Project
averaged GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for TFEC_MITF_ARNTL_BHLHE41
Z-value: 0.95
Transcription factors associated with TFEC_MITF_ARNTL_BHLHE41
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
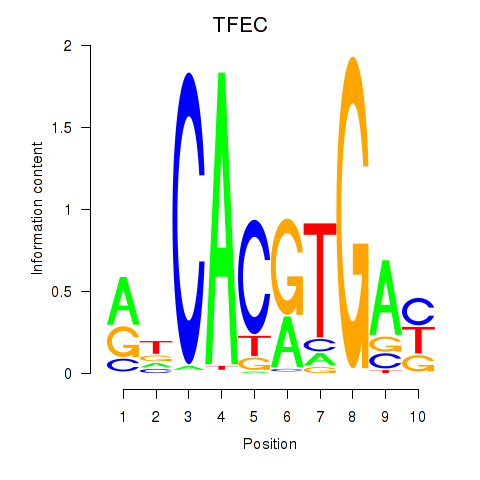
TFEC
|
ENSG00000105967.11 | transcription factor EC |
|
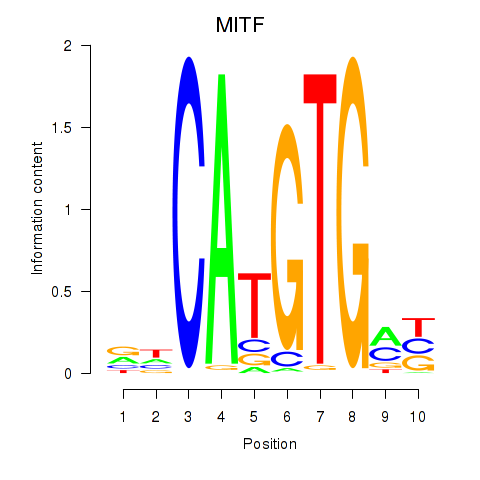
MITF
|
ENSG00000187098.10 | melanocyte inducing transcription factor |
|
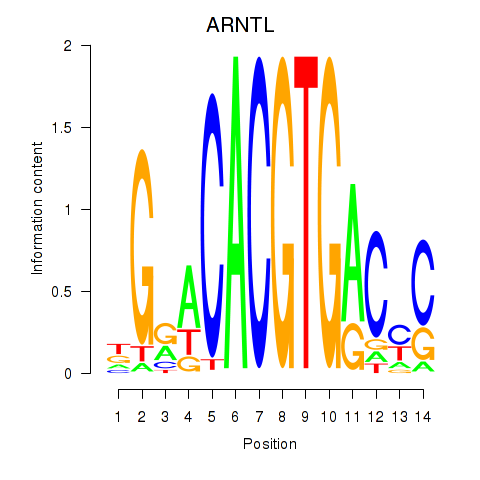
ARNTL
|
ENSG00000133794.13 | aryl hydrocarbon receptor nuclear translocator like |
|
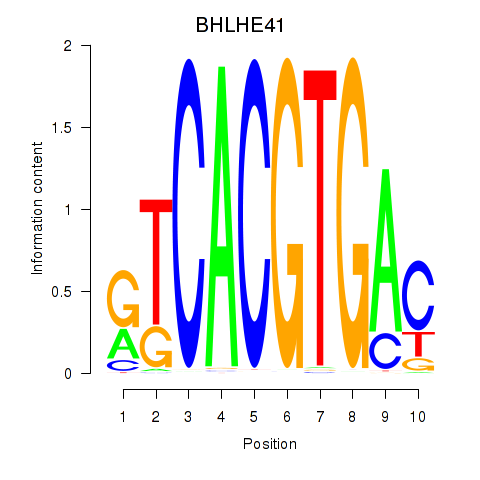
BHLHE41
|
ENSG00000123095.5 | basic helix-loop-helix family member e41 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| BHLHE41 | hg19_v2_chr12_-_26278030_26278060 | 0.34 | 1.7e-07 | Click! |
| MITF | hg19_v2_chr3_+_69788576_69788648, hg19_v2_chr3_+_69985734_69985754, hg19_v2_chr3_+_69812701_69812729, hg19_v2_chr3_+_69812877_69812962, hg19_v2_chr3_+_69928256_69928385, hg19_v2_chr3_+_69915385_69915438 | 0.31 | 2.3e-06 | Click! |
| ARNTL | hg19_v2_chr11_+_13299186_13299432 | -0.14 | 3.6e-02 | Click! |
| TFEC | hg19_v2_chr7_-_115608304_115608352, hg19_v2_chr7_-_115670804_115670867, hg19_v2_chr7_-_115670792_115670798 | -0.08 | 2.4e-01 | Click! |
Activity profile of TFEC_MITF_ARNTL_BHLHE41 motif
Sorted Z-values of TFEC_MITF_ARNTL_BHLHE41 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of TFEC_MITF_ARNTL_BHLHE41
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.2 | 20.9 | GO:0043323 | regulation of natural killer cell degranulation(GO:0043321) positive regulation of natural killer cell degranulation(GO:0043323) |
| 4.8 | 24.1 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 4.0 | 12.0 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 3.3 | 9.8 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 3.0 | 8.9 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 2.9 | 14.5 | GO:0033274 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 2.9 | 23.2 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 2.7 | 10.7 | GO:1904457 | beta-glucoside metabolic process(GO:1901804) beta-glucoside catabolic process(GO:1901805) positive regulation of neuronal action potential(GO:1904457) |
| 2.5 | 7.5 | GO:1903697 | negative regulation of microvillus assembly(GO:1903697) |
| 2.2 | 15.7 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 2.1 | 2.1 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 2.0 | 30.6 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 1.9 | 5.7 | GO:1902445 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) apoptotic process involved in embryonic digit morphogenesis(GO:1902263) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) positive regulation of apoptotic DNA fragmentation(GO:1902512) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 1.8 | 8.8 | GO:1900063 | regulation of peroxisome organization(GO:1900063) |
| 1.8 | 7.0 | GO:0019859 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 1.7 | 8.3 | GO:0009608 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 1.7 | 5.0 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 1.6 | 4.7 | GO:2000854 | positive regulation of corticosterone secretion(GO:2000854) |
| 1.5 | 9.1 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 1.5 | 16.6 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 1.5 | 9.0 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 1.5 | 30.7 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 1.4 | 4.1 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 1.2 | 6.2 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 1.2 | 4.9 | GO:1990936 | metanephric glomerular mesangial cell development(GO:0072255) reversible differentiation(GO:0090677) cell dedifferentiation involved in phenotypic switching(GO:0090678) positive regulation of phenotypic switching(GO:1900241) regulation of vascular smooth muscle cell dedifferentiation(GO:1905174) positive regulation of vascular smooth muscle cell dedifferentiation(GO:1905176) vascular smooth muscle cell dedifferentiation(GO:1990936) |
| 1.2 | 4.8 | GO:0009386 | translational attenuation(GO:0009386) |
| 1.2 | 4.8 | GO:0015808 | L-alanine transport(GO:0015808) |
| 1.2 | 4.7 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 1.1 | 5.6 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 1.1 | 4.5 | GO:0000023 | maltose metabolic process(GO:0000023) diaphragm contraction(GO:0002086) |
| 1.1 | 3.3 | GO:0051039 | histone displacement(GO:0001207) regulation of transcription involved in meiotic cell cycle(GO:0051037) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 1.1 | 3.3 | GO:0061055 | myotome development(GO:0061055) |
| 1.1 | 4.3 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 1.1 | 4.3 | GO:0043324 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) |
| 1.0 | 4.1 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 1.0 | 5.0 | GO:0000189 | MAPK import into nucleus(GO:0000189) |
| 1.0 | 3.0 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 1.0 | 3.9 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 1.0 | 3.9 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.9 | 7.6 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.9 | 19.5 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.9 | 6.4 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.9 | 26.0 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.9 | 8.9 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.9 | 2.6 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.9 | 14.5 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.8 | 2.5 | GO:1904482 | response to tetrahydrofolate(GO:1904481) cellular response to tetrahydrofolate(GO:1904482) |
| 0.8 | 6.6 | GO:0090382 | phagosome maturation(GO:0090382) |
| 0.8 | 12.7 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.8 | 0.8 | GO:0039702 | viral budding via host ESCRT complex(GO:0039702) |
| 0.8 | 3.9 | GO:1990834 | response to odorant(GO:1990834) |
| 0.7 | 8.2 | GO:0015886 | heme transport(GO:0015886) |
| 0.7 | 2.1 | GO:0050787 | antibiotic metabolic process(GO:0016999) detoxification of mercury ion(GO:0050787) response to antineoplastic agent(GO:0097327) |
| 0.7 | 10.0 | GO:0043485 | endosome to melanosome transport(GO:0035646) endosome to pigment granule transport(GO:0043485) pigment granule maturation(GO:0048757) |
| 0.7 | 2.1 | GO:0035606 | induction of programmed cell death(GO:0012502) peptidyl-cysteine S-trans-nitrosylation(GO:0035606) positive regulation of apoptotic process in other organism(GO:0044533) positive regulation by symbiont of host programmed cell death(GO:0052042) positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052330) positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052501) |
| 0.7 | 7.0 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.7 | 2.1 | GO:2000777 | positive regulation of oocyte maturation(GO:1900195) negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.7 | 8.3 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.7 | 2.0 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.7 | 2.0 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.6 | 3.8 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.6 | 0.6 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.6 | 2.5 | GO:0042335 | cuticle development(GO:0042335) |
| 0.6 | 10.0 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.6 | 1.7 | GO:0045643 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.6 | 2.3 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.6 | 5.8 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.6 | 2.8 | GO:1903564 | regulation of protein localization to cilium(GO:1903564) |
| 0.5 | 2.2 | GO:0090301 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of neural crest formation(GO:0090299) negative regulation of neural crest formation(GO:0090301) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.5 | 3.2 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.5 | 2.7 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.5 | 3.2 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.5 | 1.6 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.5 | 1.6 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.5 | 6.8 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.5 | 1.5 | GO:0021827 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) regulation of negative chemotaxis(GO:0050923) |
| 0.5 | 3.6 | GO:0006489 | dolichyl diphosphate biosynthetic process(GO:0006489) dolichyl diphosphate metabolic process(GO:0046465) |
| 0.5 | 7.0 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.5 | 1.4 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.4 | 2.7 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.4 | 6.2 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.4 | 5.3 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.4 | 1.3 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.4 | 3.0 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.4 | 3.0 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.4 | 1.3 | GO:0002276 | basophil activation involved in immune response(GO:0002276) |
| 0.4 | 2.1 | GO:0050428 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.4 | 2.4 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.4 | 1.2 | GO:2000683 | mesodermal-endodermal cell signaling(GO:0003131) programmed DNA elimination(GO:0031049) chromosome breakage(GO:0031052) histone H2A-S139 phosphorylation(GO:0035978) regulation of cellular response to X-ray(GO:2000683) positive regulation of cellular response to X-ray(GO:2000685) |
| 0.4 | 1.9 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.4 | 3.0 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.4 | 3.8 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.4 | 0.4 | GO:1902954 | regulation of early endosome to recycling endosome transport(GO:1902954) |
| 0.4 | 1.5 | GO:1905123 | regulation of endosome organization(GO:1904978) regulation of glucosylceramidase activity(GO:1905123) |
| 0.4 | 1.1 | GO:0032707 | negative regulation of interleukin-23 production(GO:0032707) |
| 0.4 | 1.5 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.4 | 4.0 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.4 | 3.9 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.3 | 3.4 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.3 | 2.4 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.3 | 0.3 | GO:0007521 | muscle cell fate determination(GO:0007521) |
| 0.3 | 1.0 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.3 | 5.9 | GO:0050812 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.3 | 3.3 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 0.3 | 2.2 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.3 | 3.1 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.3 | 5.7 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.3 | 2.1 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.3 | 2.3 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.3 | 5.2 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.3 | 0.6 | GO:0010823 | negative regulation of mitochondrion organization(GO:0010823) negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.3 | 1.9 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.3 | 1.4 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.3 | 1.9 | GO:0051126 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) negative regulation of actin nucleation(GO:0051126) |
| 0.3 | 1.6 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.3 | 1.1 | GO:0098706 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.3 | 2.9 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.3 | 0.8 | GO:1990168 | protein K33-linked deubiquitination(GO:1990168) |
| 0.3 | 1.8 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.3 | 1.0 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.3 | 1.3 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.2 | 3.2 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.2 | 1.5 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.2 | 7.9 | GO:0007628 | adult walking behavior(GO:0007628) walking behavior(GO:0090659) |
| 0.2 | 0.7 | GO:0086092 | regulation of the force of heart contraction by cardiac conduction(GO:0086092) |
| 0.2 | 2.4 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.2 | 0.7 | GO:0042636 | negative regulation of hair cycle(GO:0042636) |
| 0.2 | 0.2 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.2 | 0.7 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.2 | 1.6 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.2 | 1.1 | GO:0009181 | purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.2 | 5.9 | GO:0030220 | platelet formation(GO:0030220) |
| 0.2 | 0.7 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.2 | 1.1 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.2 | 1.5 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.2 | 0.9 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.2 | 0.4 | GO:0071284 | cellular response to lead ion(GO:0071284) |
| 0.2 | 3.7 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.2 | 1.3 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.2 | 8.8 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.2 | 1.7 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.2 | 0.2 | GO:0072229 | proximal convoluted tubule development(GO:0072019) metanephric proximal convoluted tubule development(GO:0072229) |
| 0.2 | 1.4 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.2 | 1.2 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.2 | 1.5 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.2 | 1.7 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.2 | 0.7 | GO:0090269 | fibroblast growth factor production(GO:0090269) regulation of fibroblast growth factor production(GO:0090270) |
| 0.2 | 7.5 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.2 | 1.3 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 0.2 | 1.1 | GO:1900125 | regulation of hyaluronan biosynthetic process(GO:1900125) |
| 0.2 | 1.3 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.2 | 20.8 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.2 | 0.5 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.2 | 1.1 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.2 | 3.7 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.2 | 1.5 | GO:0046689 | response to mercury ion(GO:0046689) |
| 0.2 | 0.8 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.2 | 0.2 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.2 | 1.2 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 0.2 | 1.5 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.2 | 1.8 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.2 | 0.5 | GO:1904923 | positive regulation of macromitophagy(GO:1901526) regulation of mitophagy in response to mitochondrial depolarization(GO:1904923) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.2 | 1.3 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.2 | 14.1 | GO:0043473 | pigmentation(GO:0043473) |
| 0.2 | 0.6 | GO:0044111 | development involved in symbiotic interaction(GO:0044111) |
| 0.2 | 3.3 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.2 | 5.1 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.2 | 12.4 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.2 | 2.9 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.2 | 0.2 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.2 | 0.5 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.2 | 0.5 | GO:0060535 | trachea cartilage morphogenesis(GO:0060535) |
| 0.1 | 1.5 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.1 | 0.4 | GO:0044539 | long-chain fatty acid import(GO:0044539) regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.1 | 1.0 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.1 | 1.7 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.1 | 0.9 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 3.8 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 0.4 | GO:0000967 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.1 | 1.5 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 5.2 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.1 | 0.4 | GO:0051106 | positive regulation of DNA ligation(GO:0051106) |
| 0.1 | 0.8 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.1 | 0.7 | GO:0090650 | rRNA transport(GO:0051029) response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.1 | 1.2 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.1 | 1.8 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.1 | 5.1 | GO:0030517 | negative regulation of axon extension(GO:0030517) |
| 0.1 | 0.8 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.1 | 1.2 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.1 | 1.0 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.1 | 2.4 | GO:1903206 | negative regulation of hydrogen peroxide-induced cell death(GO:1903206) |
| 0.1 | 0.5 | GO:0007501 | mesodermal cell fate specification(GO:0007501) |
| 0.1 | 0.5 | GO:0032383 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.1 | 0.6 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.1 | 4.6 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.1 | 1.2 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.1 | 0.4 | GO:0032763 | heme oxidation(GO:0006788) smooth muscle hyperplasia(GO:0014806) regulation of mast cell cytokine production(GO:0032763) |
| 0.1 | 0.6 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.1 | 2.5 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 0.4 | GO:0072076 | nephrogenic mesenchyme development(GO:0072076) |
| 0.1 | 1.6 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.1 | 0.7 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.1 | 0.5 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.1 | 0.4 | GO:1904849 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.1 | 0.6 | GO:2000255 | regulation of male germ cell proliferation(GO:2000254) negative regulation of male germ cell proliferation(GO:2000255) |
| 0.1 | 1.1 | GO:0042074 | cell migration involved in gastrulation(GO:0042074) |
| 0.1 | 0.3 | GO:1901798 | positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.1 | 0.3 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.1 | 1.7 | GO:0034384 | high-density lipoprotein particle clearance(GO:0034384) |
| 0.1 | 2.3 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 1.1 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 | 1.0 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.1 | 1.0 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 0.6 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 0.1 | 0.9 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.1 | 0.7 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.1 | 0.8 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.1 | 0.8 | GO:0007569 | cell aging(GO:0007569) |
| 0.1 | 0.7 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.1 | 3.3 | GO:1901985 | positive regulation of protein acetylation(GO:1901985) |
| 0.1 | 0.6 | GO:0002865 | negative regulation of acute inflammatory response to antigenic stimulus(GO:0002865) |
| 0.1 | 1.4 | GO:0002092 | positive regulation of receptor internalization(GO:0002092) |
| 0.1 | 0.6 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.1 | 0.3 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.1 | 4.6 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 1.6 | GO:0061458 | reproductive system development(GO:0061458) |
| 0.1 | 0.4 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.1 | 0.2 | GO:0070781 | response to biotin(GO:0070781) |
| 0.1 | 0.8 | GO:0060766 | negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.1 | 0.2 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.1 | 0.6 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.1 | 1.2 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 1.1 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.1 | 1.0 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.1 | 0.7 | GO:1904350 | regulation of protein catabolic process in the vacuole(GO:1904350) positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.1 | 1.6 | GO:0098926 | acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.1 | 2.9 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.1 | 1.0 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.1 | 0.5 | GO:0072257 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.1 | 0.5 | GO:1903358 | regulation of Golgi organization(GO:1903358) |
| 0.1 | 0.2 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.1 | 0.4 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.1 | 0.3 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.1 | 1.0 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.1 | 2.2 | GO:0051155 | positive regulation of striated muscle cell differentiation(GO:0051155) |
| 0.1 | 2.0 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 0.1 | 1.2 | GO:1903817 | diaphragm development(GO:0060539) negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.1 | 0.2 | GO:0003176 | aortic valve development(GO:0003176) aortic valve morphogenesis(GO:0003180) notochord formation(GO:0014028) |
| 0.1 | 0.5 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.1 | 0.1 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.1 | 2.0 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.1 | 1.9 | GO:0010762 | regulation of fibroblast migration(GO:0010762) |
| 0.1 | 0.3 | GO:1901660 | calcium ion export(GO:1901660) calcium ion export from cell(GO:1990034) |
| 0.1 | 0.2 | GO:0002232 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.1 | 0.3 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.1 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 1.5 | GO:1901889 | negative regulation of cell junction assembly(GO:1901889) |
| 0.1 | 0.6 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.1 | 1.0 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.1 | 0.5 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 1.1 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 0.3 | GO:0051597 | response to methylmercury(GO:0051597) |
| 0.1 | 0.4 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 0.4 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.1 | 0.7 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.1 | 0.7 | GO:0040019 | positive regulation of embryonic development(GO:0040019) |
| 0.1 | 3.0 | GO:1901016 | regulation of potassium ion transmembrane transporter activity(GO:1901016) |
| 0.1 | 6.4 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.1 | 1.3 | GO:0032607 | interferon-alpha production(GO:0032607) |
| 0.1 | 3.6 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.1 | 2.7 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.1 | 1.4 | GO:1904031 | positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.1 | 0.7 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.1 | 1.3 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.1 | 0.5 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.1 | 0.4 | GO:0006498 | N-terminal protein lipidation(GO:0006498) |
| 0.1 | 0.2 | GO:1900073 | regulation of neuromuscular synaptic transmission(GO:1900073) positive regulation of neuromuscular synaptic transmission(GO:1900075) |
| 0.1 | 0.2 | GO:0005985 | sucrose metabolic process(GO:0005985) |
| 0.1 | 0.3 | GO:0014827 | intestine smooth muscle contraction(GO:0014827) |
| 0.1 | 1.1 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.1 | 0.5 | GO:2000615 | regulation of histone H3-K9 acetylation(GO:2000615) |
| 0.1 | 0.6 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.1 | 0.1 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 | 0.3 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.0 | 0.2 | GO:0072236 | ascending thin limb development(GO:0072021) thick ascending limb development(GO:0072023) metanephric ascending thin limb development(GO:0072218) metanephric thick ascending limb development(GO:0072233) metanephric loop of Henle development(GO:0072236) |
| 0.0 | 0.2 | GO:0045713 | low-density lipoprotein particle receptor biosynthetic process(GO:0045713) regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045714) |
| 0.0 | 2.3 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.7 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.4 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 1.4 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 0.2 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.0 | 0.4 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 0.9 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.2 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 2.9 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.9 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 3.4 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 1.1 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.0 | 0.3 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.3 | GO:0031623 | receptor internalization(GO:0031623) |
| 0.0 | 0.1 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 4.2 | GO:0015698 | inorganic anion transport(GO:0015698) |
| 0.0 | 0.4 | GO:0061318 | renal filtration cell differentiation(GO:0061318) glomerular visceral epithelial cell differentiation(GO:0072112) |
| 0.0 | 1.7 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 0.1 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.8 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.0 | 0.5 | GO:0001658 | branching involved in ureteric bud morphogenesis(GO:0001658) |
| 0.0 | 1.0 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.1 | GO:1902775 | mitochondrial ribosome assembly(GO:0061668) mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.2 | GO:2000811 | negative regulation of anoikis(GO:2000811) |
| 0.0 | 0.6 | GO:0035115 | embryonic forelimb morphogenesis(GO:0035115) |
| 0.0 | 0.1 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.0 | 1.1 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.2 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.5 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.0 | 0.9 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.0 | GO:1903527 | regulation of membrane tubulation(GO:1903525) positive regulation of membrane tubulation(GO:1903527) |
| 0.0 | 1.1 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.0 | 0.2 | GO:2000111 | positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.0 | 0.0 | GO:0060168 | positive regulation of adenosine receptor signaling pathway(GO:0060168) |
| 0.0 | 0.9 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 0.2 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.0 | 1.0 | GO:0061615 | NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.4 | GO:0003197 | endocardial cushion development(GO:0003197) |
| 0.0 | 0.2 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.0 | 0.2 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.5 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.2 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.7 | GO:0006898 | receptor-mediated endocytosis(GO:0006898) |
| 0.0 | 0.2 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.4 | GO:0070233 | negative regulation of T cell apoptotic process(GO:0070233) |
| 0.0 | 1.0 | GO:0021954 | central nervous system neuron development(GO:0021954) |
| 0.0 | 0.6 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.0 | 0.1 | GO:0036109 | alpha-linolenic acid metabolic process(GO:0036109) |
| 0.0 | 0.4 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) |
| 0.0 | 1.0 | GO:0010634 | positive regulation of epithelial cell migration(GO:0010634) |
| 0.0 | 0.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.9 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.0 | 0.3 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.2 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 1.3 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.0 | 0.1 | GO:0035136 | forelimb morphogenesis(GO:0035136) |
| 0.0 | 0.4 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.0 | 0.2 | GO:0039694 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.0 | 0.2 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.4 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 0.4 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 0.3 | GO:0042921 | glucocorticoid receptor signaling pathway(GO:0042921) |
| 0.0 | 0.6 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 | 0.3 | GO:0007140 | male meiosis(GO:0007140) |
| 0.0 | 0.1 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.0 | 0.2 | GO:0030818 | negative regulation of cAMP biosynthetic process(GO:0030818) |
| 0.0 | 0.2 | GO:1903432 | regulation of TORC1 signaling(GO:1903432) |
| 0.0 | 1.1 | GO:0014066 | regulation of phosphatidylinositol 3-kinase signaling(GO:0014066) |
| 0.0 | 0.7 | GO:0031640 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) |
| 0.0 | 1.0 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.3 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
| 0.0 | 1.9 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 1.3 | GO:0008584 | male gonad development(GO:0008584) development of primary male sexual characteristics(GO:0046546) |
| 0.0 | 1.1 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.2 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.2 | GO:0048483 | autonomic nervous system development(GO:0048483) |
| 0.0 | 0.1 | GO:0060021 | palate development(GO:0060021) |
| 0.0 | 0.0 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.1 | GO:0038170 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.0 | 0.4 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.9 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.4 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.2 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.5 | 27.3 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 2.8 | 22.0 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 2.6 | 15.7 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 2.3 | 9.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 2.1 | 16.6 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 2.0 | 20.0 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 1.9 | 5.7 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 1.7 | 5.2 | GO:0019034 | viral replication complex(GO:0019034) |
| 1.4 | 12.7 | GO:0071439 | clathrin complex(GO:0071439) |
| 1.1 | 5.6 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 1.0 | 11.9 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 1.0 | 26.5 | GO:0033176 | proton-transporting V-type ATPase complex(GO:0033176) |
| 1.0 | 16.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.9 | 5.1 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.8 | 20.0 | GO:0031082 | BLOC complex(GO:0031082) |
| 0.8 | 3.3 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.8 | 3.1 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.8 | 9.4 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.8 | 7.0 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.7 | 5.6 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.6 | 1.9 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.6 | 8.9 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.6 | 2.5 | GO:0070695 | FHF complex(GO:0070695) |
| 0.6 | 6.2 | GO:0097413 | Lewy body(GO:0097413) |
| 0.6 | 4.3 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.6 | 2.4 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.6 | 10.7 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.6 | 3.4 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.6 | 4.5 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.6 | 2.2 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.5 | 3.3 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.5 | 5.8 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.5 | 6.2 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.5 | 2.7 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.4 | 5.7 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.4 | 3.0 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.4 | 6.8 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.4 | 11.5 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.4 | 5.8 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.4 | 1.6 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.4 | 10.0 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.4 | 1.1 | GO:0034667 | integrin alpha3-beta1 complex(GO:0034667) |
| 0.4 | 3.6 | GO:0097433 | dense body(GO:0097433) |
| 0.4 | 1.8 | GO:0000801 | central element(GO:0000801) |
| 0.3 | 1.4 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.3 | 18.5 | GO:0016235 | aggresome(GO:0016235) |
| 0.3 | 1.4 | GO:0089701 | U2AF(GO:0089701) |
| 0.3 | 1.6 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.2 | 0.5 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.2 | 1.4 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.2 | 0.9 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.2 | 1.4 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.2 | 2.1 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.2 | 1.6 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.2 | 1.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.2 | 0.6 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.2 | 3.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.2 | 2.4 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.2 | 2.6 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.2 | 3.4 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.2 | 5.4 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.2 | 26.3 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.2 | 0.6 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.2 | 1.8 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.2 | 2.2 | GO:0000153 | cytoplasmic ubiquitin ligase complex(GO:0000153) |
| 0.2 | 4.9 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.2 | 1.2 | GO:0042627 | chylomicron(GO:0042627) |
| 0.2 | 0.6 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 4.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 8.1 | GO:0032592 | integral component of mitochondrial membrane(GO:0032592) |
| 0.1 | 5.5 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 4.8 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 0.8 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 0.8 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 1.6 | GO:0005915 | zonula adherens(GO:0005915) catenin complex(GO:0016342) flotillin complex(GO:0016600) |
| 0.1 | 2.6 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 1.1 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 1.3 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.1 | 0.8 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.1 | 0.8 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 0.5 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.1 | 0.7 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 2.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 3.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 31.4 | GO:0005938 | cell cortex(GO:0005938) |
| 0.1 | 1.0 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 1.5 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.1 | 5.0 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.1 | 0.8 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.1 | 0.8 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.1 | 2.2 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.1 | 0.6 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.1 | 8.9 | GO:0031514 | motile cilium(GO:0031514) |
| 0.1 | 3.0 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 5.1 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 0.5 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.1 | 7.4 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 0.1 | GO:0035838 | growing cell tip(GO:0035838) |
| 0.1 | 7.1 | GO:0005770 | late endosome(GO:0005770) |
| 0.1 | 3.1 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.1 | 0.4 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 1.9 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 2.3 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.1 | 4.3 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 20.9 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 3.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 2.8 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 0.3 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 0.4 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 0.8 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 1.2 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.2 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 11.6 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 0.9 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 1.9 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 1.4 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 1.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 5.8 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 2.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 4.5 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.6 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.8 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 8.3 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 1.3 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 3.0 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 1.0 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 1.5 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 1.3 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.2 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 0.7 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 2.1 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.5 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 0.2 | GO:0042567 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 3.1 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 1.7 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.8 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 1.6 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.2 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 2.3 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.1 | GO:0044292 | dendrite terminus(GO:0044292) dendritic growth cone(GO:0044294) |
| 0.0 | 4.5 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 1.0 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 4.0 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 4.5 | GO:0005929 | cilium(GO:0005929) |
| 0.0 | 0.8 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.4 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.5 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 2.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.6 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 1.4 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 1.0 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.6 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.4 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 2.8 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.1 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.7 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.0 | 4.0 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.2 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.6 | GO:0030016 | myofibril(GO:0030016) |
| 0.0 | 3.3 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 0.2 | GO:0098791 | Golgi subcompartment(GO:0098791) |
| 0.0 | 0.1 | GO:0005859 | muscle myosin complex(GO:0005859) myosin II complex(GO:0016460) |
| 0.0 | 0.6 | GO:0031253 | cell projection membrane(GO:0031253) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 12.0 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 3.0 | 8.9 | GO:0015439 | heme-transporting ATPase activity(GO:0015439) |
| 2.9 | 14.5 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 2.8 | 11.4 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 2.7 | 10.7 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 1.9 | 5.8 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 1.9 | 30.0 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 1.9 | 5.6 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 1.6 | 44.8 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 1.5 | 4.5 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 1.4 | 4.1 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 1.3 | 29.1 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 1.3 | 15.7 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 1.3 | 3.9 | GO:0030984 | kininogen binding(GO:0030984) |
| 1.2 | 5.0 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 1.2 | 4.8 | GO:0022858 | L-alanine transmembrane transporter activity(GO:0015180) alanine transmembrane transporter activity(GO:0022858) |
| 1.2 | 7.0 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 1.1 | 8.0 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 1.0 | 3.1 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 1.0 | 20.3 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 1.0 | 5.8 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 1.0 | 4.8 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.9 | 9.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.9 | 4.3 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.8 | 3.3 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.8 | 2.4 | GO:0052810 | 1-phosphatidylinositol-5-kinase activity(GO:0052810) |
| 0.8 | 18.8 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.7 | 2.2 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.7 | 2.1 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.6 | 3.7 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.6 | 4.7 | GO:0016997 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) |
| 0.6 | 8.5 | GO:0005402 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) |
| 0.6 | 6.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.6 | 3.4 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.5 | 1.6 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.5 | 2.7 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.5 | 2.1 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) aspartic-type endopeptidase inhibitor activity(GO:0019828) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.5 | 4.7 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.5 | 5.7 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.5 | 4.0 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.5 | 1.9 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.4 | 2.7 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.4 | 18.2 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.4 | 0.4 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.4 | 3.9 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.4 | 3.9 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.4 | 1.6 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.4 | 1.2 | GO:0035501 | MH1 domain binding(GO:0035501) |
| 0.4 | 1.6 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.4 | 1.2 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.4 | 1.1 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.4 | 3.0 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.4 | 2.2 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.4 | 2.1 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.3 | 1.3 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.3 | 2.6 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.3 | 1.3 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.3 | 1.3 | GO:0008466 | glycogenin glucosyltransferase activity(GO:0008466) |
| 0.3 | 7.5 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.3 | 7.8 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.3 | 3.5 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.3 | 1.4 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.3 | 8.3 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.3 | 5.6 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.3 | 7.0 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.3 | 5.6 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.3 | 2.5 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.3 | 9.8 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.3 | 1.6 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.3 | 3.5 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.3 | 0.8 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.3 | 1.1 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.3 | 1.6 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.3 | 0.8 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.3 | 1.6 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.3 | 0.8 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.3 | 1.5 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.2 | 4.4 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.2 | 3.4 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.2 | 4.8 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.2 | 2.3 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.2 | 1.9 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.2 | 3.0 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.2 | 0.9 | GO:0032145 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.2 | 0.7 | GO:0004853 | uroporphyrinogen decarboxylase activity(GO:0004853) |
| 0.2 | 0.7 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.2 | 3.2 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.2 | 1.4 | GO:1901567 | icosanoid binding(GO:0050542) icosatetraenoic acid binding(GO:0050543) arachidonic acid binding(GO:0050544) fatty acid derivative binding(GO:1901567) |
| 0.2 | 1.3 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.2 | 1.0 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.2 | 0.6 | GO:0030375 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.2 | 0.6 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.2 | 0.6 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.2 | 1.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.2 | 0.6 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.2 | 0.6 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.2 | 0.4 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.2 | 0.5 | GO:0031177 | oleoyl-[acyl-carrier-protein] hydrolase activity(GO:0004320) myristoyl-[acyl-carrier-protein] hydrolase activity(GO:0016295) palmitoyl-[acyl-carrier-protein] hydrolase activity(GO:0016296) acyl-[acyl-carrier-protein] hydrolase activity(GO:0016297) S-acetyltransferase activity(GO:0016418) phosphopantetheine binding(GO:0031177) |
| 0.2 | 0.5 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.2 | 0.7 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.2 | 1.9 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.2 | 8.2 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.2 | 1.2 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.2 | 4.9 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.2 | 16.7 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.2 | 1.6 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.2 | 1.6 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.2 | 1.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.2 | 0.5 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.2 | 0.5 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.2 | 2.6 | GO:0015250 | water channel activity(GO:0015250) |
| 0.2 | 1.8 | GO:0035240 | dopamine binding(GO:0035240) |
| 0.2 | 7.3 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 4.1 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 13.7 | GO:0050660 | flavin adenine dinucleotide binding(GO:0050660) |
| 0.1 | 1.3 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.1 | 9.2 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 1.4 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 0.4 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.1 | 14.3 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 6.0 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 0.7 | GO:0004306 | ethanolamine-phosphate cytidylyltransferase activity(GO:0004306) |
| 0.1 | 1.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.1 | 0.5 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 1.5 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 0.3 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 7.2 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 0.4 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.1 | 1.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 0.7 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 1.5 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 0.7 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 1.4 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 0.8 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.1 | 0.6 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 0.9 | GO:0015929 | hexosaminidase activity(GO:0015929) |
| 0.1 | 0.6 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 0.3 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.1 | 5.2 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.1 | 0.4 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.1 | 2.4 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 2.5 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.1 | 1.0 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.6 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.1 | 0.2 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.1 | 1.7 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 0.6 | GO:0000982 | transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0000982) |
| 0.1 | 1.5 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.1 | 1.6 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.1 | 0.4 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.1 | 1.0 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.1 | 0.4 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 0.7 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 2.1 | GO:0005310 | dicarboxylic acid transmembrane transporter activity(GO:0005310) |
| 0.1 | 0.6 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.1 | 1.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 6.3 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.1 | 0.3 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.1 | 0.3 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.1 | 0.7 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 5.9 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 0.5 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 1.3 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.1 | 2.2 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 1.2 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 0.7 | GO:0030346 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) protein phosphatase 2B binding(GO:0030346) |
| 0.1 | 3.3 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 1.6 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 2.0 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.1 | 0.6 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 6.9 | GO:0005525 | GTP binding(GO:0005525) |
| 0.1 | 3.4 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 0.5 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.1 | 10.6 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 1.6 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 0.6 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.1 | 1.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 0.4 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.1 | 0.2 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.1 | 0.5 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 1.1 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 0.6 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 0.7 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 1.0 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 0.2 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.1 | 1.1 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 2.3 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 1.4 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.1 | 0.2 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.1 | 1.2 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 2.4 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.1 | 2.3 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 4.2 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 5.8 | GO:0043492 | ATPase activity, coupled to movement of substances(GO:0043492) |
| 0.0 | 0.1 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.0 | 0.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.7 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 11.4 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.7 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 1.1 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 1.3 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 1.5 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 2.0 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.2 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.4 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 12.3 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 0.2 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 1.0 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 1.7 | GO:0004114 | 3',5'-cyclic-nucleotide phosphodiesterase activity(GO:0004114) |
| 0.0 | 0.4 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 3.2 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.5 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 1.8 | GO:0005215 | transporter activity(GO:0005215) |
| 0.0 | 0.5 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.2 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.8 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.8 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.1 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.0 | 3.9 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.3 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 1.6 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.2 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 1.0 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 1.7 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 1.5 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 3.4 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.2 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.2 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.1 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 0.3 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 0.3 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.1 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.3 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.0 | 0.2 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0019203 | carbohydrate phosphatase activity(GO:0019203) sugar-phosphatase activity(GO:0050308) |
| 0.0 | 0.2 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 1.3 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 7.0 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.3 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.5 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 1.1 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 0.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.1 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.7 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.0 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 2.0 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 18.8 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.4 | 12.3 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.3 | 18.2 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.3 | 10.5 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.3 | 38.3 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.3 | 5.7 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.3 | 10.4 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.2 | 4.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.2 | 1.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.2 | 8.6 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.2 | 10.4 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.2 | 3.1 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 6.5 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.1 | 0.4 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 1.5 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 3.3 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 8.9 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 8.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 2.3 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 4.9 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.1 | 0.8 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 4.7 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 10.9 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 2.6 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 2.3 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 1.1 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 3.1 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.7 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.0 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.5 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 1.5 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.9 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.8 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.7 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 6.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.6 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.5 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.3 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.9 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.5 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.4 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.1 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.2 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.6 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.2 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.5 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.4 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.8 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 49.3 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.8 | 8.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.6 | 17.5 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.5 | 9.1 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.5 | 23.0 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.4 | 4.9 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.4 | 11.2 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.4 | 11.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.4 | 6.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.3 | 27.8 | REACTOME CELL DEATH SIGNALLING VIA NRAGE NRIF AND NADE | Genes involved in Cell death signalling via NRAGE, NRIF and NADE |
| 0.3 | 13.0 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.3 | 16.2 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.3 | 2.3 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.2 | 6.5 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.2 | 1.0 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.2 | 4.9 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.2 | 2.6 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.2 | 19.2 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.2 | 2.6 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.2 | 3.6 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.2 | 1.0 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 2.5 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 8.6 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 10.4 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 0.8 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 2.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 4.3 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 5.7 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.1 | 2.5 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 1.4 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 1.7 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 1.6 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 0.4 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 1.3 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 1.1 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.1 | 2.3 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 1.5 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 1.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 1.4 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 3.0 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.1 | 0.5 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.1 | 8.1 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 2.0 | REACTOME KERATAN SULFATE KERATIN METABOLISM | Genes involved in Keratan sulfate/keratin metabolism |
| 0.1 | 1.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 1.1 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 0.9 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 4.9 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 1.4 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.1 | 6.1 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 0.8 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.1 | 1.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 3.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.6 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 1.6 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.9 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.9 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.8 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 11.3 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.6 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.9 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 1.0 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.4 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.5 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.6 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.4 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 5.8 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.9 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.6 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 1.7 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.8 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 2.6 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 1.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.1 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.4 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.1 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.2 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 1.4 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.3 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.1 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |