Project
averaged GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
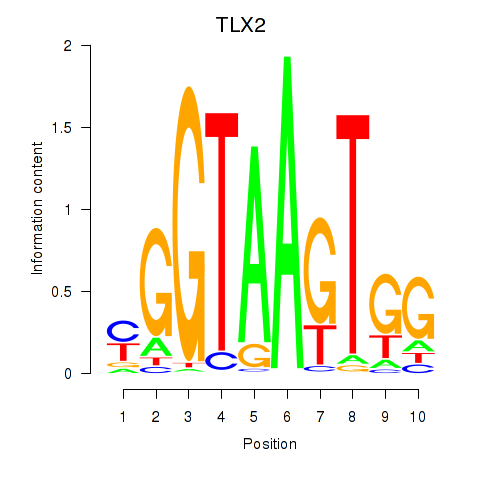
Results for TLX2
Z-value: 1.35
Transcription factors associated with TLX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TLX2
|
ENSG00000115297.9 | T cell leukemia homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TLX2 | hg19_v2_chr2_+_74741569_74741620 | 0.54 | 5.6e-18 | Click! |
Activity profile of TLX2 motif
Sorted Z-values of TLX2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of TLX2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.5 | 19.4 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 5.1 | 20.3 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 4.8 | 14.3 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 4.0 | 12.0 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 3.4 | 10.3 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 3.0 | 9.0 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 2.7 | 13.3 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 2.6 | 7.8 | GO:1905154 | negative regulation of membrane invagination(GO:1905154) |
| 2.4 | 4.8 | GO:0052042 | induction of programmed cell death(GO:0012502) positive regulation of apoptotic process in other organism(GO:0044533) positive regulation by symbiont of host programmed cell death(GO:0052042) positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052330) positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052501) |
| 2.2 | 13.1 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 1.9 | 15.3 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 1.9 | 7.5 | GO:2000537 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 1.8 | 5.5 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 1.8 | 5.5 | GO:0021778 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 1.6 | 6.5 | GO:0002884 | regulation of type IV hypersensitivity(GO:0001807) negative regulation of hypersensitivity(GO:0002884) |
| 1.5 | 4.6 | GO:0097056 | seryl-tRNA aminoacylation(GO:0006434) selenocysteinyl-tRNA(Sec) biosynthetic process(GO:0097056) |
| 1.5 | 8.9 | GO:0032119 | sequestering of zinc ion(GO:0032119) |
| 1.5 | 4.4 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 1.5 | 7.3 | GO:1903282 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) regulation of peroxidase activity(GO:2000468) positive regulation of peroxidase activity(GO:2000470) |
| 1.5 | 4.4 | GO:0002644 | negative regulation of tolerance induction(GO:0002644) interleukin-4 biosynthetic process(GO:0042097) regulation of interleukin-4 biosynthetic process(GO:0045402) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 1.4 | 8.7 | GO:0051136 | regulation of NK T cell differentiation(GO:0051136) |
| 1.4 | 10.1 | GO:0033590 | response to cobalamin(GO:0033590) |
| 1.4 | 4.3 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 1.4 | 4.3 | GO:0042938 | dipeptide transport(GO:0042938) |
| 1.4 | 4.1 | GO:1900138 | negative regulation of phospholipase A2 activity(GO:1900138) |
| 1.4 | 1.4 | GO:2000196 | positive regulation of female gonad development(GO:2000196) |
| 1.3 | 3.8 | GO:0018262 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 1.2 | 7.2 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 1.2 | 3.5 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 1.2 | 5.8 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) negative regulation of leukocyte adhesion to vascular endothelial cell(GO:1904995) |
| 1.1 | 4.5 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 1.1 | 13.5 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 1.1 | 4.5 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 1.1 | 3.3 | GO:0071934 | thiamine transport(GO:0015888) thiamine transmembrane transport(GO:0071934) |
| 1.1 | 16.3 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 1.0 | 3.0 | GO:0043163 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 1.0 | 2.0 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 1.0 | 6.9 | GO:2000546 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 1.0 | 3.0 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 1.0 | 10.7 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 1.0 | 2.9 | GO:2000583 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.9 | 4.6 | GO:0051142 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.9 | 2.8 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) negative regulation of DNA catabolic process(GO:1903625) regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 0.9 | 3.7 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.9 | 2.7 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.9 | 5.5 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.9 | 0.9 | GO:1902567 | negative regulation of eosinophil activation(GO:1902567) |
| 0.9 | 5.4 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.9 | 3.5 | GO:0044240 | activation of phospholipase A2 activity(GO:0032431) multicellular organism lipid catabolic process(GO:0044240) |
| 0.9 | 2.6 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.9 | 3.4 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.8 | 5.6 | GO:0015824 | proline transport(GO:0015824) |
| 0.8 | 6.4 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.8 | 2.4 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.8 | 2.4 | GO:0060168 | positive regulation of adenosine receptor signaling pathway(GO:0060168) |
| 0.8 | 2.3 | GO:0009726 | detection of endogenous stimulus(GO:0009726) |
| 0.8 | 4.6 | GO:0052565 | response to defense-related nitric oxide production by other organism involved in symbiotic interaction(GO:0052551) response to defense-related host nitric oxide production(GO:0052565) |
| 0.8 | 2.3 | GO:0021919 | BMP signaling pathway involved in spinal cord dorsal/ventral patterning(GO:0021919) |
| 0.8 | 5.3 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.7 | 3.7 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.7 | 4.4 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.7 | 1.5 | GO:1902573 | positive regulation of serine-type endopeptidase activity(GO:1900005) positive regulation of serine-type peptidase activity(GO:1902573) |
| 0.7 | 5.1 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.7 | 3.5 | GO:0051946 | regulation of amino acid uptake involved in synaptic transmission(GO:0051941) regulation of glutamate uptake involved in transmission of nerve impulse(GO:0051946) regulation of L-glutamate import(GO:1900920) |
| 0.7 | 2.1 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.7 | 2.8 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.7 | 4.2 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.7 | 2.1 | GO:0072093 | ureteric bud invasion(GO:0072092) metanephric renal vesicle formation(GO:0072093) |
| 0.7 | 2.0 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.7 | 4.6 | GO:0032100 | positive regulation of response to food(GO:0032097) positive regulation of appetite(GO:0032100) |
| 0.6 | 4.5 | GO:0019064 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.6 | 11.6 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.6 | 6.4 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.6 | 3.2 | GO:1903301 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.6 | 7.7 | GO:0048711 | positive regulation of astrocyte differentiation(GO:0048711) |
| 0.6 | 4.6 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.5 | 18.5 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.5 | 2.2 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.5 | 4.5 | GO:0015705 | iodide transport(GO:0015705) |
| 0.5 | 2.9 | GO:0036101 | leukotriene catabolic process(GO:0036100) leukotriene B4 catabolic process(GO:0036101) leukotriene B4 metabolic process(GO:0036102) icosanoid catabolic process(GO:1901523) fatty acid derivative catabolic process(GO:1901569) |
| 0.5 | 2.0 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.5 | 2.9 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.5 | 3.8 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.5 | 0.9 | GO:0007231 | osmosensory signaling pathway(GO:0007231) |
| 0.5 | 6.2 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.5 | 2.3 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.5 | 1.9 | GO:0042758 | long-chain fatty acid catabolic process(GO:0042758) |
| 0.5 | 3.7 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.5 | 3.2 | GO:0048703 | embryonic viscerocranium morphogenesis(GO:0048703) |
| 0.4 | 2.7 | GO:1902715 | secretory granule localization(GO:0032252) positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.4 | 0.9 | GO:0048867 | stem cell fate determination(GO:0048867) |
| 0.4 | 1.3 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.4 | 1.3 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.4 | 0.9 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.4 | 2.5 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.4 | 3.3 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.4 | 11.0 | GO:0098743 | cell aggregation(GO:0098743) |
| 0.4 | 7.0 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.4 | 1.9 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.4 | 1.9 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.4 | 1.5 | GO:0051344 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.4 | 3.0 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.4 | 9.8 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.4 | 4.1 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.4 | 16.2 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.4 | 2.1 | GO:0010663 | positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.4 | 2.5 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.3 | 1.0 | GO:0001694 | histamine biosynthetic process(GO:0001694) |
| 0.3 | 13.6 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.3 | 2.3 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.3 | 3.0 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.3 | 2.3 | GO:1904469 | positive regulation of tumor necrosis factor secretion(GO:1904469) |
| 0.3 | 4.4 | GO:0006705 | mineralocorticoid biosynthetic process(GO:0006705) mineralocorticoid metabolic process(GO:0008212) |
| 0.3 | 0.9 | GO:2001045 | closure of optic fissure(GO:0061386) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.3 | 3.4 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.3 | 3.4 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.3 | 1.1 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.3 | 7.5 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.3 | 1.1 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.3 | 4.9 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.3 | 0.8 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.3 | 0.8 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.3 | 2.3 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.2 | 4.7 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.2 | 0.7 | GO:0070781 | arginine biosynthetic process via ornithine(GO:0042450) response to biotin(GO:0070781) |
| 0.2 | 3.0 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.2 | 5.6 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.2 | 1.9 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.2 | 5.7 | GO:0048485 | sympathetic nervous system development(GO:0048485) |
| 0.2 | 1.1 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.2 | 11.5 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.2 | 4.5 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.2 | 13.9 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.2 | 4.7 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.2 | 2.9 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.2 | 7.6 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.2 | 7.2 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.2 | 5.4 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.2 | 4.1 | GO:0032011 | ARF protein signal transduction(GO:0032011) |
| 0.2 | 1.4 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.2 | 1.0 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.2 | 1.0 | GO:1903059 | positive regulation of lipoprotein metabolic process(GO:0050747) regulation of protein lipidation(GO:1903059) positive regulation of protein lipidation(GO:1903061) |
| 0.2 | 3.8 | GO:0051439 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051436) regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051439) |
| 0.2 | 1.7 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.2 | 3.0 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.2 | 2.7 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.2 | 2.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.2 | 8.0 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.2 | 5.1 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.2 | 2.6 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.2 | 4.2 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.2 | 0.8 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.2 | 1.3 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.2 | 10.0 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.2 | 1.9 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.2 | 9.0 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.2 | 2.4 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.2 | 3.2 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.1 | 0.9 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.1 | 1.0 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 | 2.4 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.1 | 1.5 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.1 | 2.1 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.1 | 9.1 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 22.0 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.1 | 1.0 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.1 | 6.1 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.1 | 3.3 | GO:0046174 | polyol catabolic process(GO:0046174) |
| 0.1 | 1.1 | GO:0034351 | regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.1 | 0.9 | GO:0071351 | interleukin-18-mediated signaling pathway(GO:0035655) cellular response to interleukin-18(GO:0071351) |
| 0.1 | 3.0 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.1 | 6.4 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.1 | 9.2 | GO:0070126 | mitochondrial translational elongation(GO:0070125) mitochondrial translational termination(GO:0070126) |
| 0.1 | 0.6 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.1 | 5.5 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.1 | 14.5 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.1 | 1.0 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 2.9 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.1 | 2.9 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 | 1.6 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 2.4 | GO:0060828 | regulation of canonical Wnt signaling pathway(GO:0060828) |
| 0.1 | 2.0 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.1 | 1.4 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.1 | 1.6 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 10.6 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.1 | 2.1 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.1 | 3.8 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 2.6 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.1 | 1.4 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.1 | 0.7 | GO:0035810 | positive regulation of urine volume(GO:0035810) |
| 0.1 | 2.7 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 0.5 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.1 | 1.2 | GO:0032988 | ribonucleoprotein complex disassembly(GO:0032988) |
| 0.1 | 11.1 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.1 | 6.0 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.1 | 1.0 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 2.3 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.1 | 0.4 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.1 | 2.3 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.1 | 0.7 | GO:0034384 | high-density lipoprotein particle clearance(GO:0034384) |
| 0.1 | 0.4 | GO:0045086 | positive regulation of interleukin-2 biosynthetic process(GO:0045086) |
| 0.1 | 2.6 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 3.5 | GO:1901264 | carbohydrate derivative transport(GO:1901264) |
| 0.1 | 2.9 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.1 | 0.2 | GO:0010193 | response to ozone(GO:0010193) negative regulation of interleukin-13 production(GO:0032696) |
| 0.1 | 4.0 | GO:0042100 | B cell proliferation(GO:0042100) |
| 0.1 | 1.0 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.1 | 1.3 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.1 | 3.1 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.1 | 1.1 | GO:1902895 | positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.1 | 0.2 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.1 | 0.5 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.4 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.1 | 7.3 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.1 | 1.6 | GO:0003016 | respiratory system process(GO:0003016) |
| 0.1 | 1.7 | GO:0072661 | protein targeting to plasma membrane(GO:0072661) |
| 0.1 | 1.8 | GO:0010390 | histone monoubiquitination(GO:0010390) |
| 0.1 | 1.3 | GO:0034204 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.1 | 11.0 | GO:0060271 | cilium morphogenesis(GO:0060271) |
| 0.1 | 1.8 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 0.4 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 3.7 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.5 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 1.1 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 3.6 | GO:0030593 | neutrophil chemotaxis(GO:0030593) |
| 0.0 | 3.5 | GO:0006898 | receptor-mediated endocytosis(GO:0006898) |
| 0.0 | 1.8 | GO:0035196 | production of miRNAs involved in gene silencing by miRNA(GO:0035196) |
| 0.0 | 2.0 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.0 | 4.5 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 1.6 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.4 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 1.1 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.8 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 6.1 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.5 | GO:0043149 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 2.5 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.0 | 0.4 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 1.9 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 2.2 | GO:0002456 | T cell mediated immunity(GO:0002456) |
| 0.0 | 5.5 | GO:0090305 | nucleic acid phosphodiester bond hydrolysis(GO:0090305) |
| 0.0 | 1.0 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 3.3 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 3.3 | GO:0000075 | cell cycle checkpoint(GO:0000075) |
| 0.0 | 0.4 | GO:0006813 | potassium ion transport(GO:0006813) |
| 0.0 | 0.9 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.8 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.1 | GO:0061325 | subthalamic nucleus development(GO:0021763) deltoid tuberosity development(GO:0035993) prolactin secreting cell differentiation(GO:0060127) left lung development(GO:0060459) left lung morphogenesis(GO:0060460) pulmonary vein morphogenesis(GO:0060577) superior vena cava morphogenesis(GO:0060578) cell proliferation involved in outflow tract morphogenesis(GO:0061325) |
| 0.0 | 0.8 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.7 | GO:0001938 | positive regulation of endothelial cell proliferation(GO:0001938) |
| 0.0 | 0.5 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.0 | 1.9 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.0 | 0.5 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 1.7 | GO:0010212 | response to ionizing radiation(GO:0010212) |
| 0.0 | 1.3 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) peptidyl-threonine modification(GO:0018210) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 7.7 | GO:0097453 | mesaxon(GO:0097453) ensheathing process(GO:1990015) |
| 2.1 | 8.4 | GO:1990745 | EARP complex(GO:1990745) |
| 1.8 | 16.4 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 1.6 | 7.8 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 1.3 | 6.4 | GO:0071546 | pi-body(GO:0071546) |
| 1.3 | 10.1 | GO:0043196 | varicosity(GO:0043196) |
| 1.2 | 9.6 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 1.2 | 4.7 | GO:0070876 | SOSS complex(GO:0070876) |
| 1.1 | 4.4 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 1.1 | 3.3 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 1.0 | 13.5 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 1.0 | 24.1 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.8 | 17.6 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.8 | 3.3 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.8 | 2.4 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.8 | 7.1 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.7 | 5.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.7 | 5.1 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.7 | 2.2 | GO:0016935 | glycine-gated chloride channel complex(GO:0016935) |
| 0.7 | 1.3 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.6 | 1.9 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.6 | 7.0 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.6 | 8.2 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.6 | 14.8 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.6 | 2.3 | GO:0031673 | H zone(GO:0031673) |
| 0.5 | 3.0 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.5 | 3.0 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.5 | 7.5 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.5 | 1.9 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.5 | 3.3 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.4 | 15.3 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.4 | 1.6 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.4 | 4.7 | GO:0036038 | MKS complex(GO:0036038) |
| 0.4 | 1.5 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.4 | 9.1 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.4 | 4.8 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.4 | 4.8 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.4 | 2.9 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.3 | 3.8 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.3 | 6.5 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.3 | 2.3 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.3 | 1.7 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.3 | 3.6 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.3 | 2.6 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 0.3 | 3.7 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.3 | 12.0 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.3 | 9.2 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.3 | 8.1 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.3 | 3.0 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.2 | 7.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.2 | 1.2 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.2 | 3.3 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.2 | 1.1 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.2 | 2.2 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.2 | 17.6 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.2 | 7.8 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.2 | 1.5 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.2 | 1.4 | GO:1903439 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.2 | 3.7 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.2 | 33.6 | GO:0030426 | growth cone(GO:0030426) |
| 0.2 | 5.0 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.2 | 3.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.2 | 2.0 | GO:0005845 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.2 | 9.1 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.2 | 9.4 | GO:0016235 | aggresome(GO:0016235) |
| 0.2 | 13.5 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.1 | 11.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 4.7 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 3.0 | GO:0044447 | axoneme part(GO:0044447) |
| 0.1 | 12.3 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.1 | 3.8 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 2.7 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.1 | 3.8 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 0.7 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 6.2 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 11.1 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 16.0 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 2.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 1.1 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 3.8 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 2.6 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 1.7 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.1 | 9.8 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 9.7 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.1 | 5.1 | GO:0043235 | receptor complex(GO:0043235) |
| 0.1 | 1.4 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.1 | 4.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 5.6 | GO:0001726 | ruffle(GO:0001726) |
| 0.1 | 4.4 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 5.3 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 3.1 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 1.9 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 4.0 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 4.9 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 0.5 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 1.4 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.1 | 1.1 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 2.5 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 2.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 4.0 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 2.4 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 2.4 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 3.4 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 3.0 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 3.8 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 2.8 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 1.3 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.5 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 4.0 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 1.4 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 3.8 | GO:0098793 | presynapse(GO:0098793) |
| 0.0 | 2.8 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.7 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 2.3 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 26.7 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 4.4 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 0.8 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.0 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.4 | 19.2 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 5.1 | 15.3 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 2.9 | 14.3 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 2.5 | 7.5 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 2.2 | 6.5 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 1.9 | 7.6 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 1.7 | 5.0 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 1.6 | 9.5 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 1.5 | 4.6 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 1.5 | 7.3 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 1.5 | 4.4 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 1.3 | 13.0 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 1.1 | 3.3 | GO:0015403 | thiamine transmembrane transporter activity(GO:0015234) thiamine uptake transmembrane transporter activity(GO:0015403) |
| 1.1 | 5.3 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 1.0 | 5.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 1.0 | 2.9 | GO:0052870 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 1.0 | 24.1 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.9 | 5.7 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.9 | 5.6 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.9 | 3.6 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.8 | 3.2 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.8 | 2.3 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.8 | 8.5 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.7 | 8.0 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.7 | 7.9 | GO:0008430 | selenium binding(GO:0008430) |
| 0.7 | 5.0 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.7 | 6.4 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.7 | 8.8 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.7 | 2.0 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.7 | 2.0 | GO:0004796 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.6 | 4.5 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.6 | 3.2 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.6 | 3.1 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.6 | 1.9 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.6 | 1.8 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.6 | 18.5 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.6 | 3.0 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.6 | 2.3 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.6 | 8.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.5 | 13.1 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.5 | 9.7 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.5 | 4.3 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.5 | 9.8 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.5 | 5.1 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.5 | 3.5 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.5 | 3.9 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.5 | 5.2 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.5 | 6.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.5 | 1.4 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.5 | 4.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.5 | 12.3 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.4 | 2.6 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.4 | 10.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.4 | 2.0 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.4 | 5.8 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.4 | 6.8 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.4 | 5.5 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.4 | 5.8 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.4 | 17.6 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.4 | 5.6 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.3 | 4.9 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.3 | 10.3 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.3 | 5.2 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.3 | 0.9 | GO:0030366 | molybdopterin synthase activity(GO:0030366) |
| 0.3 | 1.5 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.3 | 4.8 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.3 | 0.9 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.3 | 2.7 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.3 | 8.8 | GO:0043236 | laminin binding(GO:0043236) |
| 0.3 | 2.6 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.3 | 3.4 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.3 | 0.9 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.3 | 1.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.3 | 3.7 | GO:0019215 | intermediate filament binding(GO:0019215) alpha-catenin binding(GO:0045294) |
| 0.3 | 1.7 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.3 | 2.2 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.3 | 2.0 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.3 | 6.9 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.3 | 9.0 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.3 | 1.4 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.3 | 3.9 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.3 | 0.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.3 | 6.8 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.3 | 0.8 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.2 | 13.8 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.2 | 5.1 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.2 | 2.4 | GO:0015386 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.2 | 6.5 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.2 | 2.6 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.2 | 7.0 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.2 | 2.8 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.2 | 3.5 | GO:0005351 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) |
| 0.2 | 2.3 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.2 | 3.4 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.2 | 5.5 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.2 | 21.8 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.2 | 0.9 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.2 | 3.5 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.2 | 2.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.2 | 1.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.2 | 1.4 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.2 | 0.2 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.2 | 3.7 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.2 | 6.7 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.2 | 1.9 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.2 | 9.0 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.2 | 24.6 | GO:0005178 | integrin binding(GO:0005178) |
| 0.2 | 9.1 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.2 | 3.0 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.2 | 1.3 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.2 | 4.1 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.2 | 1.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.2 | 1.0 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.2 | 4.8 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 0.4 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.1 | 1.2 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 1.9 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.1 | 3.3 | GO:0016701 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) |
| 0.1 | 3.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 2.7 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 3.5 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 1.4 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.1 | 1.5 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.1 | 1.4 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 0.8 | GO:0003774 | motor activity(GO:0003774) |
| 0.1 | 2.0 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 8.0 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.1 | 2.3 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 5.1 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 6.2 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) |
| 0.1 | 4.0 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 28.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 0.9 | GO:0032356 | oxidized DNA binding(GO:0032356) |
| 0.1 | 1.5 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 1.6 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 2.2 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 8.8 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 0.7 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.1 | 3.3 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 0.4 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 2.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 1.0 | GO:0019864 | IgG binding(GO:0019864) |
| 0.1 | 2.2 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 2.4 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 1.0 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.1 | 13.6 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 0.2 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 1.3 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 0.8 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.1 | 5.1 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 5.0 | GO:0015297 | antiporter activity(GO:0015297) |
| 0.1 | 1.1 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.1 | 2.5 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 1.2 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 1.7 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 2.3 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 2.1 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 1.1 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.1 | 2.4 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 72.1 | GO:0003700 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.1 | 2.9 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 1.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 0.5 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 1.0 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 1.1 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 2.8 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.3 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 3.2 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.3 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 3.0 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 3.0 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 1.0 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 2.6 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.5 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 1.7 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 1.3 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 1.4 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 3.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 4.6 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.4 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.3 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 1.7 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 7.4 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.6 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.7 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.6 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.1 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 2.1 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 1.6 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.6 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 19.1 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.4 | 14.8 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.3 | 15.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.3 | 13.5 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.3 | 2.5 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.2 | 4.8 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.2 | 9.7 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.2 | 2.5 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.2 | 3.0 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.2 | 5.5 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 3.5 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 6.5 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 4.3 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 4.4 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 21.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 8.9 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.1 | 7.2 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.1 | 4.6 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 3.3 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 2.2 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 4.4 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 3.0 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 3.5 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 4.7 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.1 | 6.4 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 3.5 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 3.5 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.1 | 2.1 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 3.6 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 1.3 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 1.0 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.1 | 3.8 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 2.2 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 4.9 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 3.2 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.1 | 6.0 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 7.0 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 3.3 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 1.6 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 2.7 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.1 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 1.9 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 1.9 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 3.2 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 10.3 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.9 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.3 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.4 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 1.1 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.4 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 4.5 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.6 | 1.9 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.6 | 11.0 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.5 | 5.2 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.5 | 13.5 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.5 | 10.4 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.5 | 9.0 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.5 | 25.0 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.4 | 2.7 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.3 | 14.2 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.3 | 4.4 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.3 | 4.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.3 | 4.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.3 | 6.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.3 | 4.0 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.3 | 7.1 | REACTOME PROLONGED ERK ACTIVATION EVENTS | Genes involved in Prolonged ERK activation events |
| 0.3 | 9.7 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.2 | 3.4 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.2 | 2.5 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.2 | 7.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.2 | 3.5 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.2 | 7.2 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.2 | 2.2 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.2 | 3.4 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.2 | 4.8 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.2 | 4.6 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.2 | 4.8 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.2 | 3.1 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.2 | 3.3 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.2 | 3.4 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.2 | 2.9 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.2 | 4.4 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.2 | 3.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.2 | 10.0 | REACTOME IL 2 SIGNALING | Genes involved in Interleukin-2 signaling |
| 0.2 | 4.8 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.2 | 1.6 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.2 | 3.7 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.2 | 11.5 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.2 | 3.0 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 5.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 8.4 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 3.0 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 3.2 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.1 | 5.0 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.1 | 4.1 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 9.8 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 15.1 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 2.0 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 2.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 2.2 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.1 | 1.9 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 7.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 3.3 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.1 | 1.3 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 4.9 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 4.1 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.1 | 7.8 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 2.0 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 2.6 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 3.5 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 1.4 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.1 | 3.5 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 3.3 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 2.7 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 1.3 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 2.0 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 3.3 | REACTOME ION CHANNEL TRANSPORT | Genes involved in Ion channel transport |
| 0.1 | 11.4 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 1.3 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.1 | 0.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 1.1 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 3.4 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 0.7 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.1 | 2.8 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 14.8 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 0.9 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.0 | 1.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 1.1 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 1.0 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 1.9 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.0 | 1.1 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.0 | 0.6 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 1.9 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.0 | 1.0 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.2 | REACTOME PI3K EVENTS IN ERBB4 SIGNALING | Genes involved in PI3K events in ERBB4 signaling |
| 0.0 | 1.5 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.8 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 0.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |