Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for AIRE
Z-value: 0.78
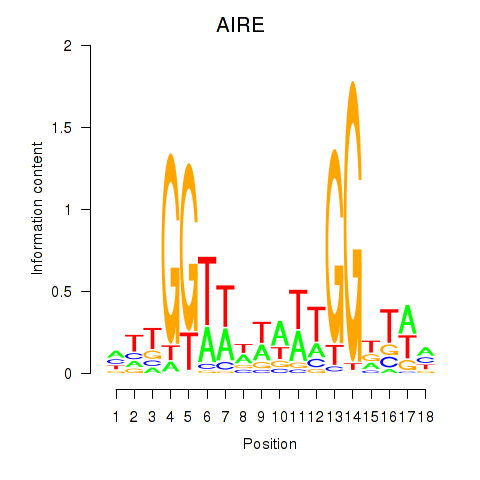
Transcription factors associated with AIRE
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
AIRE
|
ENSG00000160224.12 | autoimmune regulator |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| AIRE | hg19_v2_chr21_+_45705752_45705767 | 0.38 | 5.4e-09 | Click! |
Activity profile of AIRE motif
Sorted Z-values of AIRE motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_88450612 | 26.51 |
ENST00000418378.1
ENST00000282470.6 |
SPARCL1
|
SPARC-like 1 (hevin) |
| chr6_-_31514333 | 16.02 |
ENST00000376151.4
|
ATP6V1G2
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2 |
| chr4_-_57524061 | 15.33 |
ENST00000508121.1
|
HOPX
|
HOP homeobox |
| chr6_-_31514516 | 13.45 |
ENST00000303892.5
ENST00000483251.1 |
ATP6V1G2
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2 |
| chr19_+_18208603 | 11.73 |
ENST00000262811.6
|
MAST3
|
microtubule associated serine/threonine kinase 3 |
| chr19_-_36643329 | 11.36 |
ENST00000589154.1
|
COX7A1
|
cytochrome c oxidase subunit VIIa polypeptide 1 (muscle) |
| chrM_+_12331 | 10.91 |
ENST00000361567.2
|
MT-ND5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr10_-_69597810 | 10.66 |
ENST00000483798.2
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr20_+_44036900 | 9.85 |
ENST00000443296.1
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr20_+_44036620 | 9.71 |
ENST00000372710.3
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr3_-_49466686 | 9.34 |
ENST00000273598.3
ENST00000436744.2 |
NICN1
|
nicolin 1 |
| chr12_-_6580094 | 8.95 |
ENST00000361716.3
|
VAMP1
|
vesicle-associated membrane protein 1 (synaptobrevin 1) |
| chr17_+_41857793 | 7.99 |
ENST00000449302.3
|
C17orf105
|
chromosome 17 open reading frame 105 |
| chr12_-_6579808 | 7.45 |
ENST00000535180.1
ENST00000400911.3 |
VAMP1
|
vesicle-associated membrane protein 1 (synaptobrevin 1) |
| chr19_-_21950362 | 7.41 |
ENST00000358296.6
|
ZNF100
|
zinc finger protein 100 |
| chr7_+_150498783 | 7.07 |
ENST00000475536.1
ENST00000468689.1 |
TMEM176A
|
transmembrane protein 176A |
| chr11_-_85376121 | 7.05 |
ENST00000527447.1
|
CREBZF
|
CREB/ATF bZIP transcription factor |
| chr19_+_45417921 | 6.96 |
ENST00000252491.4
ENST00000592885.1 ENST00000589781.1 |
APOC1
|
apolipoprotein C-I |
| chr9_-_70429731 | 6.81 |
ENST00000377413.1
|
FOXD4L4
|
forkhead box D4-like 4 |
| chr7_+_20686946 | 6.70 |
ENST00000443026.2
ENST00000406935.1 |
ABCB5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr9_+_42717234 | 6.53 |
ENST00000377590.1
|
FOXD4L2
|
forkhead box D4-like 2 |
| chr7_-_36764004 | 6.26 |
ENST00000431169.1
|
AOAH
|
acyloxyacyl hydrolase (neutrophil) |
| chr7_-_137028534 | 6.20 |
ENST00000348225.2
|
PTN
|
pleiotrophin |
| chr9_-_113761720 | 6.14 |
ENST00000541779.1
ENST00000374430.2 |
LPAR1
|
lysophosphatidic acid receptor 1 |
| chr19_+_45418067 | 6.09 |
ENST00000589078.1
ENST00000586638.1 |
APOC1
|
apolipoprotein C-I |
| chr19_+_45417812 | 5.89 |
ENST00000592535.1
|
APOC1
|
apolipoprotein C-I |
| chr7_-_137028498 | 5.66 |
ENST00000393083.2
|
PTN
|
pleiotrophin |
| chr7_-_36764142 | 5.58 |
ENST00000258749.5
ENST00000535891.1 |
AOAH
|
acyloxyacyl hydrolase (neutrophil) |
| chr2_+_166150541 | 5.52 |
ENST00000283256.6
|
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit |
| chr4_+_71587669 | 5.47 |
ENST00000381006.3
ENST00000226328.4 |
RUFY3
|
RUN and FYVE domain containing 3 |
| chr9_-_70178815 | 5.05 |
ENST00000377420.1
|
FOXD4L5
|
forkhead box D4-like 5 |
| chr2_-_127977654 | 5.00 |
ENST00000409327.1
|
CYP27C1
|
cytochrome P450, family 27, subfamily C, polypeptide 1 |
| chr9_+_100615536 | 4.90 |
ENST00000375123.3
|
FOXE1
|
forkhead box E1 (thyroid transcription factor 2) |
| chr14_-_24911971 | 4.84 |
ENST00000555365.1
ENST00000399395.3 |
SDR39U1
|
short chain dehydrogenase/reductase family 39U, member 1 |
| chr12_-_113772835 | 4.66 |
ENST00000552014.1
ENST00000548186.1 ENST00000202831.3 ENST00000549181.1 |
SLC8B1
|
solute carrier family 8 (sodium/lithium/calcium exchanger), member B1 |
| chr4_-_17513851 | 4.66 |
ENST00000281243.5
|
QDPR
|
quinoid dihydropteridine reductase |
| chr5_-_107703556 | 4.63 |
ENST00000496714.1
|
FBXL17
|
F-box and leucine-rich repeat protein 17 |
| chr12_+_7282795 | 4.40 |
ENST00000266546.6
|
CLSTN3
|
calsyntenin 3 |
| chr3_+_46412345 | 4.23 |
ENST00000292303.4
|
CCR5
|
chemokine (C-C motif) receptor 5 (gene/pseudogene) |
| chr22_+_36649170 | 4.05 |
ENST00000438034.1
ENST00000427990.1 ENST00000347595.7 ENST00000397279.4 ENST00000433768.1 ENST00000440669.2 |
APOL1
|
apolipoprotein L, 1 |
| chr21_+_39644214 | 4.04 |
ENST00000438657.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr14_-_24912047 | 3.68 |
ENST00000553930.1
|
SDR39U1
|
short chain dehydrogenase/reductase family 39U, member 1 |
| chrM_+_4431 | 3.63 |
ENST00000361453.3
|
MT-ND2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr15_-_22448819 | 3.59 |
ENST00000604066.1
|
IGHV1OR15-1
|
immunoglobulin heavy variable 1/OR15-1 (non-functional) |
| chr21_+_39644305 | 3.55 |
ENST00000398930.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr22_+_25202232 | 3.55 |
ENST00000400358.4
ENST00000400359.4 |
SGSM1
|
small G protein signaling modulator 1 |
| chr4_-_186696425 | 3.54 |
ENST00000430503.1
ENST00000319454.6 ENST00000450341.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr3_+_193965426 | 3.46 |
ENST00000455821.1
|
RP11-513G11.3
|
RP11-513G11.3 |
| chr17_-_38721711 | 3.44 |
ENST00000578085.1
ENST00000246657.2 |
CCR7
|
chemokine (C-C motif) receptor 7 |
| chr12_+_20848486 | 3.23 |
ENST00000545102.1
|
SLCO1C1
|
solute carrier organic anion transporter family, member 1C1 |
| chr16_+_56672571 | 3.12 |
ENST00000290705.8
|
MT1A
|
metallothionein 1A |
| chr2_-_225811747 | 3.05 |
ENST00000409592.3
|
DOCK10
|
dedicator of cytokinesis 10 |
| chr3_+_189507460 | 3.03 |
ENST00000434928.1
|
TP63
|
tumor protein p63 |
| chr19_-_36304201 | 2.83 |
ENST00000301175.3
|
PRODH2
|
proline dehydrogenase (oxidase) 2 |
| chr16_+_3254247 | 2.80 |
ENST00000304646.2
|
OR1F1
|
olfactory receptor, family 1, subfamily F, member 1 |
| chr4_-_120243545 | 2.80 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr21_+_39644172 | 2.79 |
ENST00000398932.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr19_-_10491234 | 2.77 |
ENST00000524462.1
ENST00000531836.1 ENST00000525621.1 |
TYK2
|
tyrosine kinase 2 |
| chr2_-_166060552 | 2.56 |
ENST00000283254.7
ENST00000453007.1 |
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit |
| chr3_-_133648656 | 2.55 |
ENST00000408895.2
|
C3orf36
|
chromosome 3 open reading frame 36 |
| chr16_+_14802801 | 2.53 |
ENST00000526520.1
ENST00000531598.2 |
NPIPA3
|
nuclear pore complex interacting protein family, member A3 |
| chr21_-_44495964 | 2.41 |
ENST00000398168.1
ENST00000398165.3 |
CBS
|
cystathionine-beta-synthase |
| chr7_-_15601595 | 2.40 |
ENST00000342526.3
|
AGMO
|
alkylglycerol monooxygenase |
| chr22_+_40322623 | 2.36 |
ENST00000399090.2
|
GRAP2
|
GRB2-related adaptor protein 2 |
| chr15_-_99789736 | 2.33 |
ENST00000560235.1
ENST00000394132.2 ENST00000560860.1 ENST00000558078.1 ENST00000394136.1 ENST00000262074.4 ENST00000558613.1 ENST00000394130.1 ENST00000560772.1 |
TTC23
|
tetratricopeptide repeat domain 23 |
| chr12_+_12223867 | 2.28 |
ENST00000308721.5
|
BCL2L14
|
BCL2-like 14 (apoptosis facilitator) |
| chr14_-_106967788 | 2.28 |
ENST00000390622.2
|
IGHV1-46
|
immunoglobulin heavy variable 1-46 |
| chr1_+_206223941 | 2.24 |
ENST00000367126.4
|
AVPR1B
|
arginine vasopressin receptor 1B |
| chr2_-_166060571 | 2.18 |
ENST00000360093.3
|
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit |
| chr22_+_40322595 | 2.13 |
ENST00000420971.1
ENST00000544756.1 |
GRAP2
|
GRB2-related adaptor protein 2 |
| chr9_-_104198042 | 2.09 |
ENST00000374855.4
|
ALDOB
|
aldolase B, fructose-bisphosphate |
| chr13_-_26795840 | 2.08 |
ENST00000381570.3
ENST00000399762.2 ENST00000346166.3 |
RNF6
|
ring finger protein (C3H2C3 type) 6 |
| chr15_+_27112948 | 2.06 |
ENST00000555060.1
|
GABRA5
|
gamma-aminobutyric acid (GABA) A receptor, alpha 5 |
| chr11_+_6947647 | 2.04 |
ENST00000278319.5
|
ZNF215
|
zinc finger protein 215 |
| chr12_-_6579833 | 1.97 |
ENST00000396308.3
|
VAMP1
|
vesicle-associated membrane protein 1 (synaptobrevin 1) |
| chr11_+_64794875 | 1.95 |
ENST00000377244.3
ENST00000534637.1 ENST00000524831.1 |
SNX15
|
sorting nexin 15 |
| chr14_-_106668095 | 1.93 |
ENST00000390606.2
|
IGHV3-20
|
immunoglobulin heavy variable 3-20 |
| chrX_-_153191674 | 1.93 |
ENST00000350060.5
ENST00000370016.1 |
ARHGAP4
|
Rho GTPase activating protein 4 |
| chr12_-_15038779 | 1.93 |
ENST00000228938.5
ENST00000539261.1 |
MGP
|
matrix Gla protein |
| chr1_+_207669573 | 1.91 |
ENST00000400960.2
ENST00000534202.1 |
CR1
|
complement component (3b/4b) receptor 1 (Knops blood group) |
| chr11_-_8954491 | 1.83 |
ENST00000526227.1
ENST00000525780.1 ENST00000326053.5 |
C11orf16
|
chromosome 11 open reading frame 16 |
| chr19_-_59084922 | 1.81 |
ENST00000215057.2
ENST00000599369.1 |
MZF1
|
myeloid zinc finger 1 |
| chr16_+_72088376 | 1.78 |
ENST00000570083.1
ENST00000355906.5 ENST00000398131.2 ENST00000569639.1 ENST00000564499.1 ENST00000357763.4 ENST00000562526.1 ENST00000565574.1 ENST00000568417.2 ENST00000356967.5 |
HP
HPR
|
haptoglobin haptoglobin-related protein |
| chr20_-_43150601 | 1.76 |
ENST00000541235.1
ENST00000255175.1 ENST00000342374.4 |
SERINC3
|
serine incorporator 3 |
| chr20_+_44637526 | 1.75 |
ENST00000372330.3
|
MMP9
|
matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) |
| chr21_+_39644395 | 1.75 |
ENST00000398934.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr4_+_76649797 | 1.72 |
ENST00000538159.1
ENST00000514213.2 |
USO1
|
USO1 vesicle transport factor |
| chr8_+_105352050 | 1.62 |
ENST00000297581.2
|
DCSTAMP
|
dendrocyte expressed seven transmembrane protein |
| chr1_+_207669613 | 1.60 |
ENST00000367049.4
ENST00000529814.1 |
CR1
|
complement component (3b/4b) receptor 1 (Knops blood group) |
| chr1_-_161208013 | 1.59 |
ENST00000515452.1
ENST00000367983.4 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr1_-_167883327 | 1.54 |
ENST00000476818.2
ENST00000367851.4 ENST00000367848.1 |
ADCY10
|
adenylate cyclase 10 (soluble) |
| chrX_-_100307076 | 1.52 |
ENST00000338687.7
ENST00000545398.1 ENST00000372931.5 |
TRMT2B
|
tRNA methyltransferase 2 homolog B (S. cerevisiae) |
| chr18_+_39766626 | 1.51 |
ENST00000593234.1
ENST00000585627.1 ENST00000591199.1 ENST00000586990.1 ENST00000593051.1 ENST00000593316.1 ENST00000591381.1 ENST00000585639.1 ENST00000589068.1 |
LINC00907
|
long intergenic non-protein coding RNA 907 |
| chr2_+_204732487 | 1.46 |
ENST00000302823.3
|
CTLA4
|
cytotoxic T-lymphocyte-associated protein 4 |
| chr6_+_167525277 | 1.45 |
ENST00000400926.2
|
CCR6
|
chemokine (C-C motif) receptor 6 |
| chrX_-_100307043 | 1.43 |
ENST00000372939.1
ENST00000372935.1 ENST00000372936.3 |
TRMT2B
|
tRNA methyltransferase 2 homolog B (S. cerevisiae) |
| chr2_+_1417228 | 1.38 |
ENST00000382269.3
ENST00000337415.3 ENST00000345913.4 ENST00000346956.3 ENST00000349624.3 ENST00000539820.1 ENST00000329066.4 ENST00000382201.3 |
TPO
|
thyroid peroxidase |
| chr1_-_35497506 | 1.35 |
ENST00000317538.5
ENST00000373340.2 ENST00000357182.4 |
ZMYM6
|
zinc finger, MYM-type 6 |
| chrX_-_101694853 | 1.32 |
ENST00000372749.1
|
NXF2B
|
nuclear RNA export factor 2B |
| chr16_-_29499154 | 1.29 |
ENST00000354563.5
|
RP11-231C14.4
|
Uncharacterized protein |
| chr4_+_71588372 | 1.27 |
ENST00000536664.1
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr12_-_11463353 | 1.23 |
ENST00000279575.1
ENST00000535904.1 ENST00000445719.2 |
PRB4
|
proline-rich protein BstNI subfamily 4 |
| chr8_-_16043780 | 1.21 |
ENST00000445506.2
|
MSR1
|
macrophage scavenger receptor 1 |
| chr13_-_46756351 | 1.17 |
ENST00000323076.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr2_-_208994548 | 1.17 |
ENST00000282141.3
|
CRYGC
|
crystallin, gamma C |
| chrX_-_153191708 | 1.17 |
ENST00000393721.1
ENST00000370028.3 |
ARHGAP4
|
Rho GTPase activating protein 4 |
| chr21_-_44495919 | 1.13 |
ENST00000398158.1
|
CBS
|
cystathionine-beta-synthase |
| chr3_-_121468602 | 1.10 |
ENST00000340645.5
|
GOLGB1
|
golgin B1 |
| chr9_+_90112117 | 1.04 |
ENST00000358077.5
|
DAPK1
|
death-associated protein kinase 1 |
| chr1_-_52344471 | 1.00 |
ENST00000352171.7
ENST00000354831.7 |
NRD1
|
nardilysin (N-arginine dibasic convertase) |
| chr14_-_107283278 | 0.93 |
ENST00000390639.2
|
IGHV7-81
|
immunoglobulin heavy variable 7-81 (non-functional) |
| chr9_+_71986182 | 0.91 |
ENST00000303068.7
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chr9_+_117904097 | 0.89 |
ENST00000374016.1
|
DEC1
|
deleted in esophageal cancer 1 |
| chr14_+_58894706 | 0.85 |
ENST00000261244.5
|
KIAA0586
|
KIAA0586 |
| chr12_+_12224331 | 0.84 |
ENST00000396367.1
ENST00000266434.4 ENST00000396369.1 |
BCL2L14
|
BCL2-like 14 (apoptosis facilitator) |
| chr22_-_32767017 | 0.83 |
ENST00000400234.1
|
RFPL3S
|
RFPL3 antisense |
| chr12_+_12202774 | 0.74 |
ENST00000589718.1
|
BCL2L14
|
BCL2-like 14 (apoptosis facilitator) |
| chr15_+_52043758 | 0.63 |
ENST00000249700.4
ENST00000539962.2 |
TMOD2
|
tropomodulin 2 (neuronal) |
| chr2_+_62423242 | 0.59 |
ENST00000301998.4
|
B3GNT2
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2 |
| chr12_-_25101920 | 0.58 |
ENST00000539780.1
ENST00000546285.1 ENST00000342945.5 |
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr17_+_3323862 | 0.55 |
ENST00000291231.1
|
OR3A3
|
olfactory receptor, family 3, subfamily A, member 3 |
| chr3_-_121468513 | 0.49 |
ENST00000494517.1
ENST00000393667.3 |
GOLGB1
|
golgin B1 |
| chr12_+_12202785 | 0.45 |
ENST00000586576.1
ENST00000464885.2 |
BCL2L14
|
BCL2-like 14 (apoptosis facilitator) |
| chr19_-_48753104 | 0.43 |
ENST00000447740.2
|
CARD8
|
caspase recruitment domain family, member 8 |
| chr11_+_30252549 | 0.41 |
ENST00000254122.3
ENST00000417547.1 |
FSHB
|
follicle stimulating hormone, beta polypeptide |
| chr11_-_5173599 | 0.41 |
ENST00000328942.1
|
OR52A1
|
olfactory receptor, family 52, subfamily A, member 1 |
| chr14_+_21236586 | 0.40 |
ENST00000326783.3
|
EDDM3B
|
epididymal protein 3B |
| chr19_-_52551814 | 0.38 |
ENST00000594154.1
ENST00000598745.1 ENST00000597273.1 |
ZNF432
|
zinc finger protein 432 |
| chr4_-_123843597 | 0.34 |
ENST00000510735.1
ENST00000304430.5 |
NUDT6
|
nudix (nucleoside diphosphate linked moiety X)-type motif 6 |
| chr5_+_32788945 | 0.27 |
ENST00000326958.1
|
AC026703.1
|
AC026703.1 |
| chr1_+_152627927 | 0.20 |
ENST00000444515.1
ENST00000536536.1 |
LINC00302
|
long intergenic non-protein coding RNA 302 |
| chr1_-_158300747 | 0.17 |
ENST00000451207.1
|
CD1B
|
CD1b molecule |
| chr19_-_23869999 | 0.16 |
ENST00000601935.1
ENST00000359788.4 ENST00000600313.1 ENST00000596211.1 ENST00000599168.1 |
ZNF675
|
zinc finger protein 675 |
| chr1_+_47603109 | 0.15 |
ENST00000371890.3
ENST00000294337.3 ENST00000371891.3 |
CYP4A22
|
cytochrome P450, family 4, subfamily A, polypeptide 22 |
| chr1_-_52456352 | 0.08 |
ENST00000371655.3
|
RAB3B
|
RAB3B, member RAS oncogene family |
| chr6_-_32806506 | 0.07 |
ENST00000374897.2
ENST00000452392.2 |
TAP2
TAP2
|
transporter 2, ATP-binding cassette, sub-family B (MDR/TAP) Uncharacterized protein |
| chr6_+_26538566 | 0.06 |
ENST00000377575.2
|
HMGN4
|
high mobility group nucleosomal binding domain 4 |
| chr19_-_48752812 | 0.05 |
ENST00000359009.4
|
CARD8
|
caspase recruitment domain family, member 8 |
| chr22_+_36649056 | 0.01 |
ENST00000397278.3
ENST00000422706.1 ENST00000426053.1 ENST00000319136.4 |
APOL1
|
apolipoprotein L, 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of AIRE
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.7 | 18.9 | GO:0010900 | negative regulation of phosphatidylcholine catabolic process(GO:0010900) |
| 4.0 | 11.9 | GO:1904395 | retinal rod cell differentiation(GO:0060221) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) negative regulation of neuromuscular junction development(GO:1904397) |
| 1.7 | 6.7 | GO:0048749 | compound eye development(GO:0048749) |
| 1.6 | 4.7 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 1.5 | 6.1 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 1.2 | 4.9 | GO:0060023 | soft palate development(GO:0060023) |
| 1.2 | 3.5 | GO:0006535 | cysteine biosynthetic process from serine(GO:0006535) |
| 1.2 | 3.5 | GO:0030451 | regulation of complement activation, alternative pathway(GO:0030451) negative regulation of complement activation, alternative pathway(GO:0045957) |
| 1.1 | 3.4 | GO:2000525 | dendritic cell dendrite assembly(GO:0097026) regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.9 | 18.4 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.9 | 29.5 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.8 | 3.0 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.7 | 2.1 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.7 | 2.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.7 | 15.3 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.6 | 4.2 | GO:0044800 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.6 | 1.8 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.6 | 4.7 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.5 | 7.1 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.5 | 4.4 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.5 | 1.5 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.5 | 1.5 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.5 | 2.8 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.4 | 5.5 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.4 | 1.6 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.4 | 2.2 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.3 | 2.4 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.3 | 5.0 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.3 | 1.7 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.3 | 3.1 | GO:0010764 | negative regulation of fibroblast migration(GO:0010764) |
| 0.3 | 12.1 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.3 | 3.0 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.3 | 1.8 | GO:0009597 | detection of virus(GO:0009597) |
| 0.3 | 1.8 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
| 0.2 | 3.2 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.2 | 6.7 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.2 | 0.6 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 1.2 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.1 | 11.4 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.1 | 7.6 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 3.1 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.1 | 0.6 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 3.5 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 0.6 | GO:0009099 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.1 | 4.7 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.1 | 1.0 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.1 | 1.5 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.1 | 1.9 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.1 | 4.1 | GO:0019835 | cytolysis(GO:0019835) |
| 0.1 | 3.8 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 1.2 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 4.9 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 2.8 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.1 | 1.4 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.1 | 2.1 | GO:0060384 | innervation(GO:0060384) |
| 0.1 | 0.4 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.1 | 3.0 | GO:0001510 | RNA methylation(GO:0001510) |
| 0.1 | 6.0 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.1 | 0.5 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.1 | 17.1 | GO:0006469 | negative regulation of protein kinase activity(GO:0006469) |
| 0.1 | 24.8 | GO:0043687 | post-translational protein modification(GO:0043687) |
| 0.0 | 3.0 | GO:0045814 | negative regulation of gene expression, epigenetic(GO:0045814) |
| 0.0 | 0.1 | GO:0002476 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) |
| 0.0 | 0.2 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.0 | 1.6 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 0.1 | GO:0051582 | peptidyl-cysteine methylation(GO:0018125) positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.0 | 12.5 | GO:0006954 | inflammatory response(GO:0006954) |
| 0.0 | 0.2 | GO:0048007 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 1.6 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.8 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 29.5 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.8 | 18.9 | GO:0042627 | chylomicron(GO:0042627) |
| 0.7 | 4.7 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.6 | 10.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.5 | 4.1 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.4 | 1.8 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.4 | 37.9 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.2 | 6.7 | GO:0071437 | invadopodium(GO:0071437) |
| 0.2 | 19.0 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.1 | 11.9 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 6.1 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 1.5 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 4.9 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 1.2 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.1 | 2.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 2.1 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.1 | 30.2 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 1.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 3.5 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 0.5 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.0 | 3.0 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 1.5 | GO:0098636 | protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 1.7 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 6.0 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 5.3 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 1.6 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 1.7 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 3.5 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 4.4 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 4.3 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 2.1 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.6 | GO:0030659 | cytoplasmic vesicle membrane(GO:0030659) |
| 0.0 | 0.9 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.6 | GO:0043197 | dendritic spine(GO:0043197) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 11.8 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 3.0 | 11.9 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 2.4 | 18.9 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 1.3 | 5.0 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 1.2 | 3.5 | GO:0070025 | cystathionine beta-synthase activity(GO:0004122) oxidoreductase activity, acting on other nitrogenous compounds as donors, cytochrome as acceptor(GO:0016662) nitrite reductase (NO-forming) activity(GO:0050421) carbon monoxide binding(GO:0070025) nitrite reductase activity(GO:0098809) |
| 1.2 | 3.5 | GO:0001861 | complement component C4b receptor activity(GO:0001861) complement component C3b receptor activity(GO:0004877) |
| 1.0 | 6.7 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.9 | 4.7 | GO:0070404 | NADH binding(GO:0070404) |
| 0.7 | 2.8 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.7 | 9.1 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.7 | 6.1 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.7 | 12.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.6 | 3.0 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.5 | 4.7 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.4 | 10.3 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.4 | 3.2 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.4 | 18.4 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.4 | 11.4 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.4 | 26.5 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.3 | 2.4 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.3 | 1.8 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.3 | 1.6 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.2 | 2.2 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.2 | 3.0 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.2 | 2.8 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.2 | 1.4 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.2 | 0.6 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.2 | 29.5 | GO:0016820 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances(GO:0016820) |
| 0.2 | 2.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.2 | 0.5 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.1 | 1.8 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 7.6 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 3.5 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 2.1 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 0.6 | GO:0052655 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.1 | 4.9 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 3.8 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 7.6 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 2.8 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 1.5 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 1.2 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 11.7 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 4.1 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 1.0 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 1.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 2.1 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.6 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 1.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.2 | GO:0030883 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.0 | 24.1 | GO:0001071 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 0.2 | GO:0016713 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.0 | 1.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 11.9 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.3 | 12.1 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.3 | 2.8 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.2 | 4.5 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 28.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 1.7 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 6.6 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.1 | 3.0 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 3.1 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 4.2 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 7.3 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 2.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.0 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 1.5 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 1.5 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 29.5 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.8 | 4.2 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.7 | 18.4 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.3 | 12.1 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.2 | 6.7 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.2 | 5.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 2.8 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 1.5 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.1 | 3.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 3.5 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 4.5 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 3.8 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 1.0 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 2.1 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 3.4 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 3.1 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 1.4 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 1.8 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.5 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 2.1 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 8.4 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.4 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 5.2 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 3.0 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.6 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 1.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |