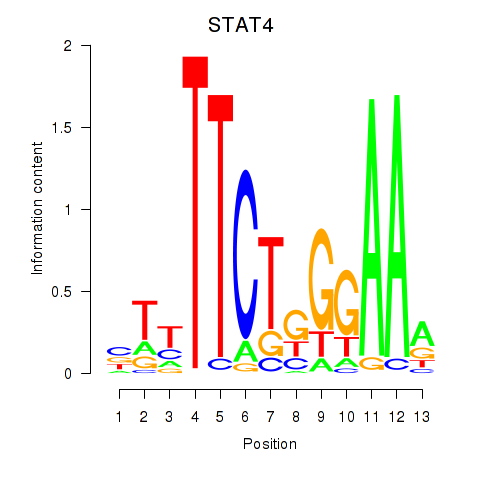
Results for STAT4
Z-value: 1.14
Transcription factors associated with STAT4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
STAT4
|
ENSG00000138378.13 | signal transducer and activator of transcription 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| STAT4 | hg19_v2_chr2_-_192016316_192016325 | 0.40 | 1.0e-09 | Click! |
Activity profile of STAT4 motif
Sorted Z-values of STAT4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_+_107288239 | 18.46 |
ENST00000217957.5
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chrX_+_107288197 | 16.77 |
ENST00000415430.3
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr4_+_158141806 | 15.34 |
ENST00000393815.2
|
GRIA2
|
glutamate receptor, ionotropic, AMPA 2 |
| chr8_-_120685608 | 15.28 |
ENST00000427067.2
|
ENPP2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr4_+_158141843 | 14.87 |
ENST00000509417.1
ENST00000296526.7 |
GRIA2
|
glutamate receptor, ionotropic, AMPA 2 |
| chr4_+_158141899 | 14.75 |
ENST00000264426.9
ENST00000506284.1 |
GRIA2
|
glutamate receptor, ionotropic, AMPA 2 |
| chr17_-_34207295 | 12.15 |
ENST00000463941.1
ENST00000293272.3 |
CCL5
|
chemokine (C-C motif) ligand 5 |
| chr6_+_32407619 | 11.69 |
ENST00000395388.2
ENST00000374982.5 |
HLA-DRA
|
major histocompatibility complex, class II, DR alpha |
| chr3_-_15469006 | 11.04 |
ENST00000443029.1
ENST00000383790.3 ENST00000383789.5 |
METTL6
|
methyltransferase like 6 |
| chr3_+_122296465 | 10.93 |
ENST00000483793.1
|
PARP15
|
poly (ADP-ribose) polymerase family, member 15 |
| chr14_-_21490958 | 10.39 |
ENST00000554104.1
|
NDRG2
|
NDRG family member 2 |
| chr19_+_42746927 | 9.92 |
ENST00000378108.1
|
AC006486.1
|
AC006486.1 |
| chr7_+_55980331 | 9.84 |
ENST00000429591.2
|
ZNF713
|
zinc finger protein 713 |
| chr8_-_27468842 | 9.77 |
ENST00000523500.1
|
CLU
|
clusterin |
| chr14_-_21270561 | 9.60 |
ENST00000412779.2
|
RNASE1
|
ribonuclease, RNase A family, 1 (pancreatic) |
| chr2_+_68962014 | 9.59 |
ENST00000467265.1
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr19_+_39881951 | 9.39 |
ENST00000315588.5
ENST00000594368.1 ENST00000599213.2 ENST00000596297.1 |
MED29
|
mediator complex subunit 29 |
| chr19_+_35225060 | 9.35 |
ENST00000599244.1
ENST00000392232.3 |
ZNF181
|
zinc finger protein 181 |
| chr3_+_15468862 | 9.25 |
ENST00000396842.2
|
EAF1
|
ELL associated factor 1 |
| chr4_+_41258786 | 9.22 |
ENST00000503431.1
ENST00000284440.4 ENST00000508768.1 ENST00000512788.1 |
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) |
| chr14_-_21490653 | 9.13 |
ENST00000449431.2
|
NDRG2
|
NDRG family member 2 |
| chr11_-_64490634 | 9.03 |
ENST00000377559.3
ENST00000265459.6 |
NRXN2
|
neurexin 2 |
| chr5_-_140013275 | 8.85 |
ENST00000512545.1
ENST00000302014.6 ENST00000401743.2 |
CD14
|
CD14 molecule |
| chr14_-_21490590 | 8.79 |
ENST00000557633.1
|
NDRG2
|
NDRG family member 2 |
| chr19_+_35168567 | 8.59 |
ENST00000457781.2
ENST00000505163.1 ENST00000505242.1 ENST00000423823.2 ENST00000507959.1 ENST00000446502.2 |
ZNF302
|
zinc finger protein 302 |
| chr19_+_35168633 | 8.40 |
ENST00000505365.2
|
ZNF302
|
zinc finger protein 302 |
| chr14_-_21490417 | 8.19 |
ENST00000556366.1
|
NDRG2
|
NDRG family member 2 |
| chr2_+_68961934 | 7.96 |
ENST00000409202.3
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr2_+_68961905 | 7.80 |
ENST00000295381.3
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr1_+_117297007 | 7.48 |
ENST00000369478.3
ENST00000369477.1 |
CD2
|
CD2 molecule |
| chrX_-_65253506 | 7.07 |
ENST00000427538.1
|
VSIG4
|
V-set and immunoglobulin domain containing 4 |
| chr1_+_233086326 | 7.03 |
ENST00000366628.5
ENST00000366627.4 |
NTPCR
|
nucleoside-triphosphatase, cancer-related |
| chr3_-_114866084 | 6.82 |
ENST00000357258.3
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr15_-_75660919 | 6.71 |
ENST00000569482.1
ENST00000565683.1 ENST00000561615.1 ENST00000563622.1 ENST00000568374.1 ENST00000566256.1 ENST00000267978.5 |
MAN2C1
|
mannosidase, alpha, class 2C, member 1 |
| chr5_-_39274617 | 6.58 |
ENST00000510188.1
|
FYB
|
FYN binding protein |
| chr5_+_140753444 | 6.22 |
ENST00000517434.1
|
PCDHGA6
|
protocadherin gamma subfamily A, 6 |
| chr18_+_61144160 | 6.03 |
ENST00000489441.1
ENST00000424602.1 |
SERPINB5
|
serpin peptidase inhibitor, clade B (ovalbumin), member 5 |
| chr3_-_57678772 | 5.86 |
ENST00000311128.5
|
DENND6A
|
DENN/MADD domain containing 6A |
| chr20_-_18774614 | 5.70 |
ENST00000412553.1
|
LINC00652
|
long intergenic non-protein coding RNA 652 |
| chr8_-_27469196 | 5.62 |
ENST00000546343.1
ENST00000560566.1 |
CLU
|
clusterin |
| chr16_+_58283814 | 5.54 |
ENST00000443128.2
ENST00000219299.4 |
CCDC113
|
coiled-coil domain containing 113 |
| chr1_-_9811600 | 5.53 |
ENST00000435891.1
|
CLSTN1
|
calsyntenin 1 |
| chr11_+_118175596 | 5.46 |
ENST00000528600.1
|
CD3E
|
CD3e molecule, epsilon (CD3-TCR complex) |
| chr5_+_140261703 | 5.45 |
ENST00000409494.1
ENST00000289272.2 |
PCDHA13
|
protocadherin alpha 13 |
| chr13_-_41240717 | 5.45 |
ENST00000379561.5
|
FOXO1
|
forkhead box O1 |
| chr14_+_24630465 | 5.44 |
ENST00000557894.1
ENST00000559284.1 ENST00000560275.1 |
IRF9
|
interferon regulatory factor 9 |
| chr13_-_45915221 | 5.15 |
ENST00000309246.5
ENST00000379060.4 ENST00000379055.1 ENST00000527226.1 ENST00000379056.1 |
TPT1
|
tumor protein, translationally-controlled 1 |
| chr19_-_59084647 | 5.08 |
ENST00000594234.1
ENST00000596039.1 |
MZF1
|
myeloid zinc finger 1 |
| chr19_-_51875894 | 4.95 |
ENST00000600427.1
ENST00000595217.1 ENST00000221978.5 |
NKG7
|
natural killer cell group 7 sequence |
| chrX_-_137793826 | 4.91 |
ENST00000315930.6
|
FGF13
|
fibroblast growth factor 13 |
| chr11_+_118175132 | 4.87 |
ENST00000361763.4
|
CD3E
|
CD3e molecule, epsilon (CD3-TCR complex) |
| chr5_-_16509101 | 4.76 |
ENST00000399793.2
|
FAM134B
|
family with sequence similarity 134, member B |
| chr10_-_6622201 | 4.46 |
ENST00000539722.1
ENST00000397176.2 |
PRKCQ
|
protein kinase C, theta |
| chr19_+_10197463 | 4.45 |
ENST00000590378.1
ENST00000397881.3 |
C19orf66
|
chromosome 19 open reading frame 66 |
| chr1_+_17906970 | 4.45 |
ENST00000375415.1
|
ARHGEF10L
|
Rho guanine nucleotide exchange factor (GEF) 10-like |
| chr12_+_58138800 | 4.38 |
ENST00000547992.1
ENST00000552816.1 ENST00000547472.1 |
TSPAN31
|
tetraspanin 31 |
| chr12_+_56324756 | 4.35 |
ENST00000331886.5
ENST00000555090.1 |
DGKA
|
diacylglycerol kinase, alpha 80kDa |
| chr5_-_111093406 | 4.30 |
ENST00000379671.3
|
NREP
|
neuronal regeneration related protein |
| chr14_-_25103472 | 4.20 |
ENST00000216341.4
ENST00000382542.1 ENST00000382540.1 |
GZMB
|
granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) |
| chr12_+_58138664 | 4.06 |
ENST00000257910.3
|
TSPAN31
|
tetraspanin 31 |
| chr16_-_20338748 | 4.06 |
ENST00000575582.1
ENST00000341642.5 ENST00000381362.4 ENST00000572347.1 ENST00000572478.1 ENST00000302555.5 |
GP2
|
glycoprotein 2 (zymogen granule membrane) |
| chr11_+_128563652 | 4.05 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr2_-_160473114 | 3.95 |
ENST00000392783.2
|
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B |
| chr5_-_111092930 | 3.88 |
ENST00000257435.7
|
NREP
|
neuronal regeneration related protein |
| chr5_+_140227048 | 3.87 |
ENST00000532602.1
|
PCDHA9
|
protocadherin alpha 9 |
| chr1_-_161279749 | 3.84 |
ENST00000533357.1
ENST00000360451.6 ENST00000336559.4 |
MPZ
|
myelin protein zero |
| chr6_-_32821599 | 3.73 |
ENST00000354258.4
|
TAP1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr3_-_155461515 | 3.68 |
ENST00000399242.2
|
AC104472.1
|
CDNA FLJ26134 fis, clone TMS03713; Uncharacterized protein |
| chr15_+_42131011 | 3.67 |
ENST00000458483.1
|
PLA2G4B
|
phospholipase A2, group IVB (cytosolic) |
| chr1_+_50571949 | 3.65 |
ENST00000357083.4
|
ELAVL4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr3_-_164875850 | 3.58 |
ENST00000472120.1
|
RP11-747D18.1
|
RP11-747D18.1 |
| chr6_+_134210243 | 3.55 |
ENST00000367882.4
|
TCF21
|
transcription factor 21 |
| chr1_-_161519682 | 3.54 |
ENST00000367969.3
ENST00000443193.1 |
FCGR3A
|
Fc fragment of IgG, low affinity IIIa, receptor (CD16a) |
| chr3_+_111260954 | 3.51 |
ENST00000283285.5
|
CD96
|
CD96 molecule |
| chr3_-_183735731 | 3.48 |
ENST00000334444.6
|
ABCC5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr17_+_41132564 | 3.46 |
ENST00000361677.1
ENST00000589705.1 |
RUNDC1
|
RUN domain containing 1 |
| chr3_+_111260856 | 3.40 |
ENST00000352690.4
|
CD96
|
CD96 molecule |
| chr10_+_17794251 | 3.38 |
ENST00000377495.1
ENST00000338221.5 |
TMEM236
|
transmembrane protein 236 |
| chr7_-_82792215 | 3.37 |
ENST00000333891.9
ENST00000423517.2 |
PCLO
|
piccolo presynaptic cytomatrix protein |
| chr12_+_56324933 | 3.32 |
ENST00000549629.1
ENST00000555218.1 |
DGKA
|
diacylglycerol kinase, alpha 80kDa |
| chr2_-_175547571 | 3.32 |
ENST00000409415.3
ENST00000359761.3 ENST00000272746.5 |
WIPF1
|
WAS/WASL interacting protein family, member 1 |
| chr19_+_57106647 | 3.15 |
ENST00000328070.6
|
ZNF71
|
zinc finger protein 71 |
| chr3_-_50649192 | 3.14 |
ENST00000443053.2
ENST00000348721.3 |
CISH
|
cytokine inducible SH2-containing protein |
| chr5_+_40909354 | 3.10 |
ENST00000313164.9
|
C7
|
complement component 7 |
| chr5_-_111093167 | 3.02 |
ENST00000446294.2
ENST00000419114.2 |
NREP
|
neuronal regeneration related protein |
| chr12_+_20848486 | 2.99 |
ENST00000545102.1
|
SLCO1C1
|
solute carrier organic anion transporter family, member 1C1 |
| chr10_-_104179682 | 2.97 |
ENST00000406432.1
|
PSD
|
pleckstrin and Sec7 domain containing |
| chr12_+_27396901 | 2.94 |
ENST00000541191.1
ENST00000389032.3 |
STK38L
|
serine/threonine kinase 38 like |
| chr22_-_31536480 | 2.94 |
ENST00000215885.3
|
PLA2G3
|
phospholipase A2, group III |
| chr2_-_177502254 | 2.94 |
ENST00000339037.3
|
AC017048.3
|
long intergenic non-protein coding RNA 1116 |
| chr17_-_58603568 | 2.77 |
ENST00000083182.3
|
APPBP2
|
amyloid beta precursor protein (cytoplasmic tail) binding protein 2 |
| chr11_+_65657875 | 2.72 |
ENST00000312579.2
|
CCDC85B
|
coiled-coil domain containing 85B |
| chr13_-_26795840 | 2.70 |
ENST00000381570.3
ENST00000399762.2 ENST00000346166.3 |
RNF6
|
ring finger protein (C3H2C3 type) 6 |
| chr19_+_44617511 | 2.62 |
ENST00000262894.6
ENST00000588926.1 ENST00000592780.1 |
ZNF225
|
zinc finger protein 225 |
| chr2_-_160472952 | 2.62 |
ENST00000541068.2
ENST00000355831.2 ENST00000343439.5 ENST00000392782.1 |
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B |
| chr14_+_65381079 | 2.52 |
ENST00000549115.1
ENST00000607599.1 ENST00000548752.2 ENST00000359118.2 ENST00000552002.2 ENST00000551947.1 ENST00000551093.1 ENST00000542227.1 ENST00000447296.2 ENST00000549987.1 |
CHURC1
FNTB
CHURC1-FNTB
|
churchill domain containing 1 farnesyltransferase, CAAX box, beta CHURC1-FNTB readthrough |
| chr2_-_37193606 | 2.52 |
ENST00000379213.2
ENST00000263918.4 |
STRN
|
striatin, calmodulin binding protein |
| chr7_-_115670792 | 2.46 |
ENST00000265440.7
ENST00000393485.1 |
TFEC
|
transcription factor EC |
| chr9_-_116163400 | 2.38 |
ENST00000277315.5
ENST00000448137.1 ENST00000409155.3 |
ALAD
|
aminolevulinate dehydratase |
| chr12_+_20848282 | 2.20 |
ENST00000545604.1
|
SLCO1C1
|
solute carrier organic anion transporter family, member 1C1 |
| chr7_-_115670804 | 2.16 |
ENST00000320239.7
|
TFEC
|
transcription factor EC |
| chr10_-_116444371 | 2.12 |
ENST00000533213.2
ENST00000369252.4 |
ABLIM1
|
actin binding LIM protein 1 |
| chr19_-_50529193 | 2.04 |
ENST00000596445.1
ENST00000599538.1 |
VRK3
|
vaccinia related kinase 3 |
| chr12_+_20848377 | 1.99 |
ENST00000540354.1
ENST00000266509.2 ENST00000381552.1 |
SLCO1C1
|
solute carrier organic anion transporter family, member 1C1 |
| chr1_+_155146318 | 1.98 |
ENST00000368385.4
ENST00000545012.1 ENST00000392451.2 ENST00000368383.3 ENST00000368382.1 ENST00000334634.4 |
TRIM46
|
tripartite motif containing 46 |
| chr6_+_167525277 | 1.92 |
ENST00000400926.2
|
CCR6
|
chemokine (C-C motif) receptor 6 |
| chr10_+_102790980 | 1.92 |
ENST00000393459.1
ENST00000224807.5 |
SFXN3
|
sideroflexin 3 |
| chr5_-_111093340 | 1.88 |
ENST00000508870.1
|
NREP
|
neuronal regeneration related protein |
| chr22_+_24551765 | 1.83 |
ENST00000337989.7
|
CABIN1
|
calcineurin binding protein 1 |
| chr1_-_120935894 | 1.83 |
ENST00000369383.4
ENST00000369384.4 |
FCGR1B
|
Fc fragment of IgG, high affinity Ib, receptor (CD64) |
| chr1_-_144994840 | 1.80 |
ENST00000369351.3
ENST00000369349.3 |
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr1_+_202431859 | 1.76 |
ENST00000391959.3
ENST00000367270.4 |
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr5_-_111093081 | 1.71 |
ENST00000453526.2
ENST00000509427.1 |
NREP
|
neuronal regeneration related protein |
| chr5_-_111092873 | 1.70 |
ENST00000509025.1
ENST00000515855.1 |
NREP
|
neuronal regeneration related protein |
| chr6_+_32821924 | 1.69 |
ENST00000374859.2
ENST00000453265.2 |
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr3_+_155838337 | 1.67 |
ENST00000490337.1
ENST00000389636.5 |
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr5_-_64777733 | 1.65 |
ENST00000381055.3
|
ADAMTS6
|
ADAM metallopeptidase with thrombospondin type 1 motif, 6 |
| chr20_+_8112824 | 1.53 |
ENST00000378641.3
|
PLCB1
|
phospholipase C, beta 1 (phosphoinositide-specific) |
| chr16_+_4674787 | 1.50 |
ENST00000262370.7
|
MGRN1
|
mahogunin ring finger 1, E3 ubiquitin protein ligase |
| chr11_-_118436707 | 1.47 |
ENST00000264020.2
ENST00000264021.3 |
IFT46
|
intraflagellar transport 46 homolog (Chlamydomonas) |
| chr1_+_149754227 | 1.45 |
ENST00000444948.1
ENST00000369168.4 |
FCGR1A
|
Fc fragment of IgG, high affinity Ia, receptor (CD64) |
| chr3_+_159570722 | 1.42 |
ENST00000482804.1
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr12_+_53443963 | 1.36 |
ENST00000546602.1
ENST00000552570.1 ENST00000549700.1 |
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2) |
| chr19_+_44669221 | 1.33 |
ENST00000590578.1
ENST00000589160.1 ENST00000337433.5 ENST00000586286.1 ENST00000585560.1 ENST00000586914.1 ENST00000588883.1 ENST00000413984.2 ENST00000588742.1 ENST00000300823.6 ENST00000585678.1 ENST00000586203.1 ENST00000590467.1 ENST00000588795.1 ENST00000588127.1 |
ZNF226
|
zinc finger protein 226 |
| chr5_+_137203465 | 1.31 |
ENST00000239926.4
|
MYOT
|
myotilin |
| chr3_-_52864680 | 1.29 |
ENST00000406595.1
ENST00000485816.1 ENST00000434759.3 ENST00000346281.5 ENST00000266041.4 |
ITIH4
|
inter-alpha-trypsin inhibitor heavy chain family, member 4 |
| chr9_-_95166841 | 1.28 |
ENST00000262551.4
|
OGN
|
osteoglycin |
| chr11_-_10590238 | 1.18 |
ENST00000256178.3
|
LYVE1
|
lymphatic vessel endothelial hyaluronan receptor 1 |
| chr3_+_171561127 | 1.17 |
ENST00000334567.5
ENST00000450693.1 |
TMEM212
|
transmembrane protein 212 |
| chr5_-_127674883 | 1.11 |
ENST00000507835.1
|
FBN2
|
fibrillin 2 |
| chr1_-_144994909 | 1.11 |
ENST00000369347.4
ENST00000369354.3 |
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr19_+_35773242 | 1.10 |
ENST00000222304.3
|
HAMP
|
hepcidin antimicrobial peptide |
| chr2_+_113885138 | 1.10 |
ENST00000409930.3
|
IL1RN
|
interleukin 1 receptor antagonist |
| chr6_+_25754927 | 1.07 |
ENST00000377905.4
ENST00000439485.2 |
SLC17A4
|
solute carrier family 17, member 4 |
| chr19_-_7812397 | 1.05 |
ENST00000593660.1
ENST00000354397.6 ENST00000593821.1 ENST00000602261.1 ENST00000315591.8 ENST00000394161.5 ENST00000204801.8 ENST00000601256.1 ENST00000601951.1 ENST00000315599.7 |
CD209
|
CD209 molecule |
| chr6_-_32152064 | 1.02 |
ENST00000375076.4
ENST00000375070.3 |
AGER
|
advanced glycosylation end product-specific receptor |
| chrX_+_135570046 | 0.95 |
ENST00000370648.3
|
BRS3
|
bombesin-like receptor 3 |
| chr19_+_50528971 | 0.95 |
ENST00000598809.1
ENST00000595661.1 ENST00000391821.2 |
ZNF473
|
zinc finger protein 473 |
| chr9_-_95166884 | 0.93 |
ENST00000375561.5
|
OGN
|
osteoglycin |
| chr7_+_117120017 | 0.92 |
ENST00000003084.6
ENST00000454343.1 |
CFTR
|
cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7) |
| chr4_-_76957214 | 0.90 |
ENST00000306621.3
|
CXCL11
|
chemokine (C-X-C motif) ligand 11 |
| chrX_-_63005405 | 0.89 |
ENST00000374878.1
ENST00000437457.2 |
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9 |
| chr12_+_60083118 | 0.85 |
ENST00000261187.4
ENST00000543448.1 |
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr12_-_49245936 | 0.83 |
ENST00000308025.3
|
DDX23
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 23 |
| chr3_+_5020801 | 0.83 |
ENST00000256495.3
|
BHLHE40
|
basic helix-loop-helix family, member e40 |
| chr16_+_4674814 | 0.70 |
ENST00000415496.1
ENST00000587747.1 ENST00000399577.5 ENST00000588994.1 ENST00000586183.1 |
MGRN1
|
mahogunin ring finger 1, E3 ubiquitin protein ligase |
| chrX_+_144908928 | 0.68 |
ENST00000408967.2
|
TMEM257
|
transmembrane protein 257 |
| chr7_+_117120106 | 0.63 |
ENST00000426809.1
|
CFTR
|
cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7) |
| chr11_-_119999611 | 0.62 |
ENST00000529044.1
|
TRIM29
|
tripartite motif containing 29 |
| chr11_-_85338311 | 0.61 |
ENST00000376104.2
|
DLG2
|
discs, large homolog 2 (Drosophila) |
| chr6_-_25874440 | 0.59 |
ENST00000361703.6
ENST00000397060.4 |
SLC17A3
|
solute carrier family 17 (organic anion transporter), member 3 |
| chr6_+_35995552 | 0.59 |
ENST00000468133.1
|
MAPK14
|
mitogen-activated protein kinase 14 |
| chr1_+_161632937 | 0.56 |
ENST00000236937.9
ENST00000367961.4 ENST00000358671.5 |
FCGR2B
|
Fc fragment of IgG, low affinity IIb, receptor (CD32) |
| chr16_+_19467772 | 0.56 |
ENST00000219821.5
ENST00000561503.1 ENST00000564959.1 |
TMC5
|
transmembrane channel-like 5 |
| chr20_-_21494654 | 0.52 |
ENST00000377142.4
|
NKX2-2
|
NK2 homeobox 2 |
| chr20_+_42136308 | 0.52 |
ENST00000434666.1
ENST00000427442.2 ENST00000439769.1 ENST00000418998.1 |
L3MBTL1
|
l(3)mbt-like 1 (Drosophila) |
| chr3_+_187871060 | 0.50 |
ENST00000448637.1
|
LPP
|
LIM domain containing preferred translocation partner in lipoma |
| chr2_-_171627269 | 0.50 |
ENST00000442456.1
|
AC007405.4
|
AC007405.4 |
| chr19_-_7812446 | 0.50 |
ENST00000394173.4
ENST00000301357.8 |
CD209
|
CD209 molecule |
| chr4_-_187476721 | 0.47 |
ENST00000307161.5
|
MTNR1A
|
melatonin receptor 1A |
| chr3_-_137851220 | 0.44 |
ENST00000236709.3
|
A4GNT
|
alpha-1,4-N-acetylglucosaminyltransferase |
| chr5_+_137203557 | 0.39 |
ENST00000515645.1
|
MYOT
|
myotilin |
| chr2_+_210444748 | 0.38 |
ENST00000392194.1
|
MAP2
|
microtubule-associated protein 2 |
| chr13_+_97928395 | 0.37 |
ENST00000445661.2
|
MBNL2
|
muscleblind-like splicing regulator 2 |
| chr1_+_9299895 | 0.36 |
ENST00000602477.1
|
H6PD
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
| chr14_-_20881579 | 0.33 |
ENST00000556935.1
ENST00000262715.5 ENST00000556549.1 |
TEP1
|
telomerase-associated protein 1 |
| chr19_-_39881669 | 0.31 |
ENST00000221266.7
|
PAF1
|
Paf1, RNA polymerase II associated factor, homolog (S. cerevisiae) |
| chrX_+_138612889 | 0.31 |
ENST00000218099.2
ENST00000394090.2 |
F9
|
coagulation factor IX |
| chr11_+_123986069 | 0.29 |
ENST00000456829.2
ENST00000361352.5 ENST00000449321.1 ENST00000392748.1 ENST00000360334.4 ENST00000392744.4 |
VWA5A
|
von Willebrand factor A domain containing 5A |
| chr3_-_194119995 | 0.28 |
ENST00000323007.3
|
GP5
|
glycoprotein V (platelet) |
| chr11_-_119999539 | 0.25 |
ENST00000541857.1
|
TRIM29
|
tripartite motif containing 29 |
| chrX_+_54834159 | 0.23 |
ENST00000375053.2
ENST00000347546.4 ENST00000375062.4 |
MAGED2
|
melanoma antigen family D, 2 |
| chr6_+_108977520 | 0.18 |
ENST00000540898.1
|
FOXO3
|
forkhead box O3 |
| chr15_-_60690932 | 0.16 |
ENST00000559818.1
|
ANXA2
|
annexin A2 |
| chr19_+_50529212 | 0.13 |
ENST00000270617.3
ENST00000445728.3 ENST00000601364.1 |
ZNF473
|
zinc finger protein 473 |
| chrX_-_154563889 | 0.12 |
ENST00000369449.2
ENST00000321926.4 |
CLIC2
|
chloride intracellular channel 2 |
| chr5_+_137203541 | 0.06 |
ENST00000421631.2
|
MYOT
|
myotilin |
| chr11_-_62752455 | 0.06 |
ENST00000360421.4
|
SLC22A6
|
solute carrier family 22 (organic anion transporter), member 6 |
| chr2_-_79386786 | 0.05 |
ENST00000393878.1
ENST00000305165.2 ENST00000409839.3 |
REG3A
|
regenerating islet-derived 3 alpha |
| chr5_-_121413974 | 0.02 |
ENST00000231004.4
|
LOX
|
lysyl oxidase |
Network of associatons between targets according to the STRING database.
First level regulatory network of STAT4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 9.2 | GO:0007412 | axon target recognition(GO:0007412) |
| 3.0 | 12.1 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 2.9 | 11.7 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 2.6 | 15.4 | GO:1902847 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 2.3 | 6.9 | GO:0002370 | natural killer cell cytokine production(GO:0002370) regulation of natural killer cell cytokine production(GO:0002727) |
| 2.3 | 36.5 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 2.2 | 8.8 | GO:0071727 | toll-like receptor TLR1:TLR2 signaling pathway(GO:0038123) response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 2.1 | 10.3 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 1.8 | 10.9 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 1.8 | 35.2 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 1.2 | 7.5 | GO:0030885 | regulation of myeloid dendritic cell activation(GO:0030885) |
| 1.2 | 3.6 | GO:0072276 | branchiomeric skeletal muscle development(GO:0014707) metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 1.1 | 9.0 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 1.1 | 4.5 | GO:2000570 | T-helper 2 cell activation(GO:0035712) regulation of T-helper 2 cell activation(GO:2000569) positive regulation of T-helper 2 cell activation(GO:2000570) |
| 1.1 | 5.5 | GO:0035947 | regulation of gluconeogenesis by regulation of transcription from RNA polymerase II promoter(GO:0035947) response to fluoride(GO:1902617) |
| 1.0 | 15.3 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 1.0 | 4.9 | GO:1990834 | response to odorant(GO:1990834) |
| 0.9 | 45.0 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.9 | 3.7 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.9 | 2.7 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.8 | 6.7 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.8 | 2.4 | GO:0070541 | response to platinum ion(GO:0070541) cellular response to lead ion(GO:0071284) |
| 0.6 | 1.9 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.6 | 4.2 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.6 | 7.2 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.5 | 5.9 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.5 | 1.6 | GO:0046968 | peptide antigen transport(GO:0046968) |
| 0.5 | 1.5 | GO:1902161 | positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) |
| 0.5 | 1.5 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.4 | 2.5 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.4 | 10.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.4 | 5.5 | GO:0060271 | cilium morphogenesis(GO:0060271) |
| 0.4 | 6.6 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.4 | 1.1 | GO:1903413 | cellular response to bile acid(GO:1903413) negative regulation of intestinal absorption(GO:1904479) response to iron ion starvation(GO:1990641) |
| 0.3 | 1.0 | GO:1905204 | regulation of connective tissue replacement involved in inflammatory response wound healing(GO:1904596) negative regulation of connective tissue replacement involved in inflammatory response wound healing(GO:1904597) regulation of advanced glycation end-product receptor activity(GO:1904603) negative regulation of advanced glycation end-product receptor activity(GO:1904604) negative regulation of connective tissue replacement(GO:1905204) |
| 0.3 | 7.7 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.3 | 0.8 | GO:0000354 | cis assembly of pre-catalytic spliceosome(GO:0000354) |
| 0.2 | 3.5 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.2 | 1.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.2 | 4.1 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.2 | 2.2 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.2 | 2.0 | GO:0099612 | protein localization to axon(GO:0099612) |
| 0.2 | 6.6 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.2 | 0.5 | GO:0021780 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.2 | 0.3 | GO:0043318 | regulation of cytotoxic T cell degranulation(GO:0043317) negative regulation of cytotoxic T cell degranulation(GO:0043318) |
| 0.1 | 3.0 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 6.8 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 9.6 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.1 | 1.1 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.1 | 21.1 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 1.1 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.1 | 2.0 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.1 | 1.7 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.1 | 2.9 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.1 | 5.2 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.1 | 2.2 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.1 | 0.9 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 0.9 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.1 | 2.2 | GO:0098743 | cell aggregation(GO:0098743) |
| 0.1 | 0.6 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 3.6 | GO:0060512 | prostate gland morphogenesis(GO:0060512) |
| 0.1 | 2.7 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.1 | 3.1 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.1 | 8.7 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.1 | 1.2 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.1 | 8.5 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.1 | 26.2 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.1 | 4.0 | GO:0035418 | protein localization to synapse(GO:0035418) |
| 0.1 | 1.1 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 0.2 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.1 | 4.1 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.1 | 1.3 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 6.9 | GO:0002433 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 0.0 | 1.8 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) DNA replication-independent nucleosome organization(GO:0034724) |
| 0.0 | 2.5 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.0 | 2.1 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.3 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 1.5 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.3 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.0 | 0.2 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.8 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 1.7 | GO:0060976 | coronary vasculature development(GO:0060976) |
| 0.0 | 10.8 | GO:0032259 | methylation(GO:0032259) |
| 0.0 | 0.5 | GO:0044705 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.0 | 0.4 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.0 | 0.3 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.1 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.0 | 1.8 | GO:0000086 | G2/M transition of mitotic cell cycle(GO:0000086) |
| 0.0 | 0.5 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) |
| 0.0 | 0.6 | GO:0006811 | ion transport(GO:0006811) |
| 0.0 | 1.2 | GO:0055072 | iron ion homeostasis(GO:0055072) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.0 | 45.0 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 1.8 | 8.8 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 1.2 | 15.4 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 1.0 | 5.2 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.9 | 10.3 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.6 | 2.5 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.6 | 3.7 | GO:0042825 | TAP complex(GO:0042825) |
| 0.6 | 11.7 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.3 | 3.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.3 | 2.0 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.3 | 1.5 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.2 | 1.7 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.2 | 7.5 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.2 | 5.5 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.2 | 9.2 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.2 | 7.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.2 | 41.4 | GO:0030426 | growth cone(GO:0030426) |
| 0.2 | 9.3 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.2 | 8.7 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 1.9 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 1.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 1.1 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 2.9 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 3.4 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.1 | 2.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 1.3 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.1 | 3.3 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.1 | 1.5 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 0.4 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 1.1 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 3.8 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 3.0 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 1.2 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 5.9 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 1.8 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.6 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 4.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 3.7 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 1.8 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 5.5 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 9.0 | GO:0098793 | presynapse(GO:0098793) |
| 0.0 | 42.9 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 2.7 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 2.8 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 1.1 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 2.0 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 1.8 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 2.1 | GO:0030027 | lamellipodium(GO:0030027) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 15.3 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 3.7 | 45.0 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 2.9 | 8.8 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 2.3 | 7.0 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 2.3 | 9.2 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 1.7 | 12.1 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 1.6 | 9.6 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.9 | 7.2 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.8 | 11.7 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.8 | 10.3 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.8 | 15.4 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.7 | 6.6 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.6 | 2.5 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.6 | 3.7 | GO:0046979 | TAP2 binding(GO:0046979) |
| 0.6 | 9.0 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.5 | 1.5 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.5 | 10.9 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.4 | 7.7 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.4 | 6.7 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.4 | 7.2 | GO:0019864 | IgG binding(GO:0019864) |
| 0.4 | 3.5 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.4 | 6.7 | GO:0005522 | profilin binding(GO:0005522) |
| 0.4 | 1.1 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.3 | 1.0 | GO:1904599 | advanced glycation end-product binding(GO:1904599) |
| 0.2 | 0.9 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.2 | 8.0 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.2 | 0.9 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.2 | 4.3 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.2 | 0.5 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.2 | 0.5 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.1 | 1.9 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 1.6 | GO:0046790 | virion binding(GO:0046790) |
| 0.1 | 1.1 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.1 | 1.7 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.1 | 0.4 | GO:0017057 | glucose-6-phosphate dehydrogenase activity(GO:0004345) 6-phosphogluconolactonase activity(GO:0017057) |
| 0.1 | 3.0 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 5.9 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 3.7 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 16.3 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 4.9 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 5.5 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 1.7 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.1 | 1.5 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 5.3 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 11.0 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.0 | 1.8 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 14.2 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 2.7 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.9 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 1.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 1.8 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 4.1 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 3.1 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 2.4 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.3 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 2.8 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 3.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.9 | GO:0015075 | ion transmembrane transporter activity(GO:0015075) |
| 0.0 | 3.8 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.6 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.4 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 4.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 12.8 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 1.8 | GO:0008047 | enzyme activator activity(GO:0008047) |
| 0.0 | 19.7 | GO:0001071 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 0.8 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 45.0 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.5 | 3.1 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.4 | 22.0 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.4 | 5.4 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.3 | 46.2 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.2 | 6.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.2 | 5.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 1.8 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 4.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 9.2 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 11.0 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.1 | 3.5 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 4.1 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.1 | 5.2 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 3.7 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 4.6 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 4.4 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 3.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 4.6 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.8 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.1 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.9 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 1.0 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 3.7 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.9 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.1 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.5 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 45.0 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.7 | 8.8 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.4 | 10.3 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.3 | 7.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.3 | 12.1 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.2 | 15.0 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.2 | 4.7 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.2 | 6.6 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.2 | 2.9 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.2 | 6.0 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.2 | 2.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 3.7 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 1.5 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 8.7 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 22.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 10.8 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 3.1 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.1 | 2.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 2.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 4.2 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.1 | 0.9 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 7.6 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 1.1 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 7.5 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.1 | 3.1 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 8.0 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 1.0 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.3 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 1.5 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 1.7 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.8 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.8 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |