Project
averaged GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
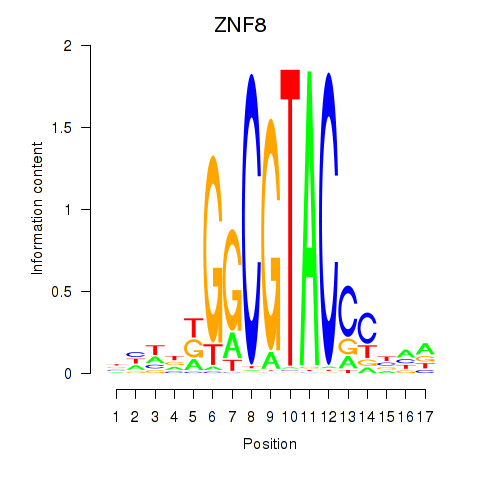
Results for ZNF8
Z-value: 0.76
Transcription factors associated with ZNF8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF8
|
ENSG00000083842.8 | zinc finger protein 8 |
|
ZNF8
|
ENSG00000273439.1 | zinc finger protein 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF8 | hg19_v2_chr19_+_58790314_58790358 | -0.56 | 3.6e-19 | Click! |
Activity profile of ZNF8 motif
Sorted Z-values of ZNF8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_46662772 | 27.10 |
ENST00000549049.1
ENST00000439706.1 ENST00000398637.5 |
SLC38A1
|
solute carrier family 38, member 1 |
| chr14_+_51706886 | 14.62 |
ENST00000457354.2
|
TMX1
|
thioredoxin-related transmembrane protein 1 |
| chr7_+_100303676 | 13.28 |
ENST00000303151.4
|
POP7
|
processing of precursor 7, ribonuclease P/MRP subunit (S. cerevisiae) |
| chr7_-_99698338 | 10.00 |
ENST00000354230.3
ENST00000425308.1 |
MCM7
|
minichromosome maintenance complex component 7 |
| chr1_-_70671216 | 9.67 |
ENST00000370952.3
|
LRRC40
|
leucine rich repeat containing 40 |
| chr1_-_235292250 | 9.09 |
ENST00000366607.4
|
TOMM20
|
translocase of outer mitochondrial membrane 20 homolog (yeast) |
| chr10_+_1034646 | 7.66 |
ENST00000360059.5
ENST00000545048.1 |
GTPBP4
|
GTP binding protein 4 |
| chr11_+_69455855 | 7.56 |
ENST00000227507.2
ENST00000536559.1 |
CCND1
|
cyclin D1 |
| chr4_-_77069573 | 7.49 |
ENST00000264883.3
|
NUP54
|
nucleoporin 54kDa |
| chr6_+_29691056 | 7.37 |
ENST00000414333.1
ENST00000334668.4 ENST00000259951.7 |
HLA-F
|
major histocompatibility complex, class I, F |
| chr16_+_21964662 | 7.11 |
ENST00000561553.1
ENST00000565331.1 |
UQCRC2
|
ubiquinol-cytochrome c reductase core protein II |
| chr6_+_29691198 | 7.02 |
ENST00000440587.2
ENST00000434407.2 |
HLA-F
|
major histocompatibility complex, class I, F |
| chr2_+_198365122 | 6.75 |
ENST00000604458.1
|
HSPE1-MOB4
|
HSPE1-MOB4 readthrough |
| chr8_-_109260897 | 6.74 |
ENST00000521297.1
ENST00000519030.1 ENST00000521440.1 ENST00000518345.1 ENST00000519627.1 ENST00000220849.5 |
EIF3E
|
eukaryotic translation initiation factor 3, subunit E |
| chr12_+_52463751 | 6.68 |
ENST00000336854.4
ENST00000550604.1 ENST00000553049.1 ENST00000548915.1 |
C12orf44
|
chromosome 12 open reading frame 44 |
| chr13_+_53029564 | 6.33 |
ENST00000468284.1
ENST00000378034.3 ENST00000258607.5 ENST00000378037.5 |
CKAP2
|
cytoskeleton associated protein 2 |
| chr6_+_106959718 | 6.30 |
ENST00000369066.3
|
AIM1
|
absent in melanoma 1 |
| chr5_-_137911049 | 5.81 |
ENST00000297185.3
|
HSPA9
|
heat shock 70kDa protein 9 (mortalin) |
| chr1_+_186344945 | 5.59 |
ENST00000419367.3
ENST00000287859.6 |
C1orf27
|
chromosome 1 open reading frame 27 |
| chr19_+_35739782 | 5.58 |
ENST00000347609.4
|
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr1_+_70671363 | 5.45 |
ENST00000370951.1
ENST00000370950.3 ENST00000405432.1 ENST00000454435.2 |
SRSF11
|
serine/arginine-rich splicing factor 11 |
| chr19_+_35739897 | 5.27 |
ENST00000605618.1
ENST00000427250.1 ENST00000601623.1 |
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr22_+_37415676 | 5.23 |
ENST00000401419.3
|
MPST
|
mercaptopyruvate sulfurtransferase |
| chr1_+_186344883 | 5.13 |
ENST00000367470.3
|
C1orf27
|
chromosome 1 open reading frame 27 |
| chrX_+_129535937 | 5.08 |
ENST00000305536.6
ENST00000370947.1 |
RBMX2
|
RNA binding motif protein, X-linked 2 |
| chr7_+_117824086 | 4.83 |
ENST00000249299.2
ENST00000424702.1 |
NAA38
|
N(alpha)-acetyltransferase 38, NatC auxiliary subunit |
| chr22_+_37415728 | 4.71 |
ENST00000404802.3
|
MPST
|
mercaptopyruvate sulfurtransferase |
| chr1_-_246670519 | 4.68 |
ENST00000388985.4
ENST00000490107.1 |
SMYD3
|
SET and MYND domain containing 3 |
| chr22_+_37415776 | 4.66 |
ENST00000341116.3
ENST00000429360.2 ENST00000404393.1 |
MPST
|
mercaptopyruvate sulfurtransferase |
| chr22_-_37415475 | 4.45 |
ENST00000403892.3
ENST00000249042.3 ENST00000438203.1 |
TST
|
thiosulfate sulfurtransferase (rhodanese) |
| chr5_+_10353780 | 4.39 |
ENST00000449913.2
ENST00000503788.1 ENST00000274140.5 |
MARCH6
|
membrane-associated ring finger (C3HC4) 6, E3 ubiquitin protein ligase |
| chr22_+_37415700 | 4.14 |
ENST00000397129.1
|
MPST
|
mercaptopyruvate sulfurtransferase |
| chr22_+_30163340 | 4.11 |
ENST00000330029.6
ENST00000401406.3 |
UQCR10
|
ubiquinol-cytochrome c reductase, complex III subunit X |
| chr11_-_58345569 | 3.92 |
ENST00000528954.1
ENST00000528489.1 |
LPXN
|
leupaxin |
| chr11_-_61560254 | 3.31 |
ENST00000543510.1
|
TMEM258
|
transmembrane protein 258 |
| chr11_+_46402583 | 3.13 |
ENST00000359803.3
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr19_-_18392422 | 2.90 |
ENST00000252818.3
|
JUND
|
jun D proto-oncogene |
| chr2_+_87144738 | 2.62 |
ENST00000559485.1
|
RGPD1
|
RANBP2-like and GRIP domain containing 1 |
| chr15_+_85523671 | 2.61 |
ENST00000310298.4
ENST00000557957.1 |
PDE8A
|
phosphodiesterase 8A |
| chr6_-_26056695 | 2.32 |
ENST00000343677.2
|
HIST1H1C
|
histone cluster 1, H1c |
| chr1_+_25664408 | 2.31 |
ENST00000374358.4
|
TMEM50A
|
transmembrane protein 50A |
| chr1_+_211500129 | 2.25 |
ENST00000427925.2
ENST00000261464.5 |
TRAF5
|
TNF receptor-associated factor 5 |
| chr13_+_45563721 | 2.20 |
ENST00000361121.2
|
GPALPP1
|
GPALPP motifs containing 1 |
| chr4_-_186347099 | 2.06 |
ENST00000505357.1
ENST00000264689.6 |
UFSP2
|
UFM1-specific peptidase 2 |
| chr8_+_61429416 | 2.01 |
ENST00000262646.7
ENST00000531289.1 |
RAB2A
|
RAB2A, member RAS oncogene family |
| chr5_+_102455968 | 2.00 |
ENST00000358359.3
|
PPIP5K2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr13_+_21750780 | 1.85 |
ENST00000309594.4
|
MRP63
|
mitochondrial ribosomal protein 63 |
| chr19_-_55652290 | 1.83 |
ENST00000589745.1
|
TNNT1
|
troponin T type 1 (skeletal, slow) |
| chr17_+_48624450 | 1.61 |
ENST00000006658.6
ENST00000356488.4 ENST00000393244.3 |
SPATA20
|
spermatogenesis associated 20 |
| chr4_+_170541660 | 1.44 |
ENST00000513761.1
ENST00000347613.4 |
CLCN3
|
chloride channel, voltage-sensitive 3 |
| chrX_+_1387693 | 1.37 |
ENST00000381529.3
ENST00000432318.2 ENST00000361536.3 ENST00000501036.2 ENST00000381524.3 ENST00000412290.1 |
CSF2RA
|
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) |
| chr2_-_105946491 | 1.35 |
ENST00000393359.2
|
TGFBRAP1
|
transforming growth factor, beta receptor associated protein 1 |
| chr14_+_105941118 | 1.24 |
ENST00000550577.1
ENST00000538259.2 |
CRIP2
|
cysteine-rich protein 2 |
| chrX_+_102631248 | 1.21 |
ENST00000361298.4
ENST00000372645.3 ENST00000372635.1 |
NGFRAP1
|
nerve growth factor receptor (TNFRSF16) associated protein 1 |
| chr17_+_75181292 | 0.96 |
ENST00000431431.2
|
SEC14L1
|
SEC14-like 1 (S. cerevisiae) |
| chr10_+_1034338 | 0.73 |
ENST00000360803.4
ENST00000538293.1 |
GTPBP4
|
GTP binding protein 4 |
| chr7_-_74489609 | 0.61 |
ENST00000329959.4
ENST00000503250.2 ENST00000543840.1 |
WBSCR16
|
Williams-Beuren syndrome chromosome region 16 |
| chr16_-_53537105 | 0.29 |
ENST00000568596.1
ENST00000570004.1 ENST00000564497.1 ENST00000300245.4 ENST00000394657.7 |
AKTIP
|
AKT interacting protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF8
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.6 | 23.2 | GO:0009439 | cyanate metabolic process(GO:0009439) cyanate catabolic process(GO:0009440) |
| 3.0 | 9.1 | GO:0035927 | RNA import into mitochondrion(GO:0035927) |
| 2.8 | 8.4 | GO:0033341 | regulation of collagen binding(GO:0033341) |
| 2.2 | 10.8 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 1.9 | 7.5 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 1.8 | 7.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 1.4 | 14.4 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 1.0 | 10.0 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.8 | 7.6 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.7 | 6.7 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.7 | 13.3 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.7 | 5.8 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.7 | 27.1 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.5 | 4.1 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.5 | 3.9 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.3 | 2.3 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.2 | 1.8 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.2 | 1.2 | GO:0030421 | defecation(GO:0030421) |
| 0.2 | 6.3 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.2 | 14.6 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.2 | 4.4 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.1 | 1.3 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 2.0 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 4.7 | GO:0071549 | cellular response to dexamethasone stimulus(GO:0071549) |
| 0.1 | 2.6 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.1 | 1.0 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.1 | 10.5 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.1 | 2.6 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 2.9 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 | 3.3 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 2.1 | GO:0033146 | regulation of intracellular estrogen receptor signaling pathway(GO:0033146) |
| 0.0 | 6.7 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 2.0 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 2.2 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 13.3 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 1.8 | 10.8 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 1.3 | 14.4 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 1.3 | 9.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 1.2 | 4.8 | GO:0031417 | NatC complex(GO:0031417) |
| 1.0 | 11.2 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.8 | 10.0 | GO:0042555 | MCM complex(GO:0042555) |
| 0.7 | 4.4 | GO:0000835 | ER ubiquitin ligase complex(GO:0000835) |
| 0.4 | 7.5 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.4 | 2.9 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.3 | 3.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.3 | 6.7 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.3 | 1.3 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.2 | 5.1 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.2 | 2.2 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 7.6 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 6.3 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 2.3 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 1.8 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.3 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 3.9 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 2.3 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 8.4 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 2.0 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 25.9 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 12.3 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 9.3 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 4.5 | GO:0005759 | mitochondrial matrix(GO:0005759) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.7 | 18.7 | GO:0016784 | 3-mercaptopyruvate sulfurtransferase activity(GO:0016784) |
| 2.4 | 14.4 | GO:0046979 | TAP2 binding(GO:0046979) |
| 2.2 | 4.5 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 1.4 | 27.1 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) |
| 0.9 | 9.1 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.8 | 13.3 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.7 | 14.6 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.6 | 4.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.5 | 4.7 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.4 | 2.0 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.4 | 4.4 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.3 | 7.5 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.2 | 7.6 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.2 | 10.0 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.2 | 1.0 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.1 | 2.2 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.1 | 1.8 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.1 | 6.7 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 2.6 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 2.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 2.1 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.1 | 7.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 5.8 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 10.4 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 1.3 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 2.3 | GO:0031490 | chromatin DNA binding(GO:0031490) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 7.6 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.2 | 10.0 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 6.7 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 2.9 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.4 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 1.2 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 1.2 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 14.4 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.7 | 27.1 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.6 | 10.0 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.3 | 7.6 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.2 | 14.9 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.2 | 7.5 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.1 | 5.4 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.1 | 2.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 4.5 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 6.7 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 1.4 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 2.3 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 1.2 | REACTOME CELL DEATH SIGNALLING VIA NRAGE NRIF AND NADE | Genes involved in Cell death signalling via NRAGE, NRIF and NADE |
| 0.0 | 2.6 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |