Project
averaged GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for VDR
Z-value: 0.86
Transcription factors associated with VDR
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
VDR
|
ENSG00000111424.6 | vitamin D receptor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| VDR | hg19_v2_chr12_-_48298785_48298828 | 0.11 | 9.2e-02 | Click! |
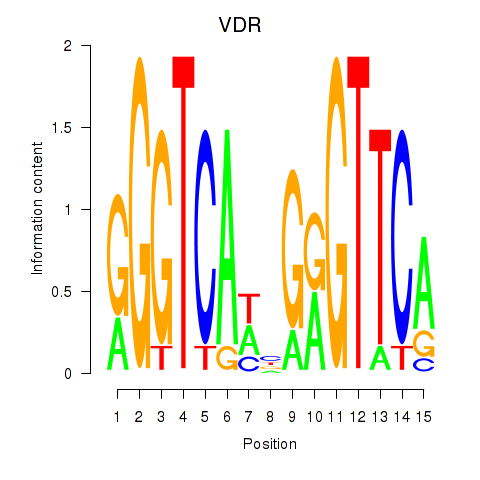
Activity profile of VDR motif
Sorted Z-values of VDR motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_34756219 | 5.38 |
ENST00000451448.2
ENST00000394359.3 |
TBC1D3C
TBC1D3H
|
TBC1 domain family, member 3C TBC1 domain family, member 3H |
| chr17_-_34808047 | 5.37 |
ENST00000592614.1
ENST00000591542.1 ENST00000330458.7 ENST00000341264.6 ENST00000592987.1 ENST00000400684.4 |
TBC1D3G
TBC1D3H
|
TBC1 domain family, member 3G TBC1 domain family, member 3H |
| chr17_-_34807272 | 5.30 |
ENST00000535592.1
ENST00000394453.1 |
TBC1D3G
|
TBC1 domain family, member 3G |
| chr17_-_36348610 | 5.29 |
ENST00000339023.4
ENST00000354664.4 |
TBC1D3
|
TBC1 domain family, member 3 |
| chr17_-_36347835 | 5.25 |
ENST00000519532.1
|
TBC1D3
|
TBC1 domain family, member 3 |
| chr17_+_36284791 | 4.95 |
ENST00000505415.1
|
TBC1D3F
|
TBC1 domain family, member 3F |
| chr22_+_18593446 | 4.89 |
ENST00000316027.6
|
TUBA8
|
tubulin, alpha 8 |
| chr5_-_68665084 | 4.72 |
ENST00000509462.1
|
TAF9
|
TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 32kDa |
| chr1_+_200993071 | 4.66 |
ENST00000446333.1
ENST00000458003.1 |
RP11-168O16.1
|
RP11-168O16.1 |
| chr2_-_85625857 | 4.48 |
ENST00000453973.1
|
CAPG
|
capping protein (actin filament), gelsolin-like |
| chr10_-_13523073 | 4.16 |
ENST00000440282.1
|
BEND7
|
BEN domain containing 7 |
| chr1_-_19426149 | 4.05 |
ENST00000429347.2
|
UBR4
|
ubiquitin protein ligase E3 component n-recognin 4 |
| chr2_-_230786619 | 3.89 |
ENST00000389045.3
ENST00000409677.1 |
TRIP12
|
thyroid hormone receptor interactor 12 |
| chr18_-_6414884 | 3.85 |
ENST00000317931.7
ENST00000284898.6 ENST00000400104.3 |
L3MBTL4
|
l(3)mbt-like 4 (Drosophila) |
| chr2_+_111490161 | 3.76 |
ENST00000340561.4
|
ACOXL
|
acyl-CoA oxidase-like |
| chr2_-_230786679 | 3.73 |
ENST00000543084.1
ENST00000343290.5 ENST00000389044.4 ENST00000283943.5 |
TRIP12
|
thyroid hormone receptor interactor 12 |
| chr5_-_179227540 | 3.70 |
ENST00000520875.1
|
MGAT4B
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme B |
| chr11_-_104817919 | 3.67 |
ENST00000533252.1
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr6_+_31802685 | 3.60 |
ENST00000375639.2
ENST00000375638.3 ENST00000375635.2 ENST00000375642.2 ENST00000395789.1 |
C6orf48
|
chromosome 6 open reading frame 48 |
| chr2_-_235405168 | 3.49 |
ENST00000339728.3
|
ARL4C
|
ADP-ribosylation factor-like 4C |
| chr3_+_54157480 | 3.42 |
ENST00000490478.1
|
CACNA2D3
|
calcium channel, voltage-dependent, alpha 2/delta subunit 3 |
| chr17_-_34591208 | 3.40 |
ENST00000336331.5
|
TBC1D3C
|
TBC1 domain family, member 3C |
| chr17_-_7082861 | 3.38 |
ENST00000269299.3
|
ASGR1
|
asialoglycoprotein receptor 1 |
| chr19_+_1407517 | 3.37 |
ENST00000336761.6
ENST00000233078.4 |
DAZAP1
|
DAZ associated protein 1 |
| chr12_-_133050726 | 3.36 |
ENST00000595994.1
|
MUC8
|
mucin 8 |
| chr10_-_43892279 | 3.31 |
ENST00000443950.2
|
HNRNPF
|
heterogeneous nuclear ribonucleoprotein F |
| chr12_+_52626898 | 3.23 |
ENST00000331817.5
|
KRT7
|
keratin 7 |
| chr15_-_59949667 | 3.18 |
ENST00000396061.1
|
GTF2A2
|
general transcription factor IIA, 2, 12kDa |
| chr10_-_43892668 | 3.16 |
ENST00000544000.1
|
HNRNPF
|
heterogeneous nuclear ribonucleoprotein F |
| chr16_-_3767551 | 3.08 |
ENST00000246957.5
|
TRAP1
|
TNF receptor-associated protein 1 |
| chr11_+_64808675 | 3.05 |
ENST00000529996.1
|
SAC3D1
|
SAC3 domain containing 1 |
| chr1_+_52082751 | 3.03 |
ENST00000447887.1
ENST00000435686.2 ENST00000428468.1 ENST00000453295.1 |
OSBPL9
|
oxysterol binding protein-like 9 |
| chr11_+_6624970 | 3.03 |
ENST00000420936.2
ENST00000528995.1 |
ILK
|
integrin-linked kinase |
| chr12_+_110906169 | 3.00 |
ENST00000377673.5
|
FAM216A
|
family with sequence similarity 216, member A |
| chr5_-_179051579 | 2.98 |
ENST00000505811.1
ENST00000515714.1 ENST00000513225.1 ENST00000503664.1 ENST00000356731.5 ENST00000523137.1 |
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 (H) |
| chr15_-_59949693 | 2.97 |
ENST00000396063.1
ENST00000396064.3 ENST00000484743.1 ENST00000559706.1 ENST00000396060.2 |
GTF2A2
|
general transcription factor IIA, 2, 12kDa |
| chr2_+_220363579 | 2.97 |
ENST00000313597.5
ENST00000373917.3 ENST00000358215.3 ENST00000373908.1 ENST00000455657.1 ENST00000435316.1 ENST00000341142.3 |
GMPPA
|
GDP-mannose pyrophosphorylase A |
| chr19_-_40331345 | 2.94 |
ENST00000597224.1
|
FBL
|
fibrillarin |
| chr12_+_10658489 | 2.92 |
ENST00000538173.1
|
EIF2S3L
|
Putative eukaryotic translation initiation factor 2 subunit 3-like protein |
| chr17_-_79805146 | 2.91 |
ENST00000415593.1
|
P4HB
|
prolyl 4-hydroxylase, beta polypeptide |
| chr9_+_138391805 | 2.80 |
ENST00000371785.1
|
MRPS2
|
mitochondrial ribosomal protein S2 |
| chr6_-_42016385 | 2.77 |
ENST00000502771.1
ENST00000508143.1 ENST00000514588.1 ENST00000510503.1 ENST00000415497.2 ENST00000372988.4 |
CCND3
|
cyclin D3 |
| chr11_+_6624955 | 2.74 |
ENST00000299421.4
ENST00000537806.1 |
ILK
|
integrin-linked kinase |
| chr14_-_35873856 | 2.66 |
ENST00000553342.1
ENST00000216797.5 ENST00000557140.1 |
NFKBIA
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha |
| chr19_+_13261216 | 2.63 |
ENST00000587885.1
ENST00000292433.3 |
IER2
|
immediate early response 2 |
| chr19_+_14672755 | 2.62 |
ENST00000594545.1
|
TECR
|
trans-2,3-enoyl-CoA reductase |
| chr11_+_6625046 | 2.60 |
ENST00000396751.2
|
ILK
|
integrin-linked kinase |
| chrX_+_153770421 | 2.52 |
ENST00000369609.5
ENST00000369607.1 |
IKBKG
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr16_-_3767506 | 2.46 |
ENST00000538171.1
|
TRAP1
|
TNF receptor-associated protein 1 |
| chr9_+_138392483 | 2.44 |
ENST00000241600.5
|
MRPS2
|
mitochondrial ribosomal protein S2 |
| chr1_+_198608146 | 2.42 |
ENST00000367376.2
ENST00000352140.3 ENST00000594404.1 ENST00000598951.1 ENST00000530727.1 ENST00000442510.2 ENST00000367367.4 ENST00000348564.6 ENST00000367364.1 ENST00000413409.2 |
PTPRC
|
protein tyrosine phosphatase, receptor type, C |
| chr3_+_180630444 | 2.38 |
ENST00000491062.1
ENST00000468861.1 ENST00000445140.2 ENST00000484958.1 |
FXR1
|
fragile X mental retardation, autosomal homolog 1 |
| chr20_+_30327063 | 2.37 |
ENST00000300403.6
ENST00000340513.4 |
TPX2
|
TPX2, microtubule-associated |
| chr16_+_69958887 | 2.33 |
ENST00000568684.1
|
WWP2
|
WW domain containing E3 ubiquitin protein ligase 2 |
| chr3_+_180630090 | 2.25 |
ENST00000357559.4
ENST00000305586.7 |
FXR1
|
fragile X mental retardation, autosomal homolog 1 |
| chr6_+_24775153 | 2.23 |
ENST00000356509.3
ENST00000230056.3 |
GMNN
|
geminin, DNA replication inhibitor |
| chr14_+_95047744 | 2.19 |
ENST00000553511.1
ENST00000554633.1 ENST00000555681.1 ENST00000554276.1 |
SERPINA5
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5 |
| chr1_+_153750622 | 2.19 |
ENST00000532853.1
|
SLC27A3
|
solute carrier family 27 (fatty acid transporter), member 3 |
| chr11_-_104840093 | 2.18 |
ENST00000417440.2
ENST00000444739.2 |
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr19_+_10765699 | 2.08 |
ENST00000590009.1
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa |
| chr2_-_203736334 | 2.02 |
ENST00000392237.2
ENST00000416760.1 ENST00000412210.1 |
ICA1L
|
islet cell autoantigen 1,69kDa-like |
| chr7_-_16844611 | 2.02 |
ENST00000401412.1
ENST00000419304.2 |
AGR2
|
anterior gradient 2 |
| chr8_-_22926623 | 1.98 |
ENST00000276431.4
|
TNFRSF10B
|
tumor necrosis factor receptor superfamily, member 10b |
| chr1_-_46152174 | 1.92 |
ENST00000290795.3
ENST00000355105.3 |
GPBP1L1
|
GC-rich promoter binding protein 1-like 1 |
| chr21_-_15918618 | 1.92 |
ENST00000400564.1
ENST00000400566.1 |
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1 |
| chr19_+_41305085 | 1.91 |
ENST00000303961.4
|
EGLN2
|
egl-9 family hypoxia-inducible factor 2 |
| chr16_+_19079215 | 1.90 |
ENST00000544894.2
ENST00000561858.1 |
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast) |
| chr19_-_54804173 | 1.90 |
ENST00000391744.3
ENST00000251390.3 |
LILRA3
|
leukocyte immunoglobulin-like receptor, subfamily A (without TM domain), member 3 |
| chr14_-_20801427 | 1.89 |
ENST00000557665.1
ENST00000358932.4 ENST00000353689.4 |
CCNB1IP1
|
cyclin B1 interacting protein 1, E3 ubiquitin protein ligase |
| chr8_+_9046503 | 1.89 |
ENST00000512942.2
|
RP11-10A14.5
|
RP11-10A14.5 |
| chr1_-_183559693 | 1.87 |
ENST00000367535.3
ENST00000413720.1 ENST00000418089.1 |
NCF2
|
neutrophil cytosolic factor 2 |
| chr9_-_139343294 | 1.80 |
ENST00000313084.5
|
SEC16A
|
SEC16 homolog A (S. cerevisiae) |
| chr9_+_71789133 | 1.76 |
ENST00000348208.4
ENST00000265384.7 |
TJP2
|
tight junction protein 2 |
| chr11_+_18433840 | 1.72 |
ENST00000541669.1
ENST00000280704.4 |
LDHC
|
lactate dehydrogenase C |
| chr13_-_38172863 | 1.68 |
ENST00000541481.1
ENST00000379743.4 ENST00000379742.4 ENST00000379749.4 ENST00000541179.1 ENST00000379747.4 |
POSTN
|
periostin, osteoblast specific factor |
| chr15_-_72563585 | 1.66 |
ENST00000287196.9
ENST00000260376.7 |
PARP6
|
poly (ADP-ribose) polymerase family, member 6 |
| chr7_-_36764004 | 1.63 |
ENST00000431169.1
|
AOAH
|
acyloxyacyl hydrolase (neutrophil) |
| chr8_-_145028013 | 1.62 |
ENST00000354958.2
|
PLEC
|
plectin |
| chr11_-_506316 | 1.61 |
ENST00000532055.1
ENST00000531540.1 |
RNH1
|
ribonuclease/angiogenin inhibitor 1 |
| chr15_-_59225758 | 1.59 |
ENST00000558486.1
ENST00000560682.1 ENST00000249736.7 ENST00000559880.1 ENST00000536328.1 |
SLTM
|
SAFB-like, transcription modulator |
| chrX_-_47518498 | 1.58 |
ENST00000335890.2
|
UXT
|
ubiquitously-expressed, prefoldin-like chaperone |
| chr16_-_28506840 | 1.56 |
ENST00000569430.1
|
CLN3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr7_+_155089486 | 1.56 |
ENST00000340368.4
ENST00000344756.4 ENST00000425172.1 ENST00000342407.5 |
INSIG1
|
insulin induced gene 1 |
| chr8_+_12809093 | 1.54 |
ENST00000528753.2
|
KIAA1456
|
KIAA1456 |
| chr19_+_41281416 | 1.47 |
ENST00000597140.1
|
MIA
|
melanoma inhibitory activity |
| chr7_-_36764142 | 1.46 |
ENST00000258749.5
ENST00000535891.1 |
AOAH
|
acyloxyacyl hydrolase (neutrophil) |
| chr1_-_32403370 | 1.45 |
ENST00000534796.1
|
PTP4A2
|
protein tyrosine phosphatase type IVA, member 2 |
| chr16_-_67281413 | 1.45 |
ENST00000258201.4
|
FHOD1
|
formin homology 2 domain containing 1 |
| chr2_+_38893047 | 1.44 |
ENST00000272252.5
|
GALM
|
galactose mutarotase (aldose 1-epimerase) |
| chr22_-_17680472 | 1.41 |
ENST00000330232.4
|
CECR1
|
cat eye syndrome chromosome region, candidate 1 |
| chr4_-_84205905 | 1.38 |
ENST00000311461.7
ENST00000311469.4 ENST00000439031.2 |
COQ2
|
coenzyme Q2 4-hydroxybenzoate polyprenyltransferase |
| chr22_+_29664248 | 1.37 |
ENST00000406548.1
ENST00000437155.2 ENST00000415761.1 ENST00000331029.7 |
EWSR1
|
EWS RNA-binding protein 1 |
| chr1_+_153232160 | 1.37 |
ENST00000368742.3
|
LOR
|
loricrin |
| chr19_-_5719860 | 1.36 |
ENST00000590729.1
|
LONP1
|
lon peptidase 1, mitochondrial |
| chr16_+_31119615 | 1.36 |
ENST00000394950.3
ENST00000287507.3 ENST00000219794.6 ENST00000561755.1 |
BCKDK
|
branched chain ketoacid dehydrogenase kinase |
| chr16_+_19078960 | 1.36 |
ENST00000568985.1
ENST00000566110.1 |
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast) |
| chr11_-_65667884 | 1.35 |
ENST00000448083.2
ENST00000531493.1 ENST00000532401.1 |
FOSL1
|
FOS-like antigen 1 |
| chr16_+_19079311 | 1.32 |
ENST00000569127.1
|
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast) |
| chr19_+_41281282 | 1.32 |
ENST00000263369.3
|
MIA
|
melanoma inhibitory activity |
| chr9_+_118950325 | 1.31 |
ENST00000534838.1
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1 |
| chr7_+_65552756 | 1.27 |
ENST00000450043.1
|
AC068533.7
|
AC068533.7 |
| chr2_+_86333301 | 1.26 |
ENST00000254630.7
|
PTCD3
|
pentatricopeptide repeat domain 3 |
| chr11_-_65667997 | 1.26 |
ENST00000312562.2
ENST00000534222.1 |
FOSL1
|
FOS-like antigen 1 |
| chr3_+_157154578 | 1.25 |
ENST00000295927.3
|
PTX3
|
pentraxin 3, long |
| chr11_-_73882029 | 1.24 |
ENST00000539061.1
|
C2CD3
|
C2 calcium-dependent domain containing 3 |
| chr1_+_36348790 | 1.22 |
ENST00000373204.4
|
AGO1
|
argonaute RISC catalytic component 1 |
| chr17_+_53828333 | 1.19 |
ENST00000268896.5
|
PCTP
|
phosphatidylcholine transfer protein |
| chr16_+_16043406 | 1.18 |
ENST00000399410.3
ENST00000399408.2 ENST00000346370.5 ENST00000351154.5 ENST00000345148.5 ENST00000349029.5 |
ABCC1
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 1 |
| chr3_+_122283064 | 1.15 |
ENST00000296161.4
|
DTX3L
|
deltex 3-like (Drosophila) |
| chr10_+_47894572 | 1.15 |
ENST00000355876.5
|
FAM21B
|
family with sequence similarity 21, member B |
| chr19_+_54694119 | 1.12 |
ENST00000456872.1
ENST00000302937.4 ENST00000429671.2 |
TSEN34
|
TSEN34 tRNA splicing endonuclease subunit |
| chr19_+_41281060 | 1.11 |
ENST00000594436.1
ENST00000597784.1 |
MIA
|
melanoma inhibitory activity |
| chr10_+_104154229 | 1.10 |
ENST00000428099.1
ENST00000369966.3 |
NFKB2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chrX_-_135962876 | 1.09 |
ENST00000431446.3
ENST00000570135.1 ENST00000320676.7 ENST00000562646.1 |
RBMX
|
RNA binding motif protein, X-linked |
| chr9_+_111624577 | 1.08 |
ENST00000333999.3
|
ACTL7A
|
actin-like 7A |
| chr16_-_87799505 | 1.08 |
ENST00000353170.5
ENST00000561825.1 ENST00000270583.5 ENST00000562261.1 ENST00000347925.5 |
KLHDC4
|
kelch domain containing 4 |
| chr7_+_155090271 | 1.07 |
ENST00000476756.1
|
INSIG1
|
insulin induced gene 1 |
| chr17_-_64225508 | 1.06 |
ENST00000205948.6
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr15_-_59225844 | 1.05 |
ENST00000380516.2
|
SLTM
|
SAFB-like, transcription modulator |
| chr14_-_54908043 | 1.04 |
ENST00000556113.1
ENST00000553660.1 ENST00000395573.4 ENST00000557690.1 ENST00000216416.4 |
CNIH1
|
cornichon family AMPA receptor auxiliary protein 1 |
| chr7_-_72722783 | 1.04 |
ENST00000428206.1
ENST00000252594.6 ENST00000310326.8 ENST00000438747.2 |
NSUN5
|
NOP2/Sun domain family, member 5 |
| chrX_-_47518527 | 1.04 |
ENST00000333119.3
|
UXT
|
ubiquitously-expressed, prefoldin-like chaperone |
| chr3_+_132036207 | 1.00 |
ENST00000336375.5
ENST00000495911.1 |
ACPP
|
acid phosphatase, prostate |
| chr3_+_151986709 | 0.98 |
ENST00000495875.2
ENST00000493459.1 ENST00000324210.5 ENST00000459747.1 |
MBNL1
|
muscleblind-like splicing regulator 1 |
| chr19_-_12595586 | 0.98 |
ENST00000397732.3
|
ZNF709
|
zinc finger protein 709 |
| chr17_+_1733276 | 0.97 |
ENST00000254719.5
|
RPA1
|
replication protein A1, 70kDa |
| chr17_+_53828381 | 0.97 |
ENST00000576183.1
|
PCTP
|
phosphatidylcholine transfer protein |
| chr12_-_25348007 | 0.97 |
ENST00000354189.5
ENST00000545133.1 ENST00000554347.1 ENST00000395987.3 ENST00000320267.9 ENST00000395990.2 ENST00000537577.1 |
CASC1
|
cancer susceptibility candidate 1 |
| chr22_+_39077947 | 0.94 |
ENST00000216034.4
|
TOMM22
|
translocase of outer mitochondrial membrane 22 homolog (yeast) |
| chr2_+_177134134 | 0.93 |
ENST00000249442.6
ENST00000392529.2 ENST00000443241.1 |
MTX2
|
metaxin 2 |
| chr3_-_122283100 | 0.91 |
ENST00000492382.1
ENST00000462315.1 |
PARP9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr9_-_115095123 | 0.89 |
ENST00000458258.1
|
PTBP3
|
polypyrimidine tract binding protein 3 |
| chr19_-_13261090 | 0.89 |
ENST00000588848.1
|
STX10
|
syntaxin 10 |
| chr19_+_57831829 | 0.89 |
ENST00000321545.4
|
ZNF543
|
zinc finger protein 543 |
| chr19_+_39936267 | 0.88 |
ENST00000359191.6
|
SUPT5H
|
suppressor of Ty 5 homolog (S. cerevisiae) |
| chr19_-_13261160 | 0.83 |
ENST00000343587.5
ENST00000591197.1 |
STX10
|
syntaxin 10 |
| chr20_-_52790512 | 0.82 |
ENST00000216862.3
|
CYP24A1
|
cytochrome P450, family 24, subfamily A, polypeptide 1 |
| chr22_+_45705728 | 0.82 |
ENST00000441876.2
|
FAM118A
|
family with sequence similarity 118, member A |
| chr8_-_22926526 | 0.79 |
ENST00000347739.3
ENST00000542226.1 |
TNFRSF10B
|
tumor necrosis factor receptor superfamily, member 10b |
| chr8_+_24298597 | 0.79 |
ENST00000380789.1
|
ADAM7
|
ADAM metallopeptidase domain 7 |
| chr12_+_3069037 | 0.78 |
ENST00000397122.2
|
TEAD4
|
TEA domain family member 4 |
| chr12_-_110906027 | 0.78 |
ENST00000537466.2
ENST00000550974.1 ENST00000228827.3 |
GPN3
|
GPN-loop GTPase 3 |
| chr1_+_92414928 | 0.78 |
ENST00000362005.3
ENST00000370389.2 ENST00000399546.2 ENST00000423434.1 ENST00000394530.3 ENST00000440509.1 |
BRDT
|
bromodomain, testis-specific |
| chr10_-_32345305 | 0.77 |
ENST00000302418.4
|
KIF5B
|
kinesin family member 5B |
| chrX_-_48776292 | 0.76 |
ENST00000376509.4
|
PIM2
|
pim-2 oncogene |
| chr14_-_91884150 | 0.76 |
ENST00000553403.1
|
CCDC88C
|
coiled-coil domain containing 88C |
| chr11_-_60719213 | 0.76 |
ENST00000227880.3
|
SLC15A3
|
solute carrier family 15 (oligopeptide transporter), member 3 |
| chr6_+_31583761 | 0.75 |
ENST00000376049.4
|
AIF1
|
allograft inflammatory factor 1 |
| chr2_-_232329186 | 0.75 |
ENST00000322723.4
|
NCL
|
nucleolin |
| chr2_+_231280908 | 0.74 |
ENST00000427101.2
ENST00000432979.1 |
SP100
|
SP100 nuclear antigen |
| chr10_+_5454505 | 0.72 |
ENST00000355029.4
|
NET1
|
neuroepithelial cell transforming 1 |
| chr8_+_24298531 | 0.71 |
ENST00000175238.6
|
ADAM7
|
ADAM metallopeptidase domain 7 |
| chr19_+_39936186 | 0.68 |
ENST00000432763.2
ENST00000402194.2 ENST00000601515.1 |
SUPT5H
|
suppressor of Ty 5 homolog (S. cerevisiae) |
| chr9_-_130617029 | 0.68 |
ENST00000373203.4
|
ENG
|
endoglin |
| chr4_-_164534657 | 0.68 |
ENST00000339875.5
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr7_+_129015484 | 0.67 |
ENST00000490911.1
|
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chr16_+_31044812 | 0.67 |
ENST00000313843.3
|
STX4
|
syntaxin 4 |
| chr1_-_241799232 | 0.64 |
ENST00000366553.1
|
CHML
|
choroideremia-like (Rab escort protein 2) |
| chr17_-_40288449 | 0.63 |
ENST00000552162.1
ENST00000550504.1 |
RAB5C
|
RAB5C, member RAS oncogene family |
| chr8_+_119294456 | 0.62 |
ENST00000366457.2
|
AC023590.1
|
Uncharacterized protein |
| chr22_+_50624323 | 0.61 |
ENST00000380909.4
ENST00000303434.4 |
TRABD
|
TraB domain containing |
| chr8_+_98900132 | 0.61 |
ENST00000520016.1
|
MATN2
|
matrilin 2 |
| chr1_+_175036966 | 0.61 |
ENST00000239462.4
|
TNN
|
tenascin N |
| chr12_+_53895364 | 0.61 |
ENST00000552817.1
ENST00000394357.2 |
TARBP2
|
TAR (HIV-1) RNA binding protein 2 |
| chrX_+_100646190 | 0.60 |
ENST00000471855.1
|
RPL36A
|
ribosomal protein L36a |
| chr7_+_120590803 | 0.60 |
ENST00000315870.5
ENST00000339121.5 ENST00000445699.1 |
ING3
|
inhibitor of growth family, member 3 |
| chr13_-_33859819 | 0.58 |
ENST00000336934.5
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13 |
| chr10_+_54074033 | 0.57 |
ENST00000373970.3
|
DKK1
|
dickkopf WNT signaling pathway inhibitor 1 |
| chr6_+_35227449 | 0.56 |
ENST00000373953.3
ENST00000440666.2 ENST00000339411.5 |
ZNF76
|
zinc finger protein 76 |
| chr1_-_27226928 | 0.56 |
ENST00000361720.5
|
GPATCH3
|
G patch domain containing 3 |
| chr5_-_145562147 | 0.55 |
ENST00000545646.1
ENST00000274562.9 ENST00000510191.1 ENST00000394434.2 |
LARS
|
leucyl-tRNA synthetase |
| chr9_-_130616915 | 0.54 |
ENST00000344849.3
|
ENG
|
endoglin |
| chr1_-_205091115 | 0.53 |
ENST00000264515.6
ENST00000367164.1 |
RBBP5
|
retinoblastoma binding protein 5 |
| chr3_+_184033135 | 0.53 |
ENST00000424196.1
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr10_-_33623564 | 0.53 |
ENST00000374875.1
ENST00000374822.4 |
NRP1
|
neuropilin 1 |
| chr5_-_82969405 | 0.53 |
ENST00000510978.1
|
HAPLN1
|
hyaluronan and proteoglycan link protein 1 |
| chr16_+_57392684 | 0.51 |
ENST00000219235.4
|
CCL22
|
chemokine (C-C motif) ligand 22 |
| chr22_-_29663954 | 0.50 |
ENST00000216085.7
|
RHBDD3
|
rhomboid domain containing 3 |
| chr14_-_23398565 | 0.49 |
ENST00000397440.4
ENST00000538452.1 ENST00000421938.2 ENST00000554867.1 ENST00000556616.1 ENST00000216350.8 ENST00000553550.1 ENST00000397441.2 ENST00000553897.1 |
PRMT5
|
protein arginine methyltransferase 5 |
| chr11_+_63998198 | 0.49 |
ENST00000321460.5
|
DNAJC4
|
DnaJ (Hsp40) homolog, subfamily C, member 4 |
| chr8_+_95653373 | 0.47 |
ENST00000358397.5
|
ESRP1
|
epithelial splicing regulatory protein 1 |
| chr4_+_144434584 | 0.45 |
ENST00000283131.3
|
SMARCA5
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5 |
| chrX_+_100645812 | 0.43 |
ENST00000427805.2
ENST00000553110.3 ENST00000392994.3 ENST00000409338.1 ENST00000409170.3 |
RPL36A
RPL36A-HNRNPH2
|
ribosomal protein L36a RPL36A-HNRNPH2 readthrough |
| chr4_+_17579110 | 0.43 |
ENST00000606142.1
|
LAP3
|
leucine aminopeptidase 3 |
| chr1_+_100111580 | 0.43 |
ENST00000605497.1
|
PALMD
|
palmdelphin |
| chr1_-_221915418 | 0.42 |
ENST00000323825.3
ENST00000366899.3 |
DUSP10
|
dual specificity phosphatase 10 |
| chr19_+_14017116 | 0.41 |
ENST00000589606.1
|
CC2D1A
|
coiled-coil and C2 domain containing 1A |
| chr5_-_39270725 | 0.41 |
ENST00000512138.1
ENST00000512982.1 ENST00000540520.1 |
FYB
|
FYN binding protein |
| chr16_-_31105870 | 0.40 |
ENST00000394971.3
|
VKORC1
|
vitamin K epoxide reductase complex, subunit 1 |
| chr8_+_23104130 | 0.40 |
ENST00000313219.7
ENST00000519984.1 |
CHMP7
|
charged multivesicular body protein 7 |
| chr5_-_7869108 | 0.39 |
ENST00000264669.5
ENST00000507572.1 ENST00000504695.1 |
FASTKD3
|
FAST kinase domains 3 |
| chr6_-_32557610 | 0.38 |
ENST00000360004.5
|
HLA-DRB1
|
major histocompatibility complex, class II, DR beta 1 |
| chr3_+_190105909 | 0.37 |
ENST00000456423.1
|
CLDN16
|
claudin 16 |
| chr19_-_49122384 | 0.37 |
ENST00000550973.1
ENST00000550645.1 ENST00000549273.1 ENST00000552588.1 |
RPL18
|
ribosomal protein L18 |
| chr4_+_142557771 | 0.36 |
ENST00000514653.1
|
IL15
|
interleukin 15 |
| chr1_-_174992544 | 0.36 |
ENST00000476371.1
|
MRPS14
|
mitochondrial ribosomal protein S14 |
| chr9_-_138391692 | 0.36 |
ENST00000429260.2
|
C9orf116
|
chromosome 9 open reading frame 116 |
| chr22_+_29664305 | 0.35 |
ENST00000414183.2
ENST00000333395.6 ENST00000455726.1 ENST00000332035.6 |
EWSR1
|
EWS RNA-binding protein 1 |
| chr1_-_75198940 | 0.34 |
ENST00000417775.1
|
CRYZ
|
crystallin, zeta (quinone reductase) |
| chr6_+_79577189 | 0.33 |
ENST00000369940.2
|
IRAK1BP1
|
interleukin-1 receptor-associated kinase 1 binding protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of VDR
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.5 | GO:0009386 | translational attenuation(GO:0009386) |
| 1.1 | 7.6 | GO:1902915 | negative regulation of histone ubiquitination(GO:0033183) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.9 | 2.6 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.7 | 2.9 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 0.6 | 4.8 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.6 | 1.7 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.5 | 5.8 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.5 | 2.4 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) bone marrow development(GO:0048539) |
| 0.5 | 1.4 | GO:0033499 | galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.5 | 1.4 | GO:0070407 | oxidation-dependent protein catabolic process(GO:0070407) |
| 0.4 | 2.7 | GO:0070417 | cellular response to cold(GO:0070417) nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.4 | 2.2 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.4 | 2.6 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.4 | 1.2 | GO:0001300 | chronological cell aging(GO:0001300) |
| 0.4 | 1.6 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.4 | 1.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.4 | 2.2 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 0.4 | 4.7 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.4 | 29.9 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.3 | 2.0 | GO:1903899 | positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.3 | 1.0 | GO:0060168 | positive regulation of adenosine receptor signaling pathway(GO:0060168) |
| 0.3 | 2.6 | GO:0042761 | fatty acid elongation(GO:0030497) very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.3 | 1.2 | GO:0021997 | neural plate axis specification(GO:0021997) |
| 0.3 | 8.4 | GO:2000178 | negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.3 | 5.7 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.3 | 1.7 | GO:0019249 | lactate biosynthetic process(GO:0019249) |
| 0.3 | 1.1 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 0.3 | 0.8 | GO:0042369 | vitamin D catabolic process(GO:0042369) |
| 0.3 | 0.8 | GO:0044376 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.3 | 3.8 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.3 | 1.0 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) |
| 0.2 | 1.2 | GO:0052405 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) negative regulation by host of symbiont molecular function(GO:0052405) |
| 0.2 | 1.2 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing by siRNA(GO:0090625) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.2 | 0.7 | GO:0043311 | positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.2 | 4.5 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.2 | 1.9 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.2 | 0.6 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.2 | 4.5 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.2 | 1.2 | GO:0035672 | oligopeptide transmembrane transport(GO:0035672) |
| 0.2 | 0.6 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.2 | 0.6 | GO:0009996 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) negative regulation of cell fate specification(GO:0009996) Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.2 | 0.5 | GO:0006425 | glutaminyl-tRNA aminoacylation(GO:0006425) valyl-tRNA aminoacylation(GO:0006438) |
| 0.2 | 4.6 | GO:0060148 | positive regulation of posttranscriptional gene silencing(GO:0060148) positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.2 | 1.8 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.2 | 1.5 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.2 | 1.0 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.2 | 0.8 | GO:0097102 | vestibulocochlear nerve structural organization(GO:0021649) positive regulation of cytokine activity(GO:0060301) ganglion morphogenesis(GO:0061552) endothelial tip cell fate specification(GO:0097102) VEGF-activated neuropilin signaling pathway involved in axon guidance(GO:1902378) dorsal root ganglion morphogenesis(GO:1904835) otic placode development(GO:1905040) |
| 0.2 | 0.9 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.2 | 0.8 | GO:0014738 | regulation of muscle hyperplasia(GO:0014738) |
| 0.1 | 2.8 | GO:0097296 | activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) |
| 0.1 | 1.4 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.1 | 3.7 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 0.5 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.1 | 0.5 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.1 | 1.7 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 0.4 | GO:0045062 | extrathymic T cell selection(GO:0045062) |
| 0.1 | 6.1 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.1 | 0.7 | GO:0002536 | respiratory burst involved in inflammatory response(GO:0002536) regulation of respiratory burst involved in inflammatory response(GO:0060264) |
| 0.1 | 1.1 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.1 | 2.6 | GO:0047497 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.1 | 1.6 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.1 | 0.6 | GO:0035087 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) siRNA loading onto RISC involved in RNA interference(GO:0035087) |
| 0.1 | 6.9 | GO:0070126 | mitochondrial translational elongation(GO:0070125) mitochondrial translational termination(GO:0070126) |
| 0.1 | 1.1 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 0.9 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.1 | 3.7 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 6.5 | GO:0035722 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.1 | 0.4 | GO:1904896 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.1 | 0.6 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.1 | 0.4 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 0.1 | 0.2 | GO:0051039 | histone displacement(GO:0001207) regulation of transcription involved in meiotic cell cycle(GO:0051037) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.1 | 0.6 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.1 | 0.2 | GO:0042427 | serotonin biosynthetic process(GO:0042427) |
| 0.1 | 2.8 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.1 | 3.0 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.1 | 0.7 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.1 | 0.2 | GO:1901836 | regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901836) |
| 0.1 | 3.4 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 2.4 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.1 | 1.9 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.1 | 0.7 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.1 | 2.3 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.1 | 1.0 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.1 | 1.6 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 1.9 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.1 | 1.3 | GO:0032354 | response to follicle-stimulating hormone(GO:0032354) |
| 0.1 | 0.4 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 0.5 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 2.5 | GO:0035666 | TRIF-dependent toll-like receptor signaling pathway(GO:0035666) |
| 0.0 | 0.3 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.0 | 1.4 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 5.0 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.4 | GO:0045333 | cellular respiration(GO:0045333) |
| 0.0 | 1.1 | GO:0010390 | histone monoubiquitination(GO:0010390) |
| 0.0 | 1.8 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 6.2 | GO:0071774 | response to fibroblast growth factor(GO:0071774) |
| 0.0 | 1.6 | GO:0006954 | inflammatory response(GO:0006954) |
| 0.0 | 0.5 | GO:0035966 | response to unfolded protein(GO:0006986) response to topologically incorrect protein(GO:0035966) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.1 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.0 | 0.4 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.0 | 2.2 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.2 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 1.0 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.6 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 2.6 | GO:0050684 | regulation of mRNA processing(GO:0050684) |
| 0.0 | 0.2 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 1.8 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.8 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.3 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.2 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.2 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.0 | 0.2 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.0 | 0.6 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.8 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 3.4 | GO:0061337 | cardiac conduction(GO:0061337) |
| 0.0 | 0.3 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 4.0 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 4.3 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.0 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
| 0.0 | 0.3 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.0 | 0.1 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 0.2 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 1.6 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 0.0 | 0.6 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.0 | 0.5 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 1.6 | GO:0006402 | mRNA catabolic process(GO:0006402) |
| 0.0 | 0.8 | GO:0016239 | positive regulation of macroautophagy(GO:0016239) |
| 0.0 | 1.5 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.3 | GO:0007249 | I-kappaB kinase/NF-kappaB signaling(GO:0007249) |
| 0.0 | 0.5 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.2 | GO:0010922 | positive regulation of phosphatase activity(GO:0010922) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.9 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 1.0 | 5.8 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 1.0 | 2.9 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.8 | 6.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.7 | 2.2 | GO:0097179 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) protease inhibitor complex(GO:0097179) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 0.7 | 2.6 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.6 | 4.7 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.4 | 1.2 | GO:0072563 | endothelial microparticle(GO:0072563) |
| 0.4 | 1.6 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.3 | 3.8 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.3 | 4.5 | GO:0008290 | F-actin capping protein complex(GO:0008290) Flemming body(GO:0090543) |
| 0.3 | 14.6 | GO:0043034 | costamere(GO:0043034) |
| 0.3 | 1.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.3 | 1.4 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.3 | 1.6 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.2 | 1.9 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.2 | 1.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.2 | 5.6 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.2 | 1.8 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.2 | 2.4 | GO:0043203 | axon hillock(GO:0043203) |
| 0.2 | 2.5 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.2 | 0.6 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.2 | 0.5 | GO:0031213 | RSF complex(GO:0031213) |
| 0.1 | 0.6 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 1.0 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 0.9 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 1.9 | GO:0001726 | ruffle(GO:0001726) |
| 0.1 | 2.6 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 1.4 | GO:0032059 | bleb(GO:0032059) |
| 0.1 | 0.7 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.1 | 3.5 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 1.0 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 1.9 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.1 | 1.1 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.1 | 6.8 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 9.3 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.1 | 11.8 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 0.8 | GO:0002116 | semaphorin receptor complex(GO:0002116) sorting endosome(GO:0097443) |
| 0.1 | 5.9 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.1 | 3.8 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 0.8 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 0.5 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 1.2 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 1.4 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.5 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.8 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 2.8 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 1.1 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.4 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 4.8 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.2 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 3.9 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 1.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.4 | GO:0044309 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 0.5 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 1.4 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 2.0 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 1.7 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 1.0 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.2 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 7.9 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 2.2 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 1.7 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 0.2 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 1.4 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.5 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 0.2 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 1.5 | GO:0005874 | microtubule(GO:0005874) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.8 | 3.8 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 0.7 | 2.6 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.6 | 2.9 | GO:0019798 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.5 | 3.7 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.5 | 2.8 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.5 | 1.4 | GO:0070361 | mitochondrial light strand promoter anti-sense binding(GO:0070361) mitochondrial heavy strand promoter anti-sense binding(GO:0070362) mitochondrial heavy strand promoter sense binding(GO:0070364) |
| 0.4 | 3.4 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.4 | 4.6 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) RNA strand annealing activity(GO:0033592) |
| 0.4 | 5.8 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.4 | 1.2 | GO:0015235 | cobalamin transporter activity(GO:0015235) |
| 0.4 | 1.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.4 | 1.5 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.3 | 4.7 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.3 | 12.6 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.3 | 1.7 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.3 | 1.0 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.3 | 1.5 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.3 | 1.9 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.3 | 0.8 | GO:0015333 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.2 | 1.7 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) L-lactate dehydrogenase activity(GO:0004459) |
| 0.2 | 2.2 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.2 | 3.4 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.2 | 2.2 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.2 | 1.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.2 | 2.9 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.2 | 34.7 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.2 | 0.8 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.2 | 1.9 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.2 | 0.5 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) valine-tRNA ligase activity(GO:0004832) |
| 0.2 | 7.6 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.2 | 2.5 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.2 | 0.7 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.2 | 0.8 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.2 | 1.6 | GO:0008428 | ribonuclease inhibitor activity(GO:0008428) |
| 0.2 | 1.3 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.2 | 2.2 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.2 | 1.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.2 | 1.2 | GO:0005534 | galactose binding(GO:0005534) |
| 0.1 | 3.0 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 1.0 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.1 | 0.7 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.1 | 0.5 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.1 | 1.4 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.1 | 5.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 0.7 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 3.5 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 0.3 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.1 | 1.4 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 0.8 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.1 | 1.0 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 2.6 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 2.4 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 3.0 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 1.0 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.1 | 2.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 0.9 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.1 | 0.3 | GO:0047057 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.1 | 1.2 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 1.4 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.1 | 2.7 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 0.3 | GO:0070404 | NADH binding(GO:0070404) |
| 0.1 | 0.6 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.1 | 1.6 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 0.6 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 3.0 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.2 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 9.0 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 1.8 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 3.1 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.2 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.7 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.2 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 0.8 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.3 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.5 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 1.9 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 7.2 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.2 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 2.2 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 2.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 2.1 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 2.3 | GO:0001190 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 2.8 | GO:0004497 | monooxygenase activity(GO:0004497) |
| 0.0 | 0.2 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.0 | 0.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 1.4 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 3.1 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.2 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 0.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.0 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.0 | 0.7 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 3.4 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 3.3 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.6 | GO:0008173 | RNA methyltransferase activity(GO:0008173) |
| 0.0 | 0.4 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.1 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.7 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 0.5 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.3 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.6 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 8.4 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.2 | 6.2 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.1 | 4.7 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 1.3 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 2.8 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 2.8 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.1 | 2.4 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.1 | 2.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 1.9 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 4.6 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.5 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 2.4 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.8 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.9 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 1.7 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 3.1 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 2.9 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 1.9 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 1.0 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 2.3 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.7 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.7 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.4 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 2.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.3 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.4 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 2.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 8.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.2 | 2.5 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.2 | 3.0 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.2 | 10.9 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.2 | 1.6 | REACTOME FORMATION OF THE HIV1 EARLY ELONGATION COMPLEX | Genes involved in Formation of the HIV-1 Early Elongation Complex |
| 0.2 | 3.8 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.2 | 3.7 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 5.8 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.1 | 2.9 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 2.6 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 2.2 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.1 | 2.8 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 2.8 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 1.8 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 1.0 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 10.4 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.1 | 0.6 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.1 | 2.8 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 1.9 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 1.6 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 3.2 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 12.8 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.9 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.0 | 0.8 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.2 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.2 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 0.8 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.3 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.8 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.4 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.5 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 1.0 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.4 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.2 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 1.7 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |