Project
averaged GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
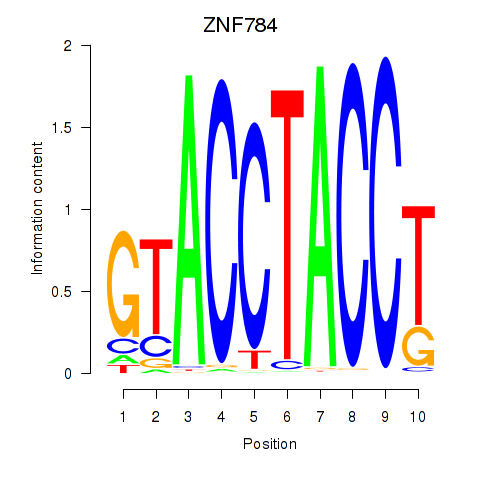
Results for ZNF784
Z-value: 2.13
Transcription factors associated with ZNF784
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF784
|
ENSG00000179922.5 | zinc finger protein 784 |
Activity profile of ZNF784 motif
Sorted Z-values of ZNF784 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_195310802 | 59.82 |
ENST00000421243.1
ENST00000453131.1 |
APOD
|
apolipoprotein D |
| chr3_-_42306248 | 43.51 |
ENST00000334681.5
|
CCK
|
cholecystokinin |
| chr11_-_5248294 | 42.88 |
ENST00000335295.4
|
HBB
|
hemoglobin, beta |
| chr19_-_42498369 | 41.85 |
ENST00000302102.5
ENST00000545399.1 |
ATP1A3
|
ATPase, Na+/K+ transporting, alpha 3 polypeptide |
| chr19_-_42498231 | 41.06 |
ENST00000602133.1
|
ATP1A3
|
ATPase, Na+/K+ transporting, alpha 3 polypeptide |
| chr4_+_158141806 | 40.43 |
ENST00000393815.2
|
GRIA2
|
glutamate receptor, ionotropic, AMPA 2 |
| chr4_+_158142750 | 40.29 |
ENST00000505888.1
ENST00000449365.1 |
GRIA2
|
glutamate receptor, ionotropic, AMPA 2 |
| chr11_+_73358594 | 40.16 |
ENST00000227214.6
ENST00000398494.4 ENST00000543085.1 |
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr8_+_80523321 | 39.85 |
ENST00000518111.1
|
STMN2
|
stathmin-like 2 |
| chr4_+_158141843 | 39.31 |
ENST00000509417.1
ENST00000296526.7 |
GRIA2
|
glutamate receptor, ionotropic, AMPA 2 |
| chr4_+_158141899 | 36.84 |
ENST00000264426.9
ENST00000506284.1 |
GRIA2
|
glutamate receptor, ionotropic, AMPA 2 |
| chr12_-_91572278 | 27.37 |
ENST00000425043.1
ENST00000420120.2 ENST00000441303.2 ENST00000456569.2 |
DCN
|
decorin |
| chr20_+_44036620 | 26.97 |
ENST00000372710.3
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr4_-_90758118 | 26.95 |
ENST00000420646.2
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr9_-_139094988 | 25.58 |
ENST00000371746.3
|
LHX3
|
LIM homeobox 3 |
| chr3_+_50712672 | 23.93 |
ENST00000266037.9
|
DOCK3
|
dedicator of cytokinesis 3 |
| chr11_-_6440283 | 23.70 |
ENST00000299402.6
ENST00000609360.1 ENST00000389906.2 ENST00000532020.2 |
APBB1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr11_-_6440624 | 23.03 |
ENST00000311051.3
|
APBB1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr10_-_93392811 | 22.99 |
ENST00000238994.5
|
PPP1R3C
|
protein phosphatase 1, regulatory subunit 3C |
| chr4_-_90758227 | 22.20 |
ENST00000506691.1
ENST00000394986.1 ENST00000506244.1 ENST00000394989.2 ENST00000394991.3 |
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr15_+_25200108 | 20.93 |
ENST00000577949.1
ENST00000338094.6 ENST00000338327.4 ENST00000579070.1 ENST00000577565.1 |
SNURF
SNRPN
|
SNRPN upstream reading frame protein small nuclear ribonucleoprotein polypeptide N |
| chr6_+_151042224 | 20.83 |
ENST00000358517.2
|
PLEKHG1
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 1 |
| chr11_-_134281812 | 20.40 |
ENST00000392580.1
ENST00000312527.4 |
B3GAT1
|
beta-1,3-glucuronyltransferase 1 (glucuronosyltransferase P) |
| chr3_-_47619623 | 20.28 |
ENST00000456150.1
|
CSPG5
|
chondroitin sulfate proteoglycan 5 (neuroglycan C) |
| chr5_-_74062930 | 20.24 |
ENST00000509430.1
ENST00000345239.2 ENST00000427854.2 ENST00000506778.1 |
GFM2
|
G elongation factor, mitochondrial 2 |
| chr8_-_9008206 | 20.03 |
ENST00000310455.3
|
PPP1R3B
|
protein phosphatase 1, regulatory subunit 3B |
| chr5_-_149535421 | 19.93 |
ENST00000261799.4
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide |
| chr2_-_85839146 | 19.56 |
ENST00000306336.5
ENST00000409734.3 |
C2orf68
|
chromosome 2 open reading frame 68 |
| chr2_-_68384603 | 19.53 |
ENST00000406245.2
ENST00000409164.1 ENST00000295121.6 |
WDR92
|
WD repeat domain 92 |
| chr12_-_66524482 | 19.26 |
ENST00000446587.2
ENST00000266604.2 |
LLPH
|
LLP homolog, long-term synaptic facilitation (Aplysia) |
| chrX_-_54384425 | 18.80 |
ENST00000375169.3
ENST00000354646.2 |
WNK3
|
WNK lysine deficient protein kinase 3 |
| chr10_+_92980517 | 18.58 |
ENST00000336126.5
|
PCGF5
|
polycomb group ring finger 5 |
| chr12_-_91573249 | 18.08 |
ENST00000550099.1
ENST00000546391.1 ENST00000551354.1 |
DCN
|
decorin |
| chr20_+_44036900 | 17.70 |
ENST00000443296.1
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr6_-_150039249 | 17.45 |
ENST00000543571.1
|
LATS1
|
large tumor suppressor kinase 1 |
| chr19_-_57988871 | 17.16 |
ENST00000596831.1
ENST00000601768.1 ENST00000356584.3 ENST00000600175.1 ENST00000425074.3 ENST00000343280.4 ENST00000427512.2 |
AC004076.9
ZNF772
|
Uncharacterized protein zinc finger protein 772 |
| chr1_+_50575292 | 16.99 |
ENST00000371821.1
ENST00000371819.1 |
ELAVL4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr18_-_21852143 | 16.89 |
ENST00000399443.3
|
OSBPL1A
|
oxysterol binding protein-like 1A |
| chr5_+_161274940 | 16.87 |
ENST00000393943.4
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr3_-_51994694 | 16.28 |
ENST00000395014.2
|
PCBP4
|
poly(rC) binding protein 4 |
| chr18_-_5419797 | 16.14 |
ENST00000542146.1
ENST00000427684.2 |
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3 |
| chr10_+_82168240 | 15.91 |
ENST00000372187.5
ENST00000372185.1 |
FAM213A
|
family with sequence similarity 213, member A |
| chr10_-_127511790 | 15.59 |
ENST00000368797.4
ENST00000420761.1 |
UROS
|
uroporphyrinogen III synthase |
| chr4_+_48833234 | 15.44 |
ENST00000510824.1
ENST00000425583.2 |
OCIAD1
|
OCIA domain containing 1 |
| chr1_+_50574585 | 15.41 |
ENST00000371824.1
ENST00000371823.4 |
ELAVL4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr11_-_62389449 | 14.76 |
ENST00000534026.1
|
B3GAT3
|
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) |
| chr2_-_11272234 | 14.06 |
ENST00000590207.1
ENST00000417697.2 ENST00000396164.1 ENST00000536743.1 ENST00000544306.1 |
AC062028.1
|
AC062028.1 |
| chr3_+_45067659 | 13.85 |
ENST00000296130.4
|
CLEC3B
|
C-type lectin domain family 3, member B |
| chr17_-_74707037 | 13.78 |
ENST00000355797.3
ENST00000375036.2 ENST00000449428.2 |
MXRA7
|
matrix-remodelling associated 7 |
| chr22_+_42196666 | 13.54 |
ENST00000402061.3
ENST00000255784.5 |
CCDC134
|
coiled-coil domain containing 134 |
| chr17_-_36956155 | 13.15 |
ENST00000269554.3
|
PIP4K2B
|
phosphatidylinositol-5-phosphate 4-kinase, type II, beta |
| chr9_-_79307096 | 13.15 |
ENST00000376717.2
ENST00000223609.6 ENST00000443509.2 |
PRUNE2
|
prune homolog 2 (Drosophila) |
| chr4_+_41362796 | 12.85 |
ENST00000508501.1
ENST00000512946.1 ENST00000313860.7 ENST00000512632.1 ENST00000512820.1 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr8_-_9009079 | 12.81 |
ENST00000519699.1
|
PPP1R3B
|
protein phosphatase 1, regulatory subunit 3B |
| chr4_-_130692631 | 12.76 |
ENST00000500092.2
ENST00000509105.1 |
RP11-519M16.1
|
RP11-519M16.1 |
| chrX_-_6146876 | 12.67 |
ENST00000381095.3
|
NLGN4X
|
neuroligin 4, X-linked |
| chr4_-_90756769 | 12.46 |
ENST00000345009.4
ENST00000505199.1 ENST00000502987.1 |
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr4_+_37455536 | 12.31 |
ENST00000381980.4
ENST00000508175.1 |
C4orf19
|
chromosome 4 open reading frame 19 |
| chr1_-_203320617 | 12.29 |
ENST00000354955.4
|
FMOD
|
fibromodulin |
| chr5_+_161275320 | 12.27 |
ENST00000437025.2
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr5_+_137225158 | 12.06 |
ENST00000290431.5
|
PKD2L2
|
polycystic kidney disease 2-like 2 |
| chr4_+_48833119 | 12.01 |
ENST00000444354.2
ENST00000509963.1 ENST00000509246.1 |
OCIAD1
|
OCIA domain containing 1 |
| chr9_-_115480303 | 11.80 |
ENST00000374234.1
ENST00000374238.1 ENST00000374236.1 ENST00000374242.4 |
INIP
|
INTS3 and NABP interacting protein |
| chr5_+_140729649 | 11.59 |
ENST00000523390.1
|
PCDHGB1
|
protocadherin gamma subfamily B, 1 |
| chr7_-_103629963 | 11.26 |
ENST00000428762.1
ENST00000343529.5 ENST00000424685.2 |
RELN
|
reelin |
| chr2_-_127864839 | 11.15 |
ENST00000409400.1
ENST00000357970.3 ENST00000393040.3 ENST00000348750.4 ENST00000259238.4 ENST00000346226.3 ENST00000393041.3 ENST00000351659.3 ENST00000352848.3 ENST00000316724.5 |
BIN1
|
bridging integrator 1 |
| chr3_-_9834375 | 11.15 |
ENST00000343450.2
ENST00000301964.2 |
TADA3
|
transcriptional adaptor 3 |
| chr3_-_122512619 | 11.11 |
ENST00000383659.1
ENST00000306103.2 |
HSPBAP1
|
HSPB (heat shock 27kDa) associated protein 1 |
| chr11_+_65154070 | 11.01 |
ENST00000317568.5
ENST00000531296.1 ENST00000533782.1 ENST00000355991.5 ENST00000416776.2 ENST00000526201.1 |
FRMD8
|
FERM domain containing 8 |
| chr4_+_6202448 | 10.90 |
ENST00000508601.1
|
RP11-586D19.1
|
RP11-586D19.1 |
| chr14_+_29241910 | 10.87 |
ENST00000399387.4
ENST00000552957.1 ENST00000548213.1 |
C14orf23
|
chromosome 14 open reading frame 23 |
| chr4_-_90757364 | 10.81 |
ENST00000508895.1
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr17_+_77020325 | 10.73 |
ENST00000311661.4
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr3_+_183993797 | 10.55 |
ENST00000359140.4
ENST00000404464.3 ENST00000357474.5 |
ECE2
|
endothelin converting enzyme 2 |
| chr8_+_85095497 | 10.55 |
ENST00000522455.1
ENST00000521695.1 |
RALYL
|
RALY RNA binding protein-like |
| chr17_+_42264556 | 10.24 |
ENST00000319511.6
ENST00000589785.1 ENST00000592825.1 ENST00000589184.1 |
TMUB2
|
transmembrane and ubiquitin-like domain containing 2 |
| chr11_-_115375107 | 10.21 |
ENST00000545380.1
ENST00000452722.3 ENST00000537058.1 ENST00000536727.1 ENST00000542447.2 ENST00000331581.6 |
CADM1
|
cell adhesion molecule 1 |
| chr5_-_174871136 | 10.09 |
ENST00000393752.2
|
DRD1
|
dopamine receptor D1 |
| chr2_-_25873079 | 10.05 |
ENST00000496972.2
|
DTNB
|
dystrobrevin, beta |
| chr4_+_71263599 | 9.98 |
ENST00000399575.2
|
PROL1
|
proline rich, lacrimal 1 |
| chrX_-_154842589 | 9.95 |
ENST00000334398.3
|
TMLHE
|
trimethyllysine hydroxylase, epsilon |
| chr4_+_62066941 | 9.81 |
ENST00000512091.2
|
LPHN3
|
latrophilin 3 |
| chr17_+_77020224 | 9.47 |
ENST00000339142.2
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr22_+_20067738 | 9.47 |
ENST00000351989.3
ENST00000383024.2 |
DGCR8
|
DGCR8 microprocessor complex subunit |
| chr8_+_11561660 | 9.44 |
ENST00000526716.1
ENST00000335135.4 ENST00000528027.1 |
GATA4
|
GATA binding protein 4 |
| chr3_+_197476621 | 9.40 |
ENST00000241502.4
|
FYTTD1
|
forty-two-three domain containing 1 |
| chr15_+_89346657 | 9.28 |
ENST00000439576.2
|
ACAN
|
aggrecan |
| chr17_+_10600894 | 9.27 |
ENST00000379774.4
|
ADPRM
|
ADP-ribose/CDP-alcohol diphosphatase, manganese-dependent |
| chr19_+_3880581 | 9.24 |
ENST00000450849.2
ENST00000301260.6 ENST00000398448.3 |
ATCAY
|
ataxia, cerebellar, Cayman type |
| chr5_-_178772424 | 9.23 |
ENST00000251582.7
ENST00000274609.5 |
ADAMTS2
|
ADAM metallopeptidase with thrombospondin type 1 motif, 2 |
| chr22_-_44708731 | 9.15 |
ENST00000381176.4
|
KIAA1644
|
KIAA1644 |
| chr8_-_57358432 | 9.10 |
ENST00000517415.1
ENST00000314922.3 |
PENK
|
proenkephalin |
| chr17_+_19281034 | 9.09 |
ENST00000308406.5
ENST00000299612.7 |
MAPK7
|
mitogen-activated protein kinase 7 |
| chr4_-_10686475 | 9.08 |
ENST00000226951.6
|
CLNK
|
cytokine-dependent hematopoietic cell linker |
| chr19_+_49622646 | 8.99 |
ENST00000334186.4
|
PPFIA3
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 |
| chr11_-_74109422 | 8.91 |
ENST00000298198.4
|
PGM2L1
|
phosphoglucomutase 2-like 1 |
| chr1_-_154155675 | 8.85 |
ENST00000330188.9
ENST00000341485.5 |
TPM3
|
tropomyosin 3 |
| chr19_+_13858593 | 8.68 |
ENST00000221554.8
|
CCDC130
|
coiled-coil domain containing 130 |
| chr7_-_97881429 | 8.62 |
ENST00000420697.1
ENST00000379795.3 ENST00000415086.1 ENST00000542604.1 ENST00000447648.2 |
TECPR1
|
tectonin beta-propeller repeat containing 1 |
| chr6_+_123100620 | 8.60 |
ENST00000368444.3
|
FABP7
|
fatty acid binding protein 7, brain |
| chr16_+_19179549 | 8.51 |
ENST00000355377.2
ENST00000568115.1 |
SYT17
|
synaptotagmin XVII |
| chr21_-_36421535 | 8.45 |
ENST00000416754.1
ENST00000437180.1 ENST00000455571.1 |
RUNX1
|
runt-related transcription factor 1 |
| chr1_-_153029980 | 8.35 |
ENST00000392653.2
|
SPRR2A
|
small proline-rich protein 2A |
| chr1_+_205012293 | 8.34 |
ENST00000331830.4
|
CNTN2
|
contactin 2 (axonal) |
| chr1_+_171283331 | 8.29 |
ENST00000367749.3
|
FMO4
|
flavin containing monooxygenase 4 |
| chr5_+_137225125 | 8.24 |
ENST00000350250.4
ENST00000508638.1 ENST00000502810.1 ENST00000508883.1 |
PKD2L2
|
polycystic kidney disease 2-like 2 |
| chr4_-_153274078 | 8.09 |
ENST00000263981.5
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr3_+_195447738 | 7.98 |
ENST00000447234.2
ENST00000320736.6 ENST00000436408.1 |
MUC20
|
mucin 20, cell surface associated |
| chr12_-_14996355 | 7.94 |
ENST00000228936.4
|
ART4
|
ADP-ribosyltransferase 4 (Dombrock blood group) |
| chr20_+_20348740 | 7.94 |
ENST00000310227.1
|
INSM1
|
insulinoma-associated 1 |
| chr1_-_85156090 | 7.94 |
ENST00000605755.1
ENST00000437941.2 |
SSX2IP
|
synovial sarcoma, X breakpoint 2 interacting protein |
| chr9_-_99381660 | 7.91 |
ENST00000375240.3
ENST00000463569.1 |
CDC14B
|
cell division cycle 14B |
| chr1_+_205473720 | 7.48 |
ENST00000429964.2
ENST00000506784.1 ENST00000360066.2 |
CDK18
|
cyclin-dependent kinase 18 |
| chr11_-_19262486 | 7.47 |
ENST00000250024.4
|
E2F8
|
E2F transcription factor 8 |
| chr22_+_25202232 | 7.46 |
ENST00000400358.4
ENST00000400359.4 |
SGSM1
|
small G protein signaling modulator 1 |
| chr11_+_5009424 | 7.38 |
ENST00000300762.1
|
MMP26
|
matrix metallopeptidase 26 |
| chr13_-_96705624 | 7.38 |
ENST00000376747.3
ENST00000376712.4 ENST00000397618.3 ENST00000376714.3 |
UGGT2
|
UDP-glucose glycoprotein glucosyltransferase 2 |
| chr21_-_36421626 | 7.38 |
ENST00000300305.3
|
RUNX1
|
runt-related transcription factor 1 |
| chrX_+_49593853 | 7.35 |
ENST00000376141.1
|
PAGE4
|
P antigen family, member 4 (prostate associated) |
| chr3_+_124303472 | 7.16 |
ENST00000291478.5
|
KALRN
|
kalirin, RhoGEF kinase |
| chr6_-_46922659 | 7.11 |
ENST00000265417.7
|
GPR116
|
G protein-coupled receptor 116 |
| chrX_+_49593888 | 7.09 |
ENST00000218068.6
|
PAGE4
|
P antigen family, member 4 (prostate associated) |
| chr9_-_99382065 | 7.09 |
ENST00000265659.2
ENST00000375241.1 ENST00000375236.1 |
CDC14B
|
cell division cycle 14B |
| chr12_+_100661156 | 6.90 |
ENST00000360820.2
|
SCYL2
|
SCY1-like 2 (S. cerevisiae) |
| chr20_-_56286479 | 6.89 |
ENST00000265626.4
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chrX_-_55024967 | 6.53 |
ENST00000545676.1
|
PFKFB1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr1_+_203651937 | 6.41 |
ENST00000341360.2
|
ATP2B4
|
ATPase, Ca++ transporting, plasma membrane 4 |
| chr19_-_15236173 | 6.34 |
ENST00000527093.1
|
ILVBL
|
ilvB (bacterial acetolactate synthase)-like |
| chr12_+_52445191 | 6.32 |
ENST00000243050.1
ENST00000394825.1 ENST00000550763.1 ENST00000394824.2 ENST00000548232.1 ENST00000562373.1 |
NR4A1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr3_-_39321512 | 6.27 |
ENST00000399220.2
|
CX3CR1
|
chemokine (C-X3-C motif) receptor 1 |
| chr7_+_119913688 | 6.14 |
ENST00000331113.4
|
KCND2
|
potassium voltage-gated channel, Shal-related subfamily, member 2 |
| chr5_+_112073544 | 5.97 |
ENST00000257430.4
ENST00000508376.2 |
APC
|
adenomatous polyposis coli |
| chr5_-_159827073 | 5.85 |
ENST00000408953.3
|
C5orf54
|
chromosome 5 open reading frame 54 |
| chr11_-_62389621 | 5.83 |
ENST00000531383.1
ENST00000265471.5 |
B3GAT3
|
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) |
| chr2_-_73869508 | 5.82 |
ENST00000272425.3
|
NAT8
|
N-acetyltransferase 8 (GCN5-related, putative) |
| chr15_-_55657428 | 5.72 |
ENST00000568543.1
|
CCPG1
|
cell cycle progression 1 |
| chr4_+_71226468 | 5.59 |
ENST00000226460.4
|
SMR3A
|
submaxillary gland androgen regulated protein 3A |
| chr16_+_2564254 | 5.49 |
ENST00000565223.1
|
ATP6V0C
|
ATPase, H+ transporting, lysosomal 16kDa, V0 subunit c |
| chr19_+_50879705 | 5.42 |
ENST00000598168.1
ENST00000411902.2 ENST00000253727.5 ENST00000597790.1 ENST00000597130.1 ENST00000599105.1 |
NR1H2
|
nuclear receptor subfamily 1, group H, member 2 |
| chr11_+_1861399 | 5.37 |
ENST00000381905.3
|
TNNI2
|
troponin I type 2 (skeletal, fast) |
| chr18_-_31802056 | 5.32 |
ENST00000538587.1
|
NOL4
|
nucleolar protein 4 |
| chr6_-_100912785 | 5.29 |
ENST00000369208.3
|
SIM1
|
single-minded family bHLH transcription factor 1 |
| chr14_+_105452094 | 5.29 |
ENST00000551606.1
ENST00000547315.1 |
C14orf79
|
chromosome 14 open reading frame 79 |
| chr18_-_31802282 | 5.20 |
ENST00000535475.1
|
NOL4
|
nucleolar protein 4 |
| chr5_-_159827033 | 5.17 |
ENST00000523213.1
|
C5orf54
|
chromosome 5 open reading frame 54 |
| chr1_+_20465805 | 5.09 |
ENST00000375102.3
|
PLA2G2F
|
phospholipase A2, group IIF |
| chr18_+_905104 | 5.05 |
ENST00000579794.1
|
ADCYAP1
|
adenylate cyclase activating polypeptide 1 (pituitary) |
| chr1_-_85156216 | 5.03 |
ENST00000342203.3
ENST00000370612.4 |
SSX2IP
|
synovial sarcoma, X breakpoint 2 interacting protein |
| chr20_+_57414795 | 4.96 |
ENST00000371098.2
ENST00000371075.3 |
GNAS
|
GNAS complex locus |
| chr1_+_165600436 | 4.91 |
ENST00000367888.4
ENST00000367885.1 ENST00000367884.2 |
MGST3
|
microsomal glutathione S-transferase 3 |
| chr2_-_70780770 | 4.90 |
ENST00000444975.1
ENST00000445399.1 ENST00000418333.2 |
TGFA
|
transforming growth factor, alpha |
| chr7_+_26332645 | 4.90 |
ENST00000396376.1
|
SNX10
|
sorting nexin 10 |
| chr1_+_3607228 | 4.84 |
ENST00000378285.1
ENST00000378280.1 ENST00000378288.4 |
TP73
|
tumor protein p73 |
| chrY_+_16636354 | 4.82 |
ENST00000339174.5
|
NLGN4Y
|
neuroligin 4, Y-linked |
| chr11_-_10590118 | 4.71 |
ENST00000529598.1
|
LYVE1
|
lymphatic vessel endothelial hyaluronan receptor 1 |
| chr3_+_124303539 | 4.71 |
ENST00000428018.2
|
KALRN
|
kalirin, RhoGEF kinase |
| chr17_-_50237343 | 4.68 |
ENST00000575181.1
ENST00000570565.1 |
CA10
|
carbonic anhydrase X |
| chr17_+_73717407 | 4.68 |
ENST00000579662.1
|
ITGB4
|
integrin, beta 4 |
| chr20_-_56285595 | 4.66 |
ENST00000395816.3
ENST00000347215.4 |
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chr3_-_48130314 | 4.64 |
ENST00000439356.1
ENST00000395734.3 ENST00000426837.2 |
MAP4
|
microtubule-associated protein 4 |
| chr4_+_6784401 | 4.57 |
ENST00000425103.1
ENST00000307659.5 |
KIAA0232
|
KIAA0232 |
| chr1_-_115238207 | 4.55 |
ENST00000520113.2
ENST00000369538.3 ENST00000353928.6 |
AMPD1
|
adenosine monophosphate deaminase 1 |
| chr1_+_32042131 | 4.54 |
ENST00000271064.7
ENST00000537531.1 |
TINAGL1
|
tubulointerstitial nephritis antigen-like 1 |
| chr9_-_127533519 | 4.52 |
ENST00000487099.2
ENST00000344523.4 ENST00000373584.3 |
NR6A1
|
nuclear receptor subfamily 6, group A, member 1 |
| chr1_-_175161890 | 4.47 |
ENST00000545251.2
ENST00000423313.1 |
KIAA0040
|
KIAA0040 |
| chr18_-_45457192 | 4.41 |
ENST00000586514.1
ENST00000591214.1 ENST00000589877.1 |
SMAD2
|
SMAD family member 2 |
| chr12_-_96390063 | 4.40 |
ENST00000541929.1
|
HAL
|
histidine ammonia-lyase |
| chr19_-_376011 | 4.34 |
ENST00000342640.4
|
THEG
|
theg spermatid protein |
| chr13_-_33760216 | 4.31 |
ENST00000255486.4
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13 |
| chr1_+_165600083 | 4.27 |
ENST00000367889.3
|
MGST3
|
microsomal glutathione S-transferase 3 |
| chr9_+_136325089 | 4.22 |
ENST00000291722.7
ENST00000316948.4 ENST00000540581.1 |
CACFD1
|
calcium channel flower domain containing 1 |
| chr19_+_39936186 | 4.15 |
ENST00000432763.2
ENST00000402194.2 ENST00000601515.1 |
SUPT5H
|
suppressor of Ty 5 homolog (S. cerevisiae) |
| chr7_+_131012605 | 4.14 |
ENST00000446815.1
ENST00000352689.6 |
MKLN1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr12_+_50497784 | 4.09 |
ENST00000548814.1
|
GPD1
|
glycerol-3-phosphate dehydrogenase 1 (soluble) |
| chr21_-_35831880 | 4.07 |
ENST00000399289.3
ENST00000432085.1 |
KCNE1
|
potassium voltage-gated channel, Isk-related family, member 1 |
| chr17_+_73717551 | 4.07 |
ENST00000450894.3
|
ITGB4
|
integrin, beta 4 |
| chr12_+_54402790 | 4.06 |
ENST00000040584.4
|
HOXC8
|
homeobox C8 |
| chr3_-_48130707 | 3.99 |
ENST00000360240.6
ENST00000383737.4 |
MAP4
|
microtubule-associated protein 4 |
| chr12_-_48119301 | 3.99 |
ENST00000545824.2
ENST00000422538.3 |
ENDOU
|
endonuclease, polyU-specific |
| chr2_-_56150910 | 3.97 |
ENST00000424836.2
ENST00000438672.1 ENST00000440439.1 ENST00000429909.1 ENST00000424207.1 ENST00000452337.1 ENST00000355426.3 ENST00000439193.1 ENST00000421664.1 |
EFEMP1
|
EGF containing fibulin-like extracellular matrix protein 1 |
| chr11_+_89867681 | 3.93 |
ENST00000534061.1
|
NAALAD2
|
N-acetylated alpha-linked acidic dipeptidase 2 |
| chr1_+_32042105 | 3.85 |
ENST00000457433.2
ENST00000441210.2 |
TINAGL1
|
tubulointerstitial nephritis antigen-like 1 |
| chrY_+_16634483 | 3.85 |
ENST00000382872.1
|
NLGN4Y
|
neuroligin 4, Y-linked |
| chr4_-_187476721 | 3.79 |
ENST00000307161.5
|
MTNR1A
|
melatonin receptor 1A |
| chr12_-_96390108 | 3.60 |
ENST00000538703.1
ENST00000261208.3 |
HAL
|
histidine ammonia-lyase |
| chr21_-_36421401 | 3.59 |
ENST00000486278.2
|
RUNX1
|
runt-related transcription factor 1 |
| chr6_+_136172820 | 3.56 |
ENST00000308191.6
|
PDE7B
|
phosphodiesterase 7B |
| chr21_+_41029235 | 3.55 |
ENST00000380618.1
|
B3GALT5
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 5 |
| chr3_+_139654018 | 3.54 |
ENST00000458420.3
|
CLSTN2
|
calsyntenin 2 |
| chr1_-_92951607 | 3.51 |
ENST00000427103.1
|
GFI1
|
growth factor independent 1 transcription repressor |
| chr4_+_71248795 | 3.49 |
ENST00000304915.3
|
SMR3B
|
submaxillary gland androgen regulated protein 3B |
| chr6_-_161695042 | 3.45 |
ENST00000366908.5
ENST00000366911.5 ENST00000366905.3 |
AGPAT4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr17_-_1394940 | 3.41 |
ENST00000570984.2
ENST00000361007.2 |
MYO1C
|
myosin IC |
| chrX_+_153686614 | 3.40 |
ENST00000369682.3
|
PLXNA3
|
plexin A3 |
| chr2_+_45878790 | 3.40 |
ENST00000306156.3
|
PRKCE
|
protein kinase C, epsilon |
| chr2_+_198669365 | 3.39 |
ENST00000428675.1
|
PLCL1
|
phospholipase C-like 1 |
| chrX_-_57163430 | 3.35 |
ENST00000374908.1
|
SPIN2A
|
spindlin family, member 2A |
| chr17_-_19281203 | 3.24 |
ENST00000487415.2
|
B9D1
|
B9 protein domain 1 |
| chr6_-_161695074 | 3.19 |
ENST00000457520.2
ENST00000366906.5 ENST00000320285.4 |
AGPAT4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr7_+_128577972 | 3.13 |
ENST00000357234.5
ENST00000477535.1 ENST00000479582.1 ENST00000464557.1 ENST00000402030.2 |
IRF5
|
interferon regulatory factor 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF784
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 19.9 | 59.8 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 15.6 | 46.7 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 14.5 | 72.4 | GO:0051945 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) regulation of peroxidase activity(GO:2000468) positive regulation of peroxidase activity(GO:2000470) |
| 10.7 | 42.9 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 10.4 | 82.9 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 10.0 | 39.9 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 6.6 | 19.9 | GO:0060437 | lung growth(GO:0060437) metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 5.4 | 43.5 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 5.1 | 20.2 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 4.7 | 18.8 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 4.3 | 25.6 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 4.1 | 12.3 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 3.9 | 15.6 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 3.9 | 15.6 | GO:0006780 | uroporphyrinogen III biosynthetic process(GO:0006780) uroporphyrinogen III metabolic process(GO:0046502) |
| 3.8 | 45.5 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 3.6 | 21.3 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 3.5 | 17.5 | GO:0001826 | inner cell mass cell differentiation(GO:0001826) |
| 3.5 | 159.7 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 3.1 | 9.4 | GO:0003285 | septum secundum development(GO:0003285) |
| 2.8 | 11.3 | GO:0097477 | NMDA glutamate receptor clustering(GO:0097114) spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 2.8 | 8.3 | GO:0031133 | regulation of axon diameter(GO:0031133) positive regulation of adenosine receptor signaling pathway(GO:0060168) |
| 2.7 | 22.0 | GO:0046959 | habituation(GO:0046959) |
| 2.7 | 8.0 | GO:0019556 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 2.6 | 7.9 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 2.6 | 10.2 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 2.5 | 20.2 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 2.5 | 7.4 | GO:0097359 | UDP-glucosylation(GO:0097359) |
| 2.3 | 32.8 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 2.3 | 16.1 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 2.3 | 9.1 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 2.1 | 6.3 | GO:0002881 | negative regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002881) |
| 2.0 | 61.3 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 2.0 | 13.8 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 1.8 | 9.1 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 1.8 | 5.4 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 1.7 | 5.1 | GO:0070253 | somatostatin secretion(GO:0070253) |
| 1.6 | 8.1 | GO:2000638 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 1.5 | 7.5 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 1.5 | 29.1 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 1.4 | 8.6 | GO:0051012 | microtubule sliding(GO:0051012) |
| 1.4 | 11.1 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 1.2 | 13.2 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 1.2 | 3.5 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 1.1 | 5.7 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 1.1 | 5.5 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 1.1 | 6.5 | GO:0033133 | fructose 2,6-bisphosphate metabolic process(GO:0006003) positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 1.0 | 4.1 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 1.0 | 15.0 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 1.0 | 9.9 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 1.0 | 6.9 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 1.0 | 11.5 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.9 | 6.3 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.9 | 9.0 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.9 | 4.3 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.9 | 11.2 | GO:0048711 | positive regulation of astrocyte differentiation(GO:0048711) |
| 0.8 | 11.7 | GO:0035878 | nail development(GO:0035878) |
| 0.8 | 2.5 | GO:0010166 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.8 | 2.4 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 0.7 | 8.9 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.7 | 12.0 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.7 | 4.6 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.6 | 30.1 | GO:0097484 | dendrite extension(GO:0097484) |
| 0.6 | 22.8 | GO:2000810 | regulation of bicellular tight junction assembly(GO:2000810) |
| 0.6 | 41.7 | GO:0007602 | phototransduction(GO:0007602) |
| 0.6 | 10.6 | GO:0010002 | cardioblast differentiation(GO:0010002) |
| 0.6 | 9.3 | GO:0034656 | nucleobase-containing small molecule catabolic process(GO:0034656) |
| 0.6 | 2.9 | GO:1900238 | positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 0.6 | 8.0 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.6 | 3.4 | GO:1902306 | negative regulation of sodium ion transport(GO:0010766) negative regulation of sodium ion transmembrane transport(GO:1902306) negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.6 | 1.7 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.6 | 5.0 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.5 | 20.3 | GO:0050982 | detection of mechanical stimulus(GO:0050982) |
| 0.5 | 2.2 | GO:1900154 | regulation of bone trabecula formation(GO:1900154) negative regulation of bone trabecula formation(GO:1900155) |
| 0.5 | 8.3 | GO:0042737 | drug catabolic process(GO:0042737) |
| 0.5 | 8.2 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.5 | 3.4 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.5 | 6.6 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.5 | 8.4 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.5 | 4.5 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.4 | 6.1 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.4 | 4.2 | GO:0097283 | keratinocyte apoptotic process(GO:0097283) regulation of keratinocyte apoptotic process(GO:1902172) |
| 0.4 | 0.8 | GO:0046066 | dGDP metabolic process(GO:0046066) |
| 0.4 | 3.4 | GO:0021637 | trigeminal nerve morphogenesis(GO:0021636) trigeminal nerve structural organization(GO:0021637) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.4 | 16.9 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.4 | 1.5 | GO:0002331 | pre-B cell differentiation(GO:0002329) pre-B cell allelic exclusion(GO:0002331) |
| 0.4 | 23.0 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.3 | 8.0 | GO:0048012 | hepatocyte growth factor receptor signaling pathway(GO:0048012) |
| 0.3 | 3.8 | GO:0007617 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.3 | 4.1 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.3 | 7.9 | GO:0006525 | arginine metabolic process(GO:0006525) |
| 0.3 | 4.8 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.3 | 9.2 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.3 | 5.0 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.3 | 8.6 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.3 | 2.2 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.3 | 9.2 | GO:1901685 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.3 | 5.1 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.3 | 5.7 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.3 | 1.3 | GO:0033686 | positive regulation of luteinizing hormone secretion(GO:0033686) |
| 0.2 | 5.7 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.2 | 0.9 | GO:0060283 | negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.2 | 7.3 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.2 | 15.9 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.2 | 2.3 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.2 | 4.7 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.2 | 13.8 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.2 | 4.9 | GO:0030316 | osteoclast differentiation(GO:0030316) |
| 0.2 | 8.4 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.2 | 2.8 | GO:0035635 | entry of bacterium into host cell(GO:0035635) |
| 0.2 | 5.8 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.1 | 1.8 | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of membrane repolarization during action potential(GO:0098903) |
| 0.1 | 0.8 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.1 | 2.3 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.1 | 9.5 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.1 | 4.1 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.1 | 2.9 | GO:0002507 | tolerance induction(GO:0002507) |
| 0.1 | 1.3 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.1 | 7.0 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.1 | 5.3 | GO:0001657 | ureteric bud development(GO:0001657) mesonephros development(GO:0001823) mesonephric epithelium development(GO:0072163) mesonephric tubule development(GO:0072164) |
| 0.1 | 1.7 | GO:0010952 | positive regulation of peptidase activity(GO:0010952) |
| 0.1 | 1.0 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.1 | 7.1 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.1 | 0.6 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 | 3.6 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 1.4 | GO:0014065 | phosphatidylinositol 3-kinase signaling(GO:0014065) |
| 0.1 | 12.7 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 11.9 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.1 | 12.8 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.1 | 9.4 | GO:0071427 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.1 | 2.0 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 3.2 | GO:0042733 | embryonic digit morphogenesis(GO:0042733) |
| 0.0 | 2.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 2.3 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.0 | 0.2 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 8.4 | GO:0016197 | endosomal transport(GO:0016197) |
| 0.0 | 2.3 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 7.8 | GO:0006469 | negative regulation of protein kinase activity(GO:0006469) |
| 0.0 | 1.7 | GO:0072583 | clathrin-mediated endocytosis(GO:0072583) |
| 0.0 | 0.5 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 1.9 | GO:0043967 | histone H4 acetylation(GO:0043967) |
| 0.0 | 2.7 | GO:0008652 | cellular amino acid biosynthetic process(GO:0008652) |
| 0.0 | 0.9 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 2.6 | GO:0048705 | skeletal system morphogenesis(GO:0048705) |
| 0.0 | 3.3 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 31.4 | 156.9 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 11.8 | 82.9 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 10.7 | 42.9 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 7.1 | 49.5 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 3.6 | 43.5 | GO:0043203 | axon hillock(GO:0043203) |
| 3.3 | 32.8 | GO:0042587 | glycogen granule(GO:0042587) |
| 3.2 | 45.5 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 3.0 | 11.8 | GO:0070876 | SOSS complex(GO:0070876) |
| 2.8 | 13.8 | GO:0001652 | granular component(GO:0001652) |
| 2.3 | 72.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 2.0 | 8.0 | GO:0032044 | DSIF complex(GO:0032044) |
| 1.5 | 9.1 | GO:0032280 | symmetric synapse(GO:0032280) |
| 1.2 | 20.9 | GO:0005687 | U4 snRNP(GO:0005687) |
| 1.2 | 8.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.9 | 16.1 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.9 | 3.4 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.8 | 29.1 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.8 | 4.9 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.8 | 19.5 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.7 | 11.1 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.7 | 4.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.6 | 12.4 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.6 | 5.5 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.5 | 44.7 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.5 | 8.8 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.4 | 51.6 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.4 | 12.0 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.4 | 20.6 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.3 | 3.4 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.3 | 23.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.3 | 18.6 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.3 | 3.2 | GO:0036038 | MKS complex(GO:0036038) |
| 0.3 | 11.7 | GO:0008305 | integrin complex(GO:0008305) |
| 0.2 | 2.2 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.2 | 8.6 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.2 | 5.4 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 7.4 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.2 | 9.0 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.2 | 39.9 | GO:0030426 | growth cone(GO:0030426) |
| 0.2 | 1.9 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.2 | 8.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 71.8 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.1 | 5.1 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 1.8 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 10.1 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 26.4 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.1 | 15.4 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 4.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 2.5 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 21.6 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.1 | 4.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 14.9 | GO:0045202 | synapse(GO:0045202) |
| 0.1 | 11.5 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 17.9 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 8.5 | GO:0098793 | presynapse(GO:0098793) |
| 0.1 | 16.8 | GO:0005770 | late endosome(GO:0005770) |
| 0.1 | 22.5 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 7.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 4.6 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 0.5 | GO:0031105 | septin complex(GO:0031105) |
| 0.0 | 6.3 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 5.3 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 1.5 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 3.5 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.4 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 5.0 | GO:0061695 | transferase complex, transferring phosphorus-containing groups(GO:0061695) |
| 0.0 | 6.5 | GO:0005768 | endosome(GO:0005768) |
| 0.0 | 49.7 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 0.4 | GO:1902555 | endoribonuclease complex(GO:1902555) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.5 | 72.4 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 13.7 | 41.0 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 13.1 | 156.9 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 7.1 | 42.9 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 6.6 | 19.9 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 5.9 | 82.9 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 5.8 | 29.1 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 5.6 | 11.3 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 4.4 | 13.2 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 4.1 | 57.9 | GO:0048156 | tau protein binding(GO:0048156) |
| 3.1 | 9.3 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 2.5 | 10.1 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 2.5 | 7.4 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 2.1 | 6.4 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 2.1 | 6.3 | GO:0016495 | C-X3-C chemokine receptor activity(GO:0016495) |
| 2.0 | 8.0 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 1.9 | 9.5 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 1.7 | 18.8 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 1.6 | 7.9 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 1.6 | 57.0 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 1.5 | 21.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 1.5 | 8.9 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 1.3 | 6.5 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 1.3 | 16.9 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 1.3 | 3.8 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 1.2 | 5.8 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 1.1 | 59.8 | GO:0015485 | cholesterol binding(GO:0015485) |
| 1.1 | 5.4 | GO:0034191 | apolipoprotein receptor binding(GO:0034190) apolipoprotein A-I receptor binding(GO:0034191) |
| 1.0 | 8.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 1.0 | 5.0 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 1.0 | 15.4 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.9 | 8.3 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.9 | 11.7 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.9 | 32.4 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.9 | 9.4 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.8 | 3.4 | GO:0035276 | ethanol binding(GO:0035276) |
| 0.8 | 2.5 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.8 | 4.1 | GO:0016901 | glycerol-3-phosphate dehydrogenase activity(GO:0004368) oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.8 | 6.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.8 | 4.6 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.7 | 20.2 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.7 | 69.2 | GO:0005518 | collagen binding(GO:0005518) |
| 0.7 | 6.3 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.6 | 13.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.5 | 3.2 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.5 | 5.4 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.5 | 55.8 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.5 | 4.6 | GO:0051430 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.5 | 4.5 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.5 | 9.1 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.5 | 5.7 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.5 | 38.8 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.4 | 4.1 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.4 | 4.8 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.4 | 11.1 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.3 | 8.9 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.3 | 21.1 | GO:0001098 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.3 | 1.7 | GO:0016453 | C-acetyltransferase activity(GO:0016453) |
| 0.3 | 6.6 | GO:0042171 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) lysophosphatidic acid acyltransferase activity(GO:0042171) lysophospholipid acyltransferase activity(GO:0071617) |
| 0.3 | 15.2 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.3 | 2.3 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.3 | 8.6 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.3 | 17.5 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.3 | 9.2 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.3 | 3.5 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.3 | 11.6 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.3 | 2.8 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.3 | 5.8 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.3 | 28.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.2 | 4.9 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.2 | 15.6 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.2 | 8.5 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.2 | 1.7 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.2 | 16.3 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.2 | 5.5 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.2 | 3.4 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.2 | 3.4 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.2 | 3.9 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.2 | 2.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.2 | 19.5 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.2 | 1.5 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.2 | 8.3 | GO:0043621 | protein self-association(GO:0043621) |
| 0.2 | 22.1 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.2 | 2.9 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.2 | 10.8 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.2 | 2.2 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.1 | 5.1 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.1 | 9.7 | GO:0016209 | antioxidant activity(GO:0016209) |
| 0.1 | 1.5 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 12.9 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 10.2 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 3.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 5.9 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 2.0 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 8.8 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 1.0 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 2.3 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 13.4 | GO:0001076 | transcription factor activity, RNA polymerase II transcription factor binding(GO:0001076) |
| 0.1 | 7.9 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.1 | 1.3 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 1.4 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 1.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 4.2 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 0.9 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 13.8 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 4.6 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 2.3 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 13.6 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 1.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 5.0 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 12.2 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.8 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 2.2 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 2.3 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 6.0 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 156.9 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 1.2 | 68.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 1.0 | 72.4 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.8 | 18.9 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.6 | 11.9 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.4 | 46.7 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.4 | 75.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.3 | 11.3 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.3 | 11.7 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.3 | 9.1 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.3 | 11.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.3 | 11.2 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.3 | 7.0 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.2 | 9.4 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.2 | 14.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.2 | 31.6 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 3.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 2.8 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 8.2 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 21.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 2.9 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 2.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 2.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 2.8 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.7 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.5 | 156.9 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 2.3 | 86.4 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 1.5 | 82.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 1.1 | 20.5 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.9 | 29.1 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.8 | 75.2 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.6 | 17.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.6 | 15.6 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.6 | 9.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.5 | 9.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.4 | 7.4 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.3 | 10.2 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.3 | 5.1 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.3 | 4.7 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.3 | 13.2 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.3 | 56.4 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.3 | 7.9 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.2 | 10.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.2 | 8.0 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.2 | 16.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 10.1 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.2 | 20.8 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.2 | 9.7 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.2 | 11.9 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.2 | 13.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.2 | 5.5 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.2 | 3.4 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.2 | 4.5 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.2 | 2.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.2 | 7.1 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.2 | 6.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 15.8 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 21.1 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 15.4 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.1 | 2.0 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 2.5 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 1.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 3.8 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 0.8 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 6.4 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 0.4 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.1 | 3.6 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 14.5 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 2.1 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |