Project
averaged GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
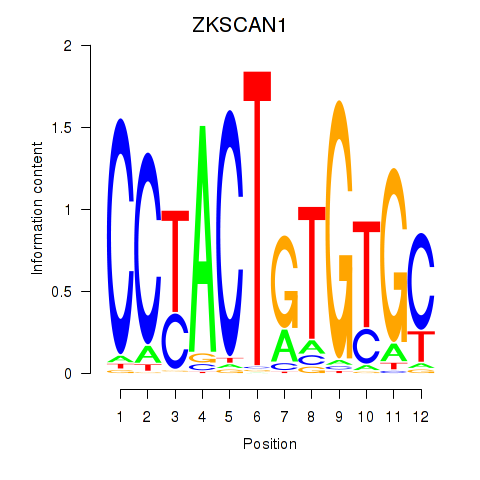
Results for ZKSCAN1
Z-value: 1.18
Transcription factors associated with ZKSCAN1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZKSCAN1
|
ENSG00000106261.12 | zinc finger with KRAB and SCAN domains 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZKSCAN1 | hg19_v2_chr7_+_99613212_99613236 | -0.13 | 5.6e-02 | Click! |
Activity profile of ZKSCAN1 motif
Sorted Z-values of ZKSCAN1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_135394840 | 53.79 |
ENST00000503087.1
|
TGFBI
|
transforming growth factor, beta-induced, 68kDa |
| chr22_+_21921994 | 36.03 |
ENST00000545681.1
|
UBE2L3
|
ubiquitin-conjugating enzyme E2L 3 |
| chr17_-_73150599 | 35.79 |
ENST00000392566.2
ENST00000581874.1 |
HN1
|
hematological and neurological expressed 1 |
| chr17_-_73150629 | 29.32 |
ENST00000356033.4
ENST00000405458.3 ENST00000409753.3 |
HN1
|
hematological and neurological expressed 1 |
| chrX_+_47441712 | 24.65 |
ENST00000218388.4
ENST00000377018.2 ENST00000456754.2 ENST00000377017.1 ENST00000441738.1 |
TIMP1
|
TIMP metallopeptidase inhibitor 1 |
| chr19_+_39138320 | 22.22 |
ENST00000424234.2
ENST00000390009.3 ENST00000589528.1 |
ACTN4
|
actinin, alpha 4 |
| chr5_-_134735568 | 21.78 |
ENST00000510038.1
ENST00000304332.4 |
H2AFY
|
H2A histone family, member Y |
| chr22_-_37640277 | 20.38 |
ENST00000401529.3
ENST00000249071.6 |
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chr20_-_48770244 | 20.00 |
ENST00000371650.5
ENST00000371652.4 ENST00000557021.1 |
TMEM189
|
transmembrane protein 189 |
| chr19_+_13051206 | 19.55 |
ENST00000586760.1
|
CALR
|
calreticulin |
| chr11_-_58343319 | 18.87 |
ENST00000395074.2
|
LPXN
|
leupaxin |
| chr1_+_62901968 | 18.41 |
ENST00000452143.1
ENST00000442679.1 ENST00000371146.1 |
USP1
|
ubiquitin specific peptidase 1 |
| chr1_-_53704157 | 17.19 |
ENST00000371466.4
ENST00000371470.3 |
MAGOH
|
mago-nashi homolog, proliferation-associated (Drosophila) |
| chr15_-_69113218 | 16.41 |
ENST00000560303.1
ENST00000465139.2 |
ANP32A
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
| chr19_+_39138271 | 15.19 |
ENST00000252699.2
|
ACTN4
|
actinin, alpha 4 |
| chr15_-_34635314 | 14.60 |
ENST00000557912.1
ENST00000328848.4 |
NOP10
|
NOP10 ribonucleoprotein |
| chr22_-_37640456 | 14.22 |
ENST00000405484.1
ENST00000441619.1 ENST00000406508.1 |
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chr11_+_64808675 | 13.71 |
ENST00000529996.1
|
SAC3D1
|
SAC3 domain containing 1 |
| chr3_+_184081213 | 13.20 |
ENST00000429568.1
|
POLR2H
|
polymerase (RNA) II (DNA directed) polypeptide H |
| chr3_+_184081175 | 13.06 |
ENST00000452961.1
ENST00000296223.3 |
POLR2H
|
polymerase (RNA) II (DNA directed) polypeptide H |
| chr17_+_75372165 | 12.16 |
ENST00000427674.2
|
SEPT9
|
septin 9 |
| chr9_-_139357413 | 11.87 |
ENST00000277537.6
|
SEC16A
|
SEC16 homolog A (S. cerevisiae) |
| chr11_+_70049269 | 11.53 |
ENST00000301838.4
|
FADD
|
Fas (TNFRSF6)-associated via death domain |
| chr3_+_184081137 | 10.96 |
ENST00000443489.1
|
POLR2H
|
polymerase (RNA) II (DNA directed) polypeptide H |
| chr7_+_73588575 | 10.95 |
ENST00000265753.8
|
EIF4H
|
eukaryotic translation initiation factor 4H |
| chr11_+_32112431 | 10.40 |
ENST00000054950.3
|
RCN1
|
reticulocalbin 1, EF-hand calcium binding domain |
| chr11_-_72070206 | 10.28 |
ENST00000544382.1
|
CLPB
|
ClpB caseinolytic peptidase B homolog (E. coli) |
| chr5_-_134734901 | 9.83 |
ENST00000312469.4
ENST00000423969.2 |
H2AFY
|
H2A histone family, member Y |
| chr19_-_19051993 | 9.55 |
ENST00000594794.1
ENST00000355887.6 ENST00000392351.3 ENST00000596482.1 |
HOMER3
|
homer homolog 3 (Drosophila) |
| chr1_+_65613340 | 9.20 |
ENST00000546702.1
|
AK4
|
adenylate kinase 4 |
| chr10_-_88281494 | 8.87 |
ENST00000298767.5
|
WAPAL
|
wings apart-like homolog (Drosophila) |
| chr8_-_131028869 | 8.62 |
ENST00000518283.1
ENST00000519110.1 |
FAM49B
|
family with sequence similarity 49, member B |
| chr7_+_73588665 | 8.58 |
ENST00000353999.6
|
EIF4H
|
eukaryotic translation initiation factor 4H |
| chr2_-_158345462 | 8.24 |
ENST00000439355.1
ENST00000540637.1 |
CYTIP
|
cytohesin 1 interacting protein |
| chr19_-_55660561 | 8.23 |
ENST00000587758.1
ENST00000356783.5 ENST00000291901.8 ENST00000588426.1 ENST00000588147.1 ENST00000536926.1 ENST00000588981.1 |
TNNT1
|
troponin T type 1 (skeletal, slow) |
| chr5_-_131562935 | 7.81 |
ENST00000379104.2
ENST00000379100.2 ENST00000428369.1 |
P4HA2
|
prolyl 4-hydroxylase, alpha polypeptide II |
| chr5_-_172756506 | 7.50 |
ENST00000265087.4
|
STC2
|
stanniocalcin 2 |
| chr6_+_32944119 | 6.58 |
ENST00000606059.1
|
BRD2
|
bromodomain containing 2 |
| chr3_+_63898275 | 6.56 |
ENST00000538065.1
|
ATXN7
|
ataxin 7 |
| chr17_+_27071002 | 6.38 |
ENST00000262395.5
ENST00000422344.1 ENST00000444415.3 ENST00000262396.6 |
TRAF4
|
TNF receptor-associated factor 4 |
| chr12_+_122326662 | 6.33 |
ENST00000261817.2
ENST00000538613.1 ENST00000542602.1 |
PSMD9
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 9 |
| chr12_+_4385230 | 6.23 |
ENST00000536537.1
|
CCND2
|
cyclin D2 |
| chr9_-_112260531 | 6.13 |
ENST00000374541.2
ENST00000262539.3 |
PTPN3
|
protein tyrosine phosphatase, non-receptor type 3 |
| chr20_+_43343517 | 6.07 |
ENST00000372865.4
|
WISP2
|
WNT1 inducible signaling pathway protein 2 |
| chr12_-_49319265 | 5.96 |
ENST00000552878.1
ENST00000453172.2 |
FKBP11
|
FK506 binding protein 11, 19 kDa |
| chr1_-_36615051 | 5.77 |
ENST00000373163.1
|
TRAPPC3
|
trafficking protein particle complex 3 |
| chr1_-_155224751 | 5.36 |
ENST00000350210.2
ENST00000368368.3 |
FAM189B
|
family with sequence similarity 189, member B |
| chrX_+_109245863 | 5.32 |
ENST00000372072.3
|
TMEM164
|
transmembrane protein 164 |
| chr1_+_236958554 | 5.24 |
ENST00000366577.5
ENST00000418145.2 |
MTR
|
5-methyltetrahydrofolate-homocysteine methyltransferase |
| chr20_+_43343476 | 5.18 |
ENST00000372868.2
|
WISP2
|
WNT1 inducible signaling pathway protein 2 |
| chr12_+_122326630 | 5.14 |
ENST00000541212.1
ENST00000340175.5 |
PSMD9
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 9 |
| chr1_-_36615065 | 4.97 |
ENST00000373166.3
ENST00000373159.1 ENST00000373162.1 |
TRAPPC3
|
trafficking protein particle complex 3 |
| chr5_-_127873659 | 4.70 |
ENST00000262464.4
|
FBN2
|
fibrillin 2 |
| chr6_-_30640811 | 4.63 |
ENST00000376442.3
|
DHX16
|
DEAH (Asp-Glu-Ala-His) box polypeptide 16 |
| chr19_-_3062881 | 4.63 |
ENST00000586742.1
|
AES
|
amino-terminal enhancer of split |
| chr10_+_112679301 | 4.55 |
ENST00000265277.5
ENST00000369452.4 |
SHOC2
|
soc-2 suppressor of clear homolog (C. elegans) |
| chr15_-_78369994 | 4.35 |
ENST00000300584.3
ENST00000409931.3 |
TBC1D2B
|
TBC1 domain family, member 2B |
| chr15_-_37390482 | 4.29 |
ENST00000559085.1
ENST00000397624.3 |
MEIS2
|
Meis homeobox 2 |
| chr2_+_101179152 | 4.27 |
ENST00000264254.6
|
PDCL3
|
phosducin-like 3 |
| chr19_-_3063099 | 4.09 |
ENST00000221561.8
|
AES
|
amino-terminal enhancer of split |
| chr10_-_112678904 | 4.00 |
ENST00000423273.1
ENST00000436562.1 ENST00000447005.1 ENST00000454061.1 |
BBIP1
|
BBSome interacting protein 1 |
| chr3_+_12329397 | 3.88 |
ENST00000397015.2
|
PPARG
|
peroxisome proliferator-activated receptor gamma |
| chr10_-_112678692 | 3.82 |
ENST00000605742.1
|
BBIP1
|
BBSome interacting protein 1 |
| chr19_-_3062465 | 3.61 |
ENST00000327141.4
|
AES
|
amino-terminal enhancer of split |
| chr12_-_25055177 | 3.34 |
ENST00000538118.1
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr3_+_12329358 | 3.31 |
ENST00000309576.6
|
PPARG
|
peroxisome proliferator-activated receptor gamma |
| chrX_-_40036520 | 3.25 |
ENST00000406200.2
ENST00000378455.4 ENST00000342274.4 |
BCOR
|
BCL6 corepressor |
| chr22_+_20861858 | 2.98 |
ENST00000414658.1
ENST00000432052.1 ENST00000425759.2 ENST00000292733.7 ENST00000542773.1 ENST00000263205.7 ENST00000406969.1 ENST00000382974.2 |
MED15
|
mediator complex subunit 15 |
| chr19_+_51630287 | 2.97 |
ENST00000599948.1
|
SIGLEC9
|
sialic acid binding Ig-like lectin 9 |
| chr15_+_75494214 | 2.85 |
ENST00000394987.4
|
C15orf39
|
chromosome 15 open reading frame 39 |
| chr1_+_65613217 | 2.80 |
ENST00000545314.1
|
AK4
|
adenylate kinase 4 |
| chr4_+_41540160 | 2.78 |
ENST00000503057.1
ENST00000511496.1 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr1_+_16370271 | 2.56 |
ENST00000375679.4
|
CLCNKB
|
chloride channel, voltage-sensitive Kb |
| chr3_+_179322481 | 2.49 |
ENST00000259037.3
|
NDUFB5
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5, 16kDa |
| chr1_+_16348497 | 2.47 |
ENST00000439316.2
|
CLCNKA
|
chloride channel, voltage-sensitive Ka |
| chr1_-_149908217 | 2.46 |
ENST00000369140.3
|
MTMR11
|
myotubularin related protein 11 |
| chr16_+_67562702 | 2.43 |
ENST00000379312.3
ENST00000042381.4 ENST00000540839.3 |
FAM65A
|
family with sequence similarity 65, member A |
| chr3_+_51663407 | 2.28 |
ENST00000432863.1
ENST00000296477.3 |
RAD54L2
|
RAD54-like 2 (S. cerevisiae) |
| chr1_-_151804314 | 2.26 |
ENST00000318247.6
|
RORC
|
RAR-related orphan receptor C |
| chr15_+_40731920 | 2.11 |
ENST00000561234.1
|
BAHD1
|
bromo adjacent homology domain containing 1 |
| chr11_+_33563821 | 2.01 |
ENST00000321505.4
ENST00000265654.5 ENST00000389726.3 |
KIAA1549L
|
KIAA1549-like |
| chr16_+_8736232 | 1.89 |
ENST00000562973.1
|
METTL22
|
methyltransferase like 22 |
| chr11_+_46366918 | 1.75 |
ENST00000528615.1
ENST00000395574.3 |
DGKZ
|
diacylglycerol kinase, zeta |
| chr16_-_11370330 | 1.67 |
ENST00000241808.4
ENST00000435245.2 |
PRM2
|
protamine 2 |
| chr1_+_16348366 | 1.61 |
ENST00000375692.1
ENST00000420078.1 |
CLCNKA
|
chloride channel, voltage-sensitive Ka |
| chr1_-_151804222 | 1.49 |
ENST00000392697.3
|
RORC
|
RAR-related orphan receptor C |
| chr10_+_118350468 | 1.41 |
ENST00000358834.4
ENST00000528052.1 ENST00000442761.1 |
PNLIPRP1
|
pancreatic lipase-related protein 1 |
| chr10_+_118350522 | 1.34 |
ENST00000530319.1
ENST00000527980.1 ENST00000471549.1 ENST00000534537.1 |
PNLIPRP1
|
pancreatic lipase-related protein 1 |
| chr10_+_23384435 | 1.34 |
ENST00000376510.3
|
MSRB2
|
methionine sulfoxide reductase B2 |
| chr19_+_13135731 | 1.32 |
ENST00000587260.1
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr6_+_35265586 | 1.19 |
ENST00000542066.1
ENST00000316637.5 |
DEF6
|
differentially expressed in FDCP 6 homolog (mouse) |
| chr20_-_62199427 | 1.07 |
ENST00000427522.2
|
HELZ2
|
helicase with zinc finger 2, transcriptional coactivator |
| chr19_+_13135790 | 0.96 |
ENST00000358552.3
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr20_+_44098385 | 0.76 |
ENST00000217425.5
ENST00000339946.3 |
WFDC2
|
WAP four-disulfide core domain 2 |
| chr17_-_26684473 | 0.69 |
ENST00000540200.1
|
POLDIP2
|
polymerase (DNA-directed), delta interacting protein 2 |
| chr10_+_89420706 | 0.62 |
ENST00000427144.2
|
PAPSS2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chr15_+_75498739 | 0.40 |
ENST00000565074.1
|
C15orf39
|
chromosome 15 open reading frame 39 |
| chr16_+_56899114 | 0.33 |
ENST00000566786.1
ENST00000438926.2 ENST00000563236.1 ENST00000262502.5 |
SLC12A3
|
solute carrier family 12 (sodium/chloride transporter), member 3 |
| chr5_+_155753745 | 0.11 |
ENST00000435422.3
ENST00000337851.4 ENST00000447401.1 |
SGCD
|
sarcoglycan, delta (35kDa dystrophin-associated glycoprotein) |
| chr2_+_1488435 | 0.03 |
ENST00000446278.1
ENST00000469607.1 |
TPO
|
thyroid peroxidase |
| chr10_-_76859247 | 0.01 |
ENST00000472493.2
ENST00000605915.1 ENST00000478873.2 |
DUSP13
|
dual specificity phosphatase 13 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZKSCAN1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.5 | 37.4 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 10.5 | 31.6 | GO:1901837 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) negative regulation of protein localization to chromosome, telomeric region(GO:1904815) |
| 5.5 | 44.2 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 4.9 | 19.5 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 3.1 | 34.6 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 2.9 | 14.6 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 2.9 | 11.5 | GO:0072683 | T cell extravasation(GO:0072683) |
| 2.4 | 18.9 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 2.3 | 11.5 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 2.0 | 12.0 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 1.8 | 7.2 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 1.5 | 18.4 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 1.4 | 8.6 | GO:2000568 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 1.1 | 12.3 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 1.1 | 3.2 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 1.1 | 4.3 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.9 | 8.2 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.8 | 37.2 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.8 | 36.0 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.6 | 6.1 | GO:0098902 | regulation of membrane depolarization during action potential(GO:0098902) |
| 0.6 | 6.6 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.6 | 4.7 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.6 | 3.3 | GO:0009099 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.6 | 12.2 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.5 | 6.4 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.5 | 4.7 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.5 | 7.8 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.4 | 9.5 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.4 | 51.4 | GO:0002062 | chondrocyte differentiation(GO:0002062) |
| 0.3 | 6.2 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.3 | 16.4 | GO:0043486 | histone exchange(GO:0043486) |
| 0.3 | 5.2 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.3 | 17.2 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.2 | 22.6 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.2 | 2.3 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.2 | 3.7 | GO:0060850 | regulation of transcription involved in cell fate commitment(GO:0060850) |
| 0.2 | 2.1 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.2 | 0.6 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.1 | 1.3 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 6.6 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.1 | 1.8 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.1 | 6.6 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 4.6 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) |
| 0.1 | 4.3 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.1 | 4.3 | GO:0008542 | visual learning(GO:0008542) |
| 0.1 | 10.3 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 1.7 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.0 | 2.5 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.7 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 10.4 | GO:0001701 | in utero embryonic development(GO:0001701) |
| 0.0 | 1.9 | GO:0018022 | peptidyl-lysine methylation(GO:0018022) |
| 0.0 | 3.0 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 7.6 | GO:0051301 | cell division(GO:0051301) |
| 0.0 | 7.7 | GO:0016567 | protein ubiquitination(GO:0016567) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.3 | 31.6 | GO:0001740 | Barr body(GO:0001740) |
| 2.4 | 19.6 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 2.4 | 14.6 | GO:0090661 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 2.1 | 37.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 1.3 | 37.4 | GO:0031143 | pseudopodium(GO:0031143) |
| 1.0 | 6.2 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.9 | 10.7 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.9 | 19.5 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.6 | 11.5 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.6 | 17.2 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.6 | 11.5 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.6 | 73.9 | GO:0005604 | basement membrane(GO:0005604) |
| 0.6 | 12.2 | GO:0031105 | septin complex(GO:0031105) |
| 0.4 | 34.6 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.3 | 8.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.3 | 18.9 | GO:0002102 | podosome(GO:0002102) |
| 0.3 | 4.6 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.3 | 2.3 | GO:0034464 | BBSome(GO:0034464) |
| 0.2 | 4.7 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.2 | 2.1 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.1 | 9.5 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.1 | 3.0 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 36.0 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.1 | 6.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 23.6 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 8.6 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 3.2 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 6.6 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 13.7 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 7.2 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 2.5 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.7 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 6.7 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 6.1 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 4.7 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 6.8 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.1 | GO:0016012 | dystroglycan complex(GO:0016011) sarcoglycan complex(GO:0016012) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.9 | 19.5 | GO:0034057 | RNA strand-exchange activity(GO:0034057) |
| 3.5 | 31.6 | GO:0000182 | rDNA binding(GO:0000182) |
| 3.0 | 36.0 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 3.0 | 12.0 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 2.4 | 19.6 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 2.4 | 14.6 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 2.1 | 37.2 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 1.6 | 7.8 | GO:0019798 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 1.4 | 8.6 | GO:0023025 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 1.3 | 11.5 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.9 | 44.6 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.9 | 4.3 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.8 | 20.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.8 | 53.8 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.7 | 5.2 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.6 | 9.5 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.6 | 8.2 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.6 | 4.7 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.6 | 3.3 | GO:0004084 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.5 | 3.7 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.4 | 6.4 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.4 | 11.2 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.3 | 1.3 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.3 | 6.6 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.2 | 22.4 | GO:0101005 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.2 | 0.6 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.2 | 10.7 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.2 | 11.5 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.2 | 6.6 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.2 | 4.6 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 1.7 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.1 | 33.2 | GO:0019887 | protein kinase regulator activity(GO:0019887) |
| 0.1 | 6.1 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 2.7 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 1.8 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 16.4 | GO:0042393 | histone binding(GO:0042393) |
| 0.1 | 20.0 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.1 | 18.8 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 4.6 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.1 | 7.5 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 21.8 | GO:0003712 | transcription cofactor activity(GO:0003712) |
| 0.0 | 2.5 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 1.9 | GO:0016278 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 2.1 | GO:0000976 | transcription regulatory region sequence-specific DNA binding(GO:0000976) |
| 0.0 | 2.3 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 4.3 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 11.2 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.3 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.0 | 5.0 | GO:0016887 | ATPase activity(GO:0016887) |
| 0.0 | 3.0 | GO:0030246 | carbohydrate binding(GO:0030246) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 53.8 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.7 | 36.0 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.5 | 29.9 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.5 | 18.4 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.4 | 24.7 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.3 | 31.6 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.3 | 11.5 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.3 | 6.2 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.2 | 7.2 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.2 | 36.5 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.2 | 12.3 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.2 | 16.4 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 6.4 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 6.6 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 3.2 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 15.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 3.0 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 1.2 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 37.2 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 1.4 | 18.4 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 1.4 | 19.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 1.2 | 37.4 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.8 | 34.6 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.7 | 11.5 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.6 | 23.6 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.6 | 53.8 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.2 | 17.2 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.2 | 27.9 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.2 | 19.5 | REACTOME ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S | Genes involved in Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S |
| 0.2 | 36.0 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 5.2 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 3.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 10.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 8.2 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 6.2 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 3.0 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 2.5 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.6 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |