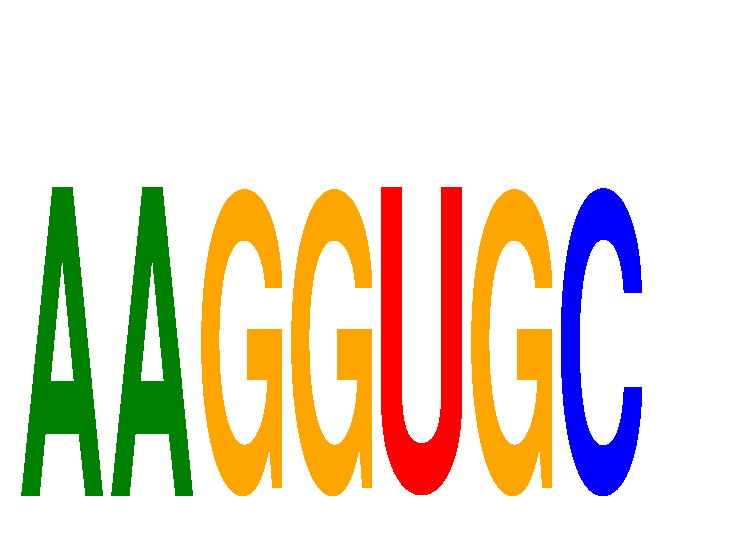Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for AAGGUGC
Z-value: 0.15
miRNA associated with seed AAGGUGC
| Name | miRBASE accession |
|---|---|
|
hsa-miR-18a-5p
|
MIMAT0000072 |
|
hsa-miR-18b-5p
|
MIMAT0001412 |
|
hsa-miR-4735-3p
|
MIMAT0019861 |
Activity profile of AAGGUGC motif
Sorted Z-values of AAGGUGC motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_235860616 | 8.48 |
ENST00000392011.2
|
SH3BP4
|
SH3-domain binding protein 4 |
| chr3_-_123603137 | 8.40 |
ENST00000360304.3
ENST00000359169.1 ENST00000346322.5 ENST00000360772.3 |
MYLK
|
myosin light chain kinase |
| chr3_+_107241783 | 6.11 |
ENST00000415149.2
ENST00000402543.1 ENST00000325805.8 ENST00000427402.1 |
BBX
|
bobby sox homolog (Drosophila) |
| chr6_-_132272504 | 4.69 |
ENST00000367976.3
|
CTGF
|
connective tissue growth factor |
| chr3_-_114790179 | 4.58 |
ENST00000462705.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr2_+_36582857 | 4.54 |
ENST00000280527.2
|
CRIM1
|
cysteine rich transmembrane BMP regulator 1 (chordin-like) |
| chr7_-_121036337 | 4.41 |
ENST00000426156.1
ENST00000359943.3 ENST00000412653.1 |
FAM3C
|
family with sequence similarity 3, member C |
| chr21_-_18985158 | 4.07 |
ENST00000339775.6
|
BTG3
|
BTG family, member 3 |
| chr15_+_39873268 | 3.74 |
ENST00000397591.2
ENST00000260356.5 |
THBS1
|
thrombospondin 1 |
| chr3_+_171758344 | 3.60 |
ENST00000336824.4
ENST00000423424.1 |
FNDC3B
|
fibronectin type III domain containing 3B |
| chr22_+_38093005 | 3.32 |
ENST00000406386.3
|
TRIOBP
|
TRIO and F-actin binding protein |
| chr3_+_187930719 | 2.83 |
ENST00000312675.4
|
LPP
|
LIM domain containing preferred translocation partner in lipoma |
| chr5_-_142783175 | 2.72 |
ENST00000231509.3
ENST00000394464.2 |
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr19_+_3572925 | 2.71 |
ENST00000333651.6
ENST00000417382.1 ENST00000453933.1 ENST00000262949.7 |
HMG20B
|
high mobility group 20B |
| chr2_-_26101374 | 2.63 |
ENST00000435504.4
|
ASXL2
|
additional sex combs like 2 (Drosophila) |
| chr6_+_39760783 | 2.47 |
ENST00000398904.2
ENST00000538976.1 |
DAAM2
|
dishevelled associated activator of morphogenesis 2 |
| chr15_-_56209306 | 2.46 |
ENST00000506154.1
ENST00000338963.2 ENST00000508342.1 |
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4, E3 ubiquitin protein ligase |
| chr4_-_157892498 | 2.43 |
ENST00000502773.1
|
PDGFC
|
platelet derived growth factor C |
| chr3_+_105085734 | 2.40 |
ENST00000306107.5
|
ALCAM
|
activated leukocyte cell adhesion molecule |
| chr7_+_66386204 | 2.26 |
ENST00000341567.4
ENST00000607045.1 |
TMEM248
|
transmembrane protein 248 |
| chr9_-_74383799 | 2.14 |
ENST00000377044.4
|
TMEM2
|
transmembrane protein 2 |
| chr2_+_12857015 | 2.03 |
ENST00000155926.4
|
TRIB2
|
tribbles pseudokinase 2 |
| chr3_-_185542817 | 2.02 |
ENST00000382199.2
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr6_-_16761678 | 1.98 |
ENST00000244769.4
ENST00000436367.1 |
ATXN1
|
ataxin 1 |
| chr17_-_40306934 | 1.89 |
ENST00000592574.1
ENST00000550406.1 ENST00000547517.1 ENST00000393860.3 ENST00000346213.4 |
CTD-2132N18.3
RAB5C
|
Uncharacterized protein RAB5C, member RAS oncogene family |
| chr2_+_85198216 | 1.84 |
ENST00000456682.1
ENST00000409785.4 |
KCMF1
|
potassium channel modulatory factor 1 |
| chr4_-_186877502 | 1.81 |
ENST00000431902.1
ENST00000284776.7 ENST00000415274.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr1_+_36621529 | 1.67 |
ENST00000316156.4
|
MAP7D1
|
MAP7 domain containing 1 |
| chrX_+_13707235 | 1.63 |
ENST00000464506.1
|
RAB9A
|
RAB9A, member RAS oncogene family |
| chr17_+_46985731 | 1.61 |
ENST00000360943.5
|
UBE2Z
|
ubiquitin-conjugating enzyme E2Z |
| chr1_-_120612240 | 1.48 |
ENST00000256646.2
|
NOTCH2
|
notch 2 |
| chr18_-_45456930 | 1.37 |
ENST00000262160.6
ENST00000587269.1 |
SMAD2
|
SMAD family member 2 |
| chr12_+_12764773 | 1.34 |
ENST00000228865.2
|
CREBL2
|
cAMP responsive element binding protein-like 2 |
| chr1_-_161993616 | 1.28 |
ENST00000294794.3
|
OLFML2B
|
olfactomedin-like 2B |
| chr3_+_152017181 | 1.26 |
ENST00000498502.1
ENST00000324196.5 ENST00000545754.1 ENST00000357472.3 |
MBNL1
|
muscleblind-like splicing regulator 1 |
| chr6_-_11232891 | 1.23 |
ENST00000379433.5
ENST00000379446.5 |
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9 |
| chr9_+_125703282 | 1.19 |
ENST00000373647.4
ENST00000402311.1 |
RABGAP1
|
RAB GTPase activating protein 1 |
| chr2_+_46926048 | 1.18 |
ENST00000306503.5
|
SOCS5
|
suppressor of cytokine signaling 5 |
| chr1_-_51425902 | 1.17 |
ENST00000396153.2
|
FAF1
|
Fas (TNFRSF6) associated factor 1 |
| chr4_+_183164574 | 1.07 |
ENST00000511685.1
|
TENM3
|
teneurin transmembrane protein 3 |
| chr10_-_735553 | 1.07 |
ENST00000280886.6
ENST00000423550.1 |
DIP2C
|
DIP2 disco-interacting protein 2 homolog C (Drosophila) |
| chr3_-_113465065 | 1.03 |
ENST00000497255.1
ENST00000478020.1 ENST00000240922.3 ENST00000493900.1 |
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr1_+_173684047 | 0.92 |
ENST00000546011.1
ENST00000209884.4 |
KLHL20
|
kelch-like family member 20 |
| chr6_+_134274322 | 0.92 |
ENST00000367871.1
ENST00000237264.4 |
TBPL1
|
TBP-like 1 |
| chr5_-_73937244 | 0.90 |
ENST00000302351.4
ENST00000510316.1 ENST00000508331.1 |
ENC1
|
ectodermal-neural cortex 1 (with BTB domain) |
| chr10_-_119806085 | 0.89 |
ENST00000355624.3
|
RAB11FIP2
|
RAB11 family interacting protein 2 (class I) |
| chr6_-_86352642 | 0.88 |
ENST00000355238.6
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr1_-_20812690 | 0.84 |
ENST00000375078.3
|
CAMK2N1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1 |
| chr12_-_44200052 | 0.83 |
ENST00000548315.1
ENST00000552521.1 ENST00000546662.1 ENST00000548403.1 ENST00000546506.1 |
TWF1
|
twinfilin actin-binding protein 1 |
| chr1_+_87794150 | 0.78 |
ENST00000370544.5
|
LMO4
|
LIM domain only 4 |
| chr20_+_56884752 | 0.70 |
ENST00000244040.3
|
RAB22A
|
RAB22A, member RAS oncogene family |
| chr1_-_95007193 | 0.64 |
ENST00000370207.4
ENST00000334047.7 |
F3
|
coagulation factor III (thromboplastin, tissue factor) |
| chr3_+_37903432 | 0.63 |
ENST00000443503.2
|
CTDSPL
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
| chr10_-_81205373 | 0.62 |
ENST00000372336.3
|
ZCCHC24
|
zinc finger, CCHC domain containing 24 |
| chr12_+_56360550 | 0.60 |
ENST00000266970.4
|
CDK2
|
cyclin-dependent kinase 2 |
| chr1_+_84543734 | 0.59 |
ENST00000370689.2
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr9_-_127952032 | 0.57 |
ENST00000456642.1
ENST00000373546.3 ENST00000373547.4 |
PPP6C
|
protein phosphatase 6, catalytic subunit |
| chr14_+_52456327 | 0.55 |
ENST00000556760.1
|
C14orf166
|
chromosome 14 open reading frame 166 |
| chr16_-_85722530 | 0.51 |
ENST00000253462.3
|
GINS2
|
GINS complex subunit 2 (Psf2 homolog) |
| chr20_+_30865429 | 0.47 |
ENST00000375712.3
|
KIF3B
|
kinesin family member 3B |
| chr12_+_109535373 | 0.47 |
ENST00000242576.2
|
UNG
|
uracil-DNA glycosylase |
| chr9_+_91003271 | 0.43 |
ENST00000375859.3
ENST00000541629.1 |
SPIN1
|
spindlin 1 |
| chr4_+_154125565 | 0.42 |
ENST00000338700.5
|
TRIM2
|
tripartite motif containing 2 |
| chr6_-_111136513 | 0.41 |
ENST00000368911.3
|
CDK19
|
cyclin-dependent kinase 19 |
| chr9_+_96338860 | 0.40 |
ENST00000375376.4
|
PHF2
|
PHD finger protein 2 |
| chr10_-_71930222 | 0.38 |
ENST00000458634.2
ENST00000373239.2 ENST00000373242.2 ENST00000373241.4 |
SAR1A
|
SAR1 homolog A (S. cerevisiae) |
| chr17_+_48423453 | 0.38 |
ENST00000017003.2
ENST00000509778.1 ENST00000507602.1 |
XYLT2
|
xylosyltransferase II |
| chr10_+_72575643 | 0.37 |
ENST00000373202.3
|
SGPL1
|
sphingosine-1-phosphate lyase 1 |
| chr17_-_62340581 | 0.32 |
ENST00000258991.3
ENST00000583738.1 ENST00000584379.1 |
TEX2
|
testis expressed 2 |
| chr1_-_109506036 | 0.32 |
ENST00000369976.1
ENST00000356970.2 ENST00000369971.2 ENST00000415331.1 ENST00000357393.4 |
CLCC1
AKNAD1
|
chloride channel CLIC-like 1 AKNA domain containing 1 |
| chr11_+_94822968 | 0.29 |
ENST00000278505.4
|
ENDOD1
|
endonuclease domain containing 1 |
| chr1_-_38325256 | 0.26 |
ENST00000373036.4
|
MTF1
|
metal-regulatory transcription factor 1 |
| chr11_-_34379546 | 0.22 |
ENST00000435224.2
|
ABTB2
|
ankyrin repeat and BTB (POZ) domain containing 2 |
| chr14_+_55518349 | 0.18 |
ENST00000395468.4
|
MAPK1IP1L
|
mitogen-activated protein kinase 1 interacting protein 1-like |
| chr4_+_6784401 | 0.18 |
ENST00000425103.1
ENST00000307659.5 |
KIAA0232
|
KIAA0232 |
| chr17_-_1395954 | 0.18 |
ENST00000359786.5
|
MYO1C
|
myosin IC |
| chr3_-_33481835 | 0.17 |
ENST00000283629.3
|
UBP1
|
upstream binding protein 1 (LBP-1a) |
| chr16_-_17564738 | 0.16 |
ENST00000261381.6
|
XYLT1
|
xylosyltransferase I |
| chr4_-_185395672 | 0.16 |
ENST00000393593.3
|
IRF2
|
interferon regulatory factor 2 |
| chr6_+_16238786 | 0.15 |
ENST00000259727.4
|
GMPR
|
guanosine monophosphate reductase |
| chr1_+_22379120 | 0.13 |
ENST00000400259.1
ENST00000344548.3 |
CDC42
|
cell division cycle 42 |
| chr18_+_20513782 | 0.11 |
ENST00000399722.2
ENST00000399725.2 ENST00000399721.2 ENST00000583594.1 |
RBBP8
|
retinoblastoma binding protein 8 |
| chr1_+_100435315 | 0.11 |
ENST00000370155.3
ENST00000465289.1 |
SLC35A3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3 |
| chr16_-_74640986 | 0.10 |
ENST00000422840.2
ENST00000565260.1 ENST00000447066.2 ENST00000205061.5 |
GLG1
|
golgi glycoprotein 1 |
| chr12_+_49212514 | 0.09 |
ENST00000301050.2
ENST00000548279.1 ENST00000547230.1 |
CACNB3
|
calcium channel, voltage-dependent, beta 3 subunit |
| chr7_+_90225796 | 0.09 |
ENST00000380050.3
|
CDK14
|
cyclin-dependent kinase 14 |
| chr2_+_28615669 | 0.08 |
ENST00000379619.1
ENST00000264716.4 |
FOSL2
|
FOS-like antigen 2 |
| chr4_+_55524085 | 0.08 |
ENST00000412167.2
ENST00000288135.5 |
KIT
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog |
| chr16_+_30960375 | 0.06 |
ENST00000318663.4
ENST00000566237.1 ENST00000562699.1 |
ORAI3
|
ORAI calcium release-activated calcium modulator 3 |
| chr8_-_18871159 | 0.06 |
ENST00000327040.8
ENST00000440756.2 |
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chr5_+_102594403 | 0.04 |
ENST00000319933.2
|
C5orf30
|
chromosome 5 open reading frame 30 |
| chrX_+_108780062 | 0.02 |
ENST00000372106.1
|
NXT2
|
nuclear transport factor 2-like export factor 2 |
| chr7_-_111846435 | 0.01 |
ENST00000437633.1
ENST00000428084.1 |
DOCK4
|
dedicator of cytokinesis 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of AAGGUGC
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.7 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 1.2 | 3.7 | GO:0002605 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) negative regulation of dendritic cell antigen processing and presentation(GO:0002605) |
| 1.0 | 8.4 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.8 | 4.2 | GO:0030047 | actin modification(GO:0030047) |
| 0.6 | 2.5 | GO:0044111 | development involved in symbiotic interaction(GO:0044111) |
| 0.5 | 1.4 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.4 | 2.0 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.3 | 2.7 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.3 | 1.6 | GO:0052405 | negative regulation by host of symbiont molecular function(GO:0052405) |
| 0.2 | 2.6 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.2 | 0.9 | GO:0009212 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) |
| 0.2 | 8.5 | GO:0043090 | amino acid import(GO:0043090) |
| 0.2 | 0.6 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.2 | 1.2 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.2 | 1.0 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.1 | 1.5 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 1.9 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.1 | 2.7 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.1 | 0.4 | GO:0061188 | negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.1 | 0.9 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.1 | 0.6 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.1 | 0.6 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.1 | 0.9 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 2.4 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.1 | 4.6 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 0.8 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.1 | 4.4 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.1 | 0.8 | GO:0042659 | regulation of cell fate specification(GO:0042659) |
| 0.1 | 2.4 | GO:1902284 | axon extension involved in axon guidance(GO:0048846) neuron projection extension involved in neuron projection guidance(GO:1902284) |
| 0.1 | 2.1 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.1 | 1.2 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.1 | 0.5 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.1 | 1.8 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.5 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.0 | 0.6 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.9 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.4 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.5 | GO:0045008 | depyrimidination(GO:0045008) |
| 0.0 | 0.4 | GO:0030202 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.0 | 0.2 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 6.3 | GO:0060348 | bone development(GO:0060348) |
| 0.0 | 1.3 | GO:0046326 | positive regulation of glucose import(GO:0046326) |
| 0.0 | 4.7 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.0 | 0.4 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 2.0 | GO:0042035 | regulation of cytokine biosynthetic process(GO:0042035) |
| 0.0 | 0.9 | GO:0003091 | renal water homeostasis(GO:0003091) |
| 0.0 | 1.4 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.0 | 1.2 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.1 | GO:0045002 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) double-strand break repair via single-strand annealing(GO:0045002) |
| 0.0 | 0.1 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 1.1 | GO:0045445 | myoblast differentiation(GO:0045445) |
| 0.0 | 0.2 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.0 | 0.4 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.1 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
| 0.0 | 0.2 | GO:0097237 | cellular response to toxic substance(GO:0097237) |
| 0.0 | 4.0 | GO:0045930 | negative regulation of mitotic cell cycle(GO:0045930) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.0 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.3 | 3.7 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.2 | 4.4 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.2 | 0.6 | GO:0097134 | Y chromosome(GO:0000806) cyclin A2-CDK2 complex(GO:0097124) cyclin E1-CDK2 complex(GO:0097134) |
| 0.2 | 1.4 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.2 | 0.5 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.1 | 1.0 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 0.5 | GO:0000811 | GINS complex(GO:0000811) |
| 0.1 | 0.9 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.1 | 8.4 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 1.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 0.6 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 4.7 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 8.5 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 2.4 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.1 | 2.5 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.2 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.0 | 0.1 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.0 | 0.6 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.6 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 2.3 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 1.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.2 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.0 | 1.9 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 3.5 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 5.3 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 0.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.4 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.6 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 1.2 | GO:0005875 | microtubule associated complex(GO:0005875) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 8.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 1.2 | 3.7 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.7 | 4.2 | GO:0038049 | transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) |
| 0.4 | 1.3 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.4 | 2.0 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.4 | 2.5 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.3 | 8.5 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.3 | 0.9 | GO:0019961 | interferon binding(GO:0019961) |
| 0.3 | 3.3 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.2 | 9.2 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.2 | 2.1 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.2 | 2.6 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.2 | 0.5 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.1 | 0.8 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.1 | 2.0 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.1 | 2.4 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 1.4 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.1 | 1.0 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 1.8 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 2.0 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 0.5 | GO:0004844 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.1 | 4.2 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.0 | 2.3 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.6 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 1.6 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.5 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.9 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 1.0 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 2.9 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 4.6 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.2 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.0 | 0.4 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.6 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 0.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 1.1 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.6 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.4 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 4.4 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.2 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 1.8 | GO:0017137 | Rab GTPase binding(GO:0017137) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 8.5 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 4.7 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 3.7 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 2.7 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 1.4 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 2.5 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 2.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.9 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.6 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 1.6 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 4.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.6 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.8 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 1.2 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.4 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.6 | PID BMP PATHWAY | BMP receptor signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 8.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 2.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 4.6 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 1.8 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 1.4 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.5 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 3.7 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 4.2 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 2.5 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 1.9 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.9 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.6 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 2.1 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |