Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
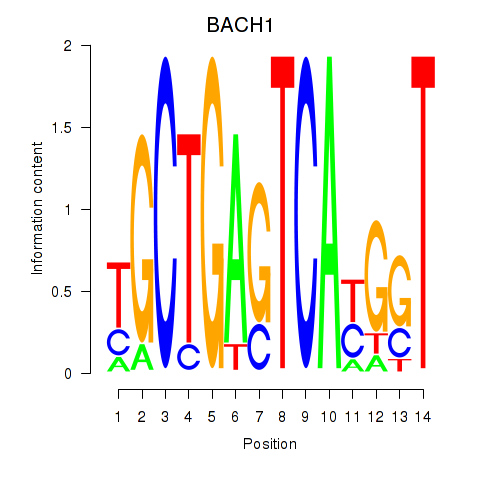
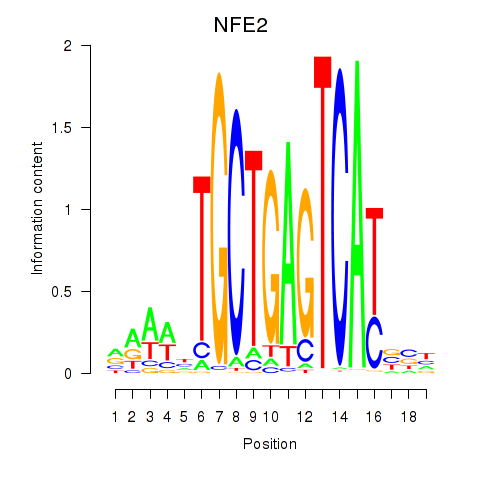
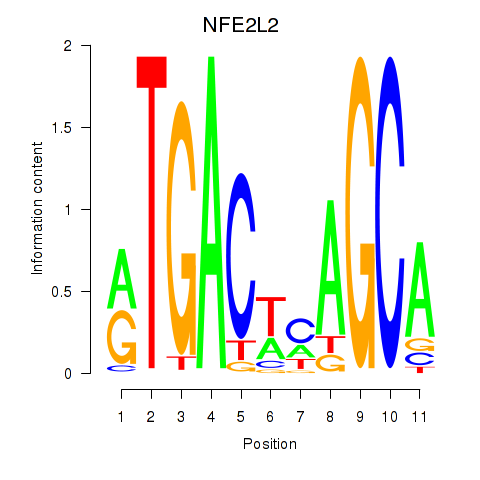
Results for BACH1_NFE2_NFE2L2
Z-value: 1.14
Transcription factors associated with BACH1_NFE2_NFE2L2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BACH1
|
ENSG00000156273.11 | BTB domain and CNC homolog 1 |
|
NFE2
|
ENSG00000123405.9 | nuclear factor, erythroid 2 |
|
NFE2L2
|
ENSG00000116044.11 | nuclear factor, erythroid 2 like 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NFE2L2 | hg19_v2_chr2_-_178257401_178257432, hg19_v2_chr2_-_178129853_178129860, hg19_v2_chr2_-_178128528_178128574, hg19_v2_chr2_-_178128250_178128300 | 0.27 | 4.9e-05 | Click! |
| BACH1 | hg19_v2_chr21_+_30671189_30671207, hg19_v2_chr21_+_30671690_30671762 | -0.20 | 2.8e-03 | Click! |
| NFE2 | hg19_v2_chr12_-_54689532_54689551, hg19_v2_chr12_-_54694807_54694905, hg19_v2_chr12_-_54694758_54694805 | 0.02 | 7.5e-01 | Click! |
Activity profile of BACH1_NFE2_NFE2L2 motif
Sorted Z-values of BACH1_NFE2_NFE2L2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_58711539 | 119.09 |
ENST00000216455.4
ENST00000412908.2 ENST00000557508.1 |
PSMA3
|
proteasome (prosome, macropain) subunit, alpha type, 3 |
| chr9_-_113018835 | 88.19 |
ENST00000374517.5
|
TXN
|
thioredoxin |
| chr16_-_69760409 | 78.18 |
ENST00000561500.1
ENST00000439109.2 ENST00000564043.1 ENST00000379046.2 ENST00000379047.3 |
NQO1
|
NAD(P)H dehydrogenase, quinone 1 |
| chr17_-_73150629 | 72.97 |
ENST00000356033.4
ENST00000405458.3 ENST00000409753.3 |
HN1
|
hematological and neurological expressed 1 |
| chr1_-_45987526 | 67.45 |
ENST00000372079.1
ENST00000262746.1 ENST00000447184.1 ENST00000319248.8 |
PRDX1
|
peroxiredoxin 1 |
| chr16_+_89988259 | 63.39 |
ENST00000554444.1
ENST00000556565.1 |
TUBB3
|
Tubulin beta-3 chain |
| chr9_-_113018746 | 62.94 |
ENST00000374515.5
|
TXN
|
thioredoxin |
| chr1_-_109968973 | 53.77 |
ENST00000271308.4
ENST00000538610.1 |
PSMA5
|
proteasome (prosome, macropain) subunit, alpha type, 5 |
| chr12_+_104680659 | 49.52 |
ENST00000526691.1
ENST00000531691.1 ENST00000388854.3 ENST00000354940.6 ENST00000526390.1 ENST00000531689.1 |
TXNRD1
|
thioredoxin reductase 1 |
| chrX_-_15511438 | 49.42 |
ENST00000380420.5
|
PIR
|
pirin (iron-binding nuclear protein) |
| chr16_+_30077055 | 48.88 |
ENST00000564595.2
ENST00000569798.1 |
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr18_+_158513 | 47.71 |
ENST00000400266.3
ENST00000580410.1 ENST00000383589.2 ENST00000261601.7 |
USP14
|
ubiquitin specific peptidase 14 (tRNA-guanine transglycosylase) |
| chr1_-_225616515 | 46.80 |
ENST00000338179.2
ENST00000425080.1 |
LBR
|
lamin B receptor |
| chr7_+_112063192 | 46.41 |
ENST00000005558.4
|
IFRD1
|
interferon-related developmental regulator 1 |
| chr10_-_127505167 | 44.36 |
ENST00000368786.1
|
UROS
|
uroporphyrinogen III synthase |
| chr1_-_94374946 | 43.71 |
ENST00000370238.3
|
GCLM
|
glutamate-cysteine ligase, modifier subunit |
| chr16_+_74330673 | 40.40 |
ENST00000219313.4
ENST00000540379.1 ENST00000567958.1 ENST00000568615.2 |
PSMD7
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 7 |
| chr11_-_14541872 | 39.82 |
ENST00000419365.2
ENST00000530457.1 ENST00000532256.1 ENST00000533068.1 |
PSMA1
|
proteasome (prosome, macropain) subunit, alpha type, 1 |
| chr14_+_35761580 | 38.58 |
ENST00000553809.1
ENST00000555764.1 ENST00000556506.1 |
PSMA6
|
proteasome (prosome, macropain) subunit, alpha type, 6 |
| chr10_+_85899196 | 37.76 |
ENST00000372134.3
|
GHITM
|
growth hormone inducible transmembrane protein |
| chr6_+_44214824 | 37.66 |
ENST00000371646.5
ENST00000353801.3 |
HSP90AB1
|
heat shock protein 90kDa alpha (cytosolic), class B member 1 |
| chrX_+_153770421 | 37.51 |
ENST00000369609.5
ENST00000369607.1 |
IKBKG
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr6_-_3157760 | 36.90 |
ENST00000333628.3
|
TUBB2A
|
tubulin, beta 2A class IIa |
| chr21_+_33031935 | 34.19 |
ENST00000270142.6
ENST00000389995.4 |
SOD1
|
superoxide dismutase 1, soluble |
| chr14_+_35761540 | 34.18 |
ENST00000261479.4
|
PSMA6
|
proteasome (prosome, macropain) subunit, alpha type, 6 |
| chr16_+_30077098 | 32.02 |
ENST00000395240.3
ENST00000566846.1 |
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr1_+_165796753 | 31.59 |
ENST00000367879.4
|
UCK2
|
uridine-cytidine kinase 2 |
| chr11_+_67351019 | 30.96 |
ENST00000398606.3
|
GSTP1
|
glutathione S-transferase pi 1 |
| chr5_-_137911049 | 30.59 |
ENST00000297185.3
|
HSPA9
|
heat shock 70kDa protein 9 (mortalin) |
| chr17_+_30771279 | 30.47 |
ENST00000261712.3
ENST00000578213.1 ENST00000457654.2 ENST00000579451.1 |
PSMD11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
| chr10_+_106013923 | 29.58 |
ENST00000539281.1
|
GSTO1
|
glutathione S-transferase omega 1 |
| chr6_-_84140757 | 29.19 |
ENST00000541327.1
ENST00000369705.3 ENST00000543031.1 |
ME1
|
malic enzyme 1, NADP(+)-dependent, cytosolic |
| chr6_-_35888905 | 28.01 |
ENST00000510290.1
ENST00000423325.2 ENST00000373822.1 |
SRPK1
|
SRSF protein kinase 1 |
| chr8_-_30585439 | 27.08 |
ENST00000221130.5
|
GSR
|
glutathione reductase |
| chr22_-_19466683 | 26.14 |
ENST00000399523.1
ENST00000421968.2 ENST00000447868.1 |
UFD1L
|
ubiquitin fusion degradation 1 like (yeast) |
| chr3_+_100053625 | 26.12 |
ENST00000497785.1
|
NIT2
|
nitrilase family, member 2 |
| chr8_-_30585294 | 24.86 |
ENST00000546342.1
ENST00000541648.1 ENST00000537535.1 |
GSR
|
glutathione reductase |
| chr18_-_54305658 | 24.83 |
ENST00000586262.1
ENST00000217515.6 |
TXNL1
|
thioredoxin-like 1 |
| chr2_+_231921574 | 23.99 |
ENST00000308696.6
ENST00000373635.4 ENST00000440838.1 ENST00000409643.1 |
PSMD1
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 1 |
| chr22_-_19466732 | 23.86 |
ENST00000263202.10
ENST00000360834.4 |
UFD1L
|
ubiquitin fusion degradation 1 like (yeast) |
| chr14_+_103801140 | 23.52 |
ENST00000561325.1
ENST00000392715.2 ENST00000559130.1 ENST00000559532.1 ENST00000558506.1 |
EIF5
|
eukaryotic translation initiation factor 5 |
| chr15_+_78833071 | 22.87 |
ENST00000559365.1
|
PSMA4
|
proteasome (prosome, macropain) subunit, alpha type, 4 |
| chr3_+_184038073 | 22.77 |
ENST00000428387.1
ENST00000434061.2 |
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr10_+_106014468 | 22.72 |
ENST00000369710.4
ENST00000369713.5 ENST00000445155.1 |
GSTO1
|
glutathione S-transferase omega 1 |
| chr3_+_100053542 | 22.53 |
ENST00000394140.4
|
NIT2
|
nitrilase family, member 2 |
| chr16_-_28222797 | 22.00 |
ENST00000569951.1
ENST00000565698.1 |
XPO6
|
exportin 6 |
| chr11_+_118958689 | 21.33 |
ENST00000535253.1
ENST00000392841.1 |
HMBS
|
hydroxymethylbilane synthase |
| chr15_+_78833105 | 20.69 |
ENST00000558341.1
ENST00000559437.1 |
PSMA4
|
proteasome (prosome, macropain) subunit, alpha type, 4 |
| chr15_+_78832747 | 20.59 |
ENST00000560217.1
ENST00000044462.7 ENST00000559082.1 ENST00000559948.1 ENST00000413382.2 ENST00000559146.1 ENST00000558281.1 |
PSMA4
|
proteasome (prosome, macropain) subunit, alpha type, 4 |
| chr6_+_37012607 | 19.91 |
ENST00000423336.1
|
COX6A1P2
|
cytochrome c oxidase subunit VIa polypeptide 1 pseudogene 2 |
| chr1_-_150208498 | 19.17 |
ENST00000314136.8
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr10_-_75173785 | 19.04 |
ENST00000535178.1
ENST00000372921.5 ENST00000372919.4 |
ANXA7
|
annexin A7 |
| chr22_+_45072958 | 18.93 |
ENST00000403581.1
|
PRR5
|
proline rich 5 (renal) |
| chr14_-_23504087 | 18.92 |
ENST00000493471.2
ENST00000460922.2 |
PSMB5
|
proteasome (prosome, macropain) subunit, beta type, 5 |
| chr22_+_45072925 | 18.85 |
ENST00000006251.7
|
PRR5
|
proline rich 5 (renal) |
| chr20_+_60878005 | 18.32 |
ENST00000253003.2
|
ADRM1
|
adhesion regulating molecule 1 |
| chr1_-_150208412 | 18.13 |
ENST00000532744.1
ENST00000369114.5 ENST00000369115.2 ENST00000369116.4 |
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr12_-_57081940 | 18.08 |
ENST00000436399.2
|
PTGES3
|
prostaglandin E synthase 3 (cytosolic) |
| chr11_-_47447767 | 17.77 |
ENST00000530651.1
ENST00000524447.2 ENST00000531051.2 ENST00000526993.1 ENST00000602866.1 |
PSMC3
|
proteasome (prosome, macropain) 26S subunit, ATPase, 3 |
| chr17_-_65362678 | 17.55 |
ENST00000357146.4
ENST00000356126.3 |
PSMD12
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 12 |
| chr17_+_61905058 | 17.29 |
ENST00000375812.4
ENST00000581882.1 |
PSMC5
|
proteasome (prosome, macropain) 26S subunit, ATPase, 5 |
| chr2_-_10588630 | 17.15 |
ENST00000234111.4
|
ODC1
|
ornithine decarboxylase 1 |
| chr1_+_76251879 | 17.13 |
ENST00000535300.1
ENST00000319942.3 |
RABGGTB
|
Rab geranylgeranyltransferase, beta subunit |
| chr8_-_30585217 | 16.99 |
ENST00000520888.1
ENST00000414019.1 |
GSR
|
glutathione reductase |
| chr17_+_18625336 | 16.44 |
ENST00000395671.4
ENST00000571542.1 ENST00000395672.2 ENST00000414850.2 ENST00000424146.2 |
TRIM16L
|
tripartite motif containing 16-like |
| chr1_-_150208363 | 16.43 |
ENST00000436748.2
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr6_-_31704282 | 16.40 |
ENST00000375784.3
ENST00000375779.2 |
CLIC1
|
chloride intracellular channel 1 |
| chrX_+_56590002 | 16.03 |
ENST00000338222.5
|
UBQLN2
|
ubiquilin 2 |
| chr19_+_18699535 | 15.62 |
ENST00000358607.6
|
C19orf60
|
chromosome 19 open reading frame 60 |
| chr7_+_102988082 | 15.60 |
ENST00000292644.3
ENST00000544811.1 |
PSMC2
|
proteasome (prosome, macropain) 26S subunit, ATPase, 2 |
| chr1_+_26605618 | 15.49 |
ENST00000270792.5
|
SH3BGRL3
|
SH3 domain binding glutamic acid-rich protein like 3 |
| chrX_-_109683446 | 15.47 |
ENST00000372057.1
|
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 |
| chr11_-_47447970 | 15.23 |
ENST00000298852.3
ENST00000530912.1 |
PSMC3
|
proteasome (prosome, macropain) 26S subunit, ATPase, 3 |
| chr7_-_42971759 | 15.10 |
ENST00000538645.1
ENST00000445517.1 ENST00000223321.4 |
PSMA2
|
proteasome (prosome, macropain) subunit, alpha type, 2 |
| chr5_+_68530668 | 14.77 |
ENST00000506563.1
|
CDK7
|
cyclin-dependent kinase 7 |
| chr22_+_19466980 | 14.70 |
ENST00000407835.1
ENST00000438587.1 |
CDC45
|
cell division cycle 45 |
| chr19_-_10613421 | 14.45 |
ENST00000393623.2
|
KEAP1
|
kelch-like ECH-associated protein 1 |
| chr3_+_184037466 | 14.45 |
ENST00000441154.1
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr11_-_2924720 | 14.34 |
ENST00000455942.2
|
SLC22A18AS
|
solute carrier family 22 (organic cation transporter), member 18 antisense |
| chr2_-_209118974 | 14.20 |
ENST00000415913.1
ENST00000415282.1 ENST00000446179.1 |
IDH1
|
isocitrate dehydrogenase 1 (NADP+), soluble |
| chr17_+_61904766 | 14.02 |
ENST00000581842.1
ENST00000582130.1 ENST00000584320.1 ENST00000585123.1 ENST00000580864.1 |
PSMC5
|
proteasome (prosome, macropain) 26S subunit, ATPase, 5 |
| chr18_-_55253989 | 13.63 |
ENST00000262093.5
|
FECH
|
ferrochelatase |
| chr11_-_62474803 | 13.48 |
ENST00000533982.1
ENST00000360796.5 |
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr11_+_67351213 | 13.38 |
ENST00000398603.1
|
GSTP1
|
glutathione S-transferase pi 1 |
| chr17_-_79881408 | 13.31 |
ENST00000392366.3
|
MAFG
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog G |
| chr5_+_179247759 | 12.92 |
ENST00000389805.4
ENST00000504627.1 ENST00000402874.3 ENST00000510187.1 |
SQSTM1
|
sequestosome 1 |
| chr7_+_134212312 | 12.86 |
ENST00000359579.4
|
AKR1B10
|
aldo-keto reductase family 1, member B10 (aldose reductase) |
| chr1_+_156756667 | 12.56 |
ENST00000526188.1
ENST00000454659.1 |
PRCC
|
papillary renal cell carcinoma (translocation-associated) |
| chr6_-_35888824 | 12.44 |
ENST00000361690.3
ENST00000512445.1 |
SRPK1
|
SRSF protein kinase 1 |
| chr1_+_26869597 | 12.35 |
ENST00000530003.1
|
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1 |
| chr12_-_53343602 | 12.28 |
ENST00000546897.1
ENST00000552551.1 |
KRT8
|
keratin 8 |
| chr1_-_150208291 | 12.18 |
ENST00000533654.1
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr4_+_17616253 | 12.14 |
ENST00000237380.7
|
MED28
|
mediator complex subunit 28 |
| chr6_+_44215603 | 11.91 |
ENST00000371554.1
|
HSP90AB1
|
heat shock protein 90kDa alpha (cytosolic), class B member 1 |
| chr17_+_30677136 | 11.67 |
ENST00000394670.4
ENST00000321233.6 ENST00000394673.2 ENST00000341711.6 ENST00000579634.1 ENST00000580759.1 ENST00000342555.6 ENST00000577908.1 ENST00000394679.5 ENST00000582165.1 |
ZNF207
|
zinc finger protein 207 |
| chr3_+_120315160 | 11.56 |
ENST00000485064.1
ENST00000492739.1 |
NDUFB4
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 4, 15kDa |
| chr3_+_120315149 | 11.32 |
ENST00000184266.2
|
NDUFB4
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 4, 15kDa |
| chr2_-_220119280 | 11.23 |
ENST00000392088.2
|
TUBA4A
|
tubulin, alpha 4a |
| chr19_+_38865398 | 11.15 |
ENST00000585598.1
ENST00000602911.1 ENST00000592561.1 |
PSMD8
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 8 |
| chr5_+_68530697 | 10.97 |
ENST00000256443.3
ENST00000514676.1 |
CDK7
|
cyclin-dependent kinase 7 |
| chr17_-_61905005 | 10.86 |
ENST00000584574.1
ENST00000585145.1 ENST00000427159.2 |
FTSJ3
|
FtsJ homolog 3 (E. coli) |
| chr12_-_57082060 | 10.68 |
ENST00000448157.2
ENST00000414274.3 ENST00000262033.6 ENST00000456859.2 |
PTGES3
|
prostaglandin E synthase 3 (cytosolic) |
| chr18_+_3252265 | 10.47 |
ENST00000580887.1
ENST00000536605.1 |
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr19_-_59023348 | 10.22 |
ENST00000601355.1
ENST00000263093.2 |
SLC27A5
|
solute carrier family 27 (fatty acid transporter), member 5 |
| chr19_+_38865176 | 10.00 |
ENST00000215071.4
|
PSMD8
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 8 |
| chr3_+_69134080 | 9.73 |
ENST00000273258.3
|
ARL6IP5
|
ADP-ribosylation-like factor 6 interacting protein 5 |
| chr3_-_81792780 | 9.50 |
ENST00000489715.1
|
GBE1
|
glucan (1,4-alpha-), branching enzyme 1 |
| chr22_-_45608324 | 9.46 |
ENST00000496226.1
ENST00000251993.7 |
KIAA0930
|
KIAA0930 |
| chr16_+_57662419 | 9.43 |
ENST00000388812.4
ENST00000538815.1 ENST00000456916.1 ENST00000567154.1 ENST00000388813.5 ENST00000562558.1 ENST00000566271.2 |
GPR56
|
G protein-coupled receptor 56 |
| chr18_-_55253871 | 9.42 |
ENST00000382873.3
|
FECH
|
ferrochelatase |
| chr12_+_75874460 | 9.41 |
ENST00000266659.3
|
GLIPR1
|
GLI pathogenesis-related 1 |
| chr19_+_18699599 | 9.20 |
ENST00000450195.2
|
C19orf60
|
chromosome 19 open reading frame 60 |
| chr5_+_179125368 | 9.05 |
ENST00000502296.1
ENST00000504734.1 ENST00000415618.2 |
CANX
|
calnexin |
| chr17_-_28618948 | 8.99 |
ENST00000261714.6
|
BLMH
|
bleomycin hydrolase |
| chr16_+_22308717 | 8.98 |
ENST00000299853.5
ENST00000564209.1 ENST00000565358.1 ENST00000418581.2 ENST00000564883.1 ENST00000359210.4 ENST00000563024.1 |
POLR3E
|
polymerase (RNA) III (DNA directed) polypeptide E (80kD) |
| chr2_+_122513109 | 8.82 |
ENST00000389682.3
ENST00000536142.1 |
TSN
|
translin |
| chr3_+_140660634 | 8.82 |
ENST00000446041.2
ENST00000507429.1 ENST00000324194.6 |
SLC25A36
|
solute carrier family 25 (pyrimidine nucleotide carrier ), member 36 |
| chr19_+_18496957 | 8.74 |
ENST00000252809.3
|
GDF15
|
growth differentiation factor 15 |
| chr14_-_65409502 | 8.55 |
ENST00000389614.5
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal) |
| chr17_+_18759612 | 8.54 |
ENST00000432893.2
ENST00000414602.1 ENST00000574522.1 ENST00000570450.1 ENST00000419071.2 |
PRPSAP2
|
phosphoribosyl pyrophosphate synthetase-associated protein 2 |
| chr16_+_67261008 | 8.34 |
ENST00000304800.9
ENST00000563953.1 ENST00000565201.1 |
TMEM208
|
transmembrane protein 208 |
| chr17_+_75372165 | 8.17 |
ENST00000427674.2
|
SEPT9
|
septin 9 |
| chr3_+_140660743 | 8.01 |
ENST00000453248.2
|
SLC25A36
|
solute carrier family 25 (pyrimidine nucleotide carrier ), member 36 |
| chr11_-_2924970 | 7.94 |
ENST00000533594.1
|
SLC22A18AS
|
solute carrier family 22 (organic cation transporter), member 18 antisense |
| chr11_+_35160709 | 7.94 |
ENST00000415148.2
ENST00000433354.2 ENST00000449691.2 ENST00000437706.2 ENST00000360158.4 ENST00000428726.2 ENST00000526669.2 ENST00000433892.2 ENST00000278386.6 ENST00000434472.2 ENST00000352818.4 ENST00000442151.2 |
CD44
|
CD44 molecule (Indian blood group) |
| chr16_+_57662138 | 7.86 |
ENST00000562414.1
ENST00000561969.1 ENST00000562631.1 ENST00000563445.1 ENST00000565338.1 ENST00000567702.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr9_+_71819927 | 7.70 |
ENST00000535702.1
|
TJP2
|
tight junction protein 2 |
| chr3_-_98241358 | 7.68 |
ENST00000503004.1
ENST00000506575.1 ENST00000513452.1 ENST00000515620.1 |
CLDND1
|
claudin domain containing 1 |
| chrX_+_51636629 | 7.48 |
ENST00000375722.1
ENST00000326587.7 ENST00000375695.2 |
MAGED1
|
melanoma antigen family D, 1 |
| chr11_-_66103867 | 7.42 |
ENST00000424433.2
|
RIN1
|
Ras and Rab interactor 1 |
| chr9_+_71820057 | 7.41 |
ENST00000539225.1
|
TJP2
|
tight junction protein 2 |
| chr11_+_63448918 | 7.34 |
ENST00000341307.2
ENST00000356000.3 ENST00000542238.1 |
RTN3
|
reticulon 3 |
| chr7_+_101460882 | 7.32 |
ENST00000292535.7
ENST00000549414.2 ENST00000550008.2 ENST00000546411.2 ENST00000556210.1 |
CUX1
|
cut-like homeobox 1 |
| chr14_+_52456193 | 7.29 |
ENST00000261700.3
|
C14orf166
|
chromosome 14 open reading frame 166 |
| chr1_+_45241109 | 7.21 |
ENST00000396651.3
|
RPS8
|
ribosomal protein S8 |
| chrX_-_153718953 | 7.17 |
ENST00000369649.4
ENST00000393586.1 |
SLC10A3
|
solute carrier family 10, member 3 |
| chr5_+_135385202 | 6.99 |
ENST00000514554.1
|
TGFBI
|
transforming growth factor, beta-induced, 68kDa |
| chr11_-_102668879 | 6.98 |
ENST00000315274.6
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase) |
| chr7_+_22766766 | 6.98 |
ENST00000426291.1
ENST00000401651.1 ENST00000258743.5 ENST00000420258.2 ENST00000407492.1 ENST00000401630.3 ENST00000406575.1 |
IL6
|
interleukin 6 (interferon, beta 2) |
| chr11_+_62649158 | 6.97 |
ENST00000539891.1
ENST00000536981.1 |
SLC3A2
|
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr6_-_24721054 | 6.90 |
ENST00000378119.4
|
C6orf62
|
chromosome 6 open reading frame 62 |
| chr7_+_130794846 | 6.89 |
ENST00000421797.2
|
MKLN1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr7_-_102985035 | 6.76 |
ENST00000426036.2
ENST00000249270.7 ENST00000454277.1 ENST00000412522.1 |
DNAJC2
|
DnaJ (Hsp40) homolog, subfamily C, member 2 |
| chr14_-_75536182 | 6.72 |
ENST00000555463.1
|
ACYP1
|
acylphosphatase 1, erythrocyte (common) type |
| chr6_-_170862322 | 6.71 |
ENST00000262193.6
|
PSMB1
|
proteasome (prosome, macropain) subunit, beta type, 1 |
| chr7_-_86849883 | 6.69 |
ENST00000433078.1
|
TMEM243
|
transmembrane protein 243, mitochondrial |
| chr3_+_69134124 | 6.68 |
ENST00000478935.1
|
ARL6IP5
|
ADP-ribosylation-like factor 6 interacting protein 5 |
| chr1_+_84609944 | 6.61 |
ENST00000370685.3
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr10_+_45495898 | 6.57 |
ENST00000298299.3
|
ZNF22
|
zinc finger protein 22 |
| chr8_-_145013711 | 6.52 |
ENST00000345136.3
|
PLEC
|
plectin |
| chr2_-_190044480 | 6.44 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr22_-_32058166 | 6.40 |
ENST00000435900.1
ENST00000336566.4 |
PISD
|
phosphatidylserine decarboxylase |
| chr20_-_34330129 | 6.39 |
ENST00000397370.3
ENST00000528062.3 ENST00000407261.4 ENST00000374038.3 ENST00000361162.6 |
RBM39
|
RNA binding motif protein 39 |
| chr1_-_150208320 | 6.25 |
ENST00000534220.1
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr22_-_39268192 | 6.03 |
ENST00000216083.6
|
CBX6
|
chromobox homolog 6 |
| chr19_+_17865011 | 6.01 |
ENST00000596462.1
ENST00000596865.1 ENST00000598960.1 ENST00000539407.1 |
FCHO1
|
FCH domain only 1 |
| chr6_+_84569359 | 6.01 |
ENST00000369681.5
ENST00000369679.4 |
CYB5R4
|
cytochrome b5 reductase 4 |
| chr16_+_29819446 | 5.99 |
ENST00000568282.1
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr8_-_59572301 | 5.95 |
ENST00000038176.3
|
NSMAF
|
neutral sphingomyelinase (N-SMase) activation associated factor |
| chr3_-_149293990 | 5.95 |
ENST00000472417.1
|
WWTR1
|
WW domain containing transcription regulator 1 |
| chr17_-_74733404 | 5.87 |
ENST00000508921.3
ENST00000583836.1 ENST00000358156.6 ENST00000392485.2 ENST00000359995.5 |
SRSF2
|
serine/arginine-rich splicing factor 2 |
| chr15_-_59225844 | 5.81 |
ENST00000380516.2
|
SLTM
|
SAFB-like, transcription modulator |
| chr18_+_11857439 | 5.58 |
ENST00000602628.1
|
GNAL
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr16_+_30211181 | 5.56 |
ENST00000395138.2
|
SULT1A3
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3 |
| chr17_+_21191341 | 5.51 |
ENST00000526076.2
ENST00000361818.5 ENST00000316920.6 |
MAP2K3
|
mitogen-activated protein kinase kinase 3 |
| chr19_+_6531010 | 5.51 |
ENST00000245817.3
|
TNFSF9
|
tumor necrosis factor (ligand) superfamily, member 9 |
| chr3_-_98241760 | 5.49 |
ENST00000507874.1
ENST00000502299.1 ENST00000508659.1 ENST00000510545.1 ENST00000511667.1 ENST00000394185.2 ENST00000394181.2 ENST00000508902.1 ENST00000341181.6 ENST00000437922.1 ENST00000394180.2 |
CLDND1
|
claudin domain containing 1 |
| chr1_+_45241186 | 5.44 |
ENST00000372209.3
|
RPS8
|
ribosomal protein S8 |
| chr4_-_987217 | 5.37 |
ENST00000361661.2
ENST00000398516.2 |
SLC26A1
|
solute carrier family 26 (anion exchanger), member 1 |
| chr18_+_3252206 | 5.18 |
ENST00000578562.2
|
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr12_+_75874580 | 5.16 |
ENST00000456650.3
|
GLIPR1
|
GLI pathogenesis-related 1 |
| chr16_-_11036300 | 4.94 |
ENST00000331808.4
|
DEXI
|
Dexi homolog (mouse) |
| chr1_-_39339777 | 4.88 |
ENST00000397572.2
|
MYCBP
|
MYC binding protein |
| chr2_+_109237717 | 4.87 |
ENST00000409441.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr9_+_140083099 | 4.53 |
ENST00000322310.5
|
SSNA1
|
Sjogren syndrome nuclear autoantigen 1 |
| chr12_-_120687948 | 4.44 |
ENST00000458477.2
|
PXN
|
paxillin |
| chr1_+_81771806 | 4.24 |
ENST00000370721.1
ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2
|
latrophilin 2 |
| chr17_-_76899275 | 4.11 |
ENST00000322630.2
ENST00000586713.1 |
DDC8
|
Protein DDC8 homolog |
| chr20_-_43280361 | 4.10 |
ENST00000372874.4
|
ADA
|
adenosine deaminase |
| chr6_-_138428613 | 4.03 |
ENST00000421351.3
|
PERP
|
PERP, TP53 apoptosis effector |
| chr6_+_30034966 | 3.96 |
ENST00000376769.2
|
PPP1R11
|
protein phosphatase 1, regulatory (inhibitor) subunit 11 |
| chrX_-_72434628 | 3.86 |
ENST00000536638.1
ENST00000373517.3 |
NAP1L2
|
nucleosome assembly protein 1-like 2 |
| chr7_-_104909435 | 3.77 |
ENST00000357311.3
|
SRPK2
|
SRSF protein kinase 2 |
| chr20_-_43280325 | 3.76 |
ENST00000537820.1
|
ADA
|
adenosine deaminase |
| chr22_+_22730353 | 3.73 |
ENST00000390296.2
|
IGLV5-45
|
immunoglobulin lambda variable 5-45 |
| chr19_+_926000 | 3.67 |
ENST00000263620.3
|
ARID3A
|
AT rich interactive domain 3A (BRIGHT-like) |
| chr12_-_125401885 | 3.67 |
ENST00000542416.1
|
UBC
|
ubiquitin C |
| chr6_+_30034865 | 3.62 |
ENST00000376772.3
|
PPP1R11
|
protein phosphatase 1, regulatory (inhibitor) subunit 11 |
| chrX_-_119445263 | 3.50 |
ENST00000309720.5
|
TMEM255A
|
transmembrane protein 255A |
| chrX_-_119445306 | 3.44 |
ENST00000371369.4
ENST00000440464.1 ENST00000519908.1 |
TMEM255A
|
transmembrane protein 255A |
| chr4_+_183164574 | 3.39 |
ENST00000511685.1
|
TENM3
|
teneurin transmembrane protein 3 |
| chr18_-_12377283 | 3.35 |
ENST00000269143.3
|
AFG3L2
|
AFG3-like AAA ATPase 2 |
| chr11_+_65339820 | 3.31 |
ENST00000316409.2
ENST00000449319.2 ENST00000530349.1 |
FAM89B
|
family with sequence similarity 89, member B |
| chr16_+_12059091 | 3.01 |
ENST00000562385.1
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr15_+_40531621 | 2.98 |
ENST00000560346.1
|
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6 |
| chr17_+_74372662 | 2.94 |
ENST00000591651.1
ENST00000545180.1 |
SPHK1
|
sphingosine kinase 1 |
| chr17_-_79604075 | 2.90 |
ENST00000374747.5
ENST00000539314.1 ENST00000331134.6 |
NPLOC4
|
nuclear protein localization 4 homolog (S. cerevisiae) |
| chr9_-_74383799 | 2.84 |
ENST00000377044.4
|
TMEM2
|
transmembrane protein 2 |
| chr16_-_73093597 | 2.83 |
ENST00000397992.5
|
ZFHX3
|
zinc finger homeobox 3 |
| chr11_+_46402297 | 2.75 |
ENST00000405308.2
|
MDK
|
midkine (neurite growth-promoting factor 2) |
Network of associatons between targets according to the STRING database.
First level regulatory network of BACH1_NFE2_NFE2L2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 16.8 | 151.1 | GO:0030091 | protein repair(GO:0030091) |
| 15.9 | 47.7 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 15.5 | 46.4 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 12.4 | 49.6 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 11.1 | 44.4 | GO:0006780 | uroporphyrinogen III biosynthetic process(GO:0006780) uroporphyrinogen III metabolic process(GO:0046502) |
| 11.1 | 44.3 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 10.9 | 43.7 | GO:1904386 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 8.7 | 52.3 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 7.9 | 31.6 | GO:0009224 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 7.7 | 23.1 | GO:0070541 | response to platinum ion(GO:0070541) |
| 7.1 | 14.2 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 6.8 | 34.2 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 6.7 | 699.1 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 6.1 | 48.7 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 5.6 | 16.8 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 5.5 | 49.5 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 4.9 | 14.7 | GO:0031938 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) regulation of chromatin silencing at telomere(GO:0031938) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 4.9 | 29.2 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 4.6 | 18.3 | GO:0043248 | proteasome assembly(GO:0043248) |
| 4.5 | 80.9 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 4.5 | 40.4 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 4.3 | 12.9 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 4.1 | 37.2 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 4.0 | 16.0 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 3.8 | 30.6 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 3.6 | 14.4 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 3.4 | 37.8 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 2.8 | 8.3 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 2.6 | 7.9 | GO:0046102 | hypoxanthine salvage(GO:0043103) inosine metabolic process(GO:0046102) |
| 2.3 | 7.0 | GO:0002384 | hepatic immune response(GO:0002384) response to prolactin(GO:1990637) regulation of STAT protein import into nucleus(GO:2000364) positive regulation of STAT protein import into nucleus(GO:2000366) |
| 2.3 | 176.8 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 2.3 | 6.8 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 2.1 | 17.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 2.1 | 21.3 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 2.1 | 28.8 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 2.1 | 16.4 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 1.8 | 19.3 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 1.6 | 6.4 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) |
| 1.6 | 9.6 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 1.5 | 15.1 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 1.5 | 10.2 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 1.4 | 7.0 | GO:0060356 | leucine import(GO:0060356) |
| 1.4 | 5.5 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 1.4 | 59.9 | GO:0043486 | histone exchange(GO:0043486) |
| 1.3 | 6.6 | GO:0097338 | response to clozapine(GO:0097338) |
| 1.2 | 7.3 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 1.1 | 37.5 | GO:0002479 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent(GO:0002479) |
| 1.1 | 4.5 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 1.1 | 12.3 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 1.1 | 12.9 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 1.1 | 5.4 | GO:0050428 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 1.0 | 7.9 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 1.0 | 2.9 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.9 | 7.3 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.9 | 10.9 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.9 | 9.1 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.9 | 2.7 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.8 | 49.4 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.8 | 3.3 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.8 | 2.4 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.8 | 5.6 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.8 | 3.9 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) regulation of stem cell division(GO:2000035) |
| 0.8 | 21.2 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.8 | 4.5 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.8 | 3.8 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.7 | 2.8 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.7 | 2.1 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.7 | 5.5 | GO:0035897 | proteolysis in other organism(GO:0035897) |
| 0.7 | 8.8 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.7 | 5.9 | GO:0033197 | response to vitamin E(GO:0033197) |
| 0.6 | 7.3 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.6 | 6.6 | GO:0072257 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.6 | 13.5 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.6 | 25.7 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.6 | 3.4 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.5 | 2.5 | GO:1904977 | lymphatic endothelial cell migration(GO:1904977) |
| 0.5 | 10.2 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.5 | 43.8 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) |
| 0.5 | 2.7 | GO:0030421 | defecation(GO:0030421) |
| 0.4 | 12.3 | GO:0072575 | hepatocyte proliferation(GO:0072574) epithelial cell proliferation involved in liver morphogenesis(GO:0072575) |
| 0.4 | 11.9 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.4 | 1.6 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.4 | 1.2 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 0.4 | 1.6 | GO:1901628 | positive regulation of postsynaptic membrane organization(GO:1901628) |
| 0.4 | 7.6 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.4 | 8.2 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.4 | 4.0 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.4 | 2.2 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.4 | 22.9 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.3 | 7.5 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.3 | 30.4 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.3 | 12.7 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.3 | 11.7 | GO:0007094 | mitotic spindle assembly checkpoint(GO:0007094) spindle assembly checkpoint(GO:0071173) |
| 0.3 | 4.2 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.3 | 0.8 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.3 | 2.9 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.3 | 18.3 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.3 | 3.6 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.2 | 3.4 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.2 | 7.0 | GO:0032461 | positive regulation of protein oligomerization(GO:0032461) |
| 0.2 | 1.7 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.2 | 2.5 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.2 | 2.8 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.2 | 7.3 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.2 | 4.4 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.2 | 13.3 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.2 | 0.4 | GO:0071109 | superior temporal gyrus development(GO:0071109) |
| 0.2 | 9.0 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.2 | 4.9 | GO:0009165 | nucleotide biosynthetic process(GO:0009165) |
| 0.2 | 47.9 | GO:0007411 | axon guidance(GO:0007411) |
| 0.1 | 4.2 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 6.0 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 4.9 | GO:1900026 | positive regulation of focal adhesion assembly(GO:0051894) positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) positive regulation of adherens junction organization(GO:1903393) |
| 0.1 | 0.4 | GO:0038188 | cholecystokinin signaling pathway(GO:0038188) |
| 0.1 | 1.9 | GO:0050855 | regulation of B cell receptor signaling pathway(GO:0050855) |
| 0.1 | 0.4 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.1 | 0.6 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.1 | 5.2 | GO:0002260 | lymphocyte homeostasis(GO:0002260) |
| 0.1 | 1.1 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.1 | 0.9 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 2.9 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 6.0 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 6.5 | GO:0002062 | chondrocyte differentiation(GO:0002062) |
| 0.1 | 1.4 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.1 | 0.7 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 1.1 | GO:0043065 | positive regulation of apoptotic process(GO:0043065) |
| 0.1 | 6.6 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.1 | 1.7 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.1 | 1.9 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 8.2 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.1 | 0.2 | GO:0045425 | regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045423) positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.1 | 0.5 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.1 | 1.3 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.6 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.0 | 5.8 | GO:0050684 | regulation of mRNA processing(GO:0050684) |
| 0.0 | 6.3 | GO:0010389 | regulation of G2/M transition of mitotic cell cycle(GO:0010389) |
| 0.0 | 2.4 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 1.2 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.2 | GO:0070857 | regulation of bile acid biosynthetic process(GO:0070857) |
| 0.0 | 2.2 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.0 | 0.8 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 6.4 | GO:0008380 | RNA splicing(GO:0008380) |
| 0.0 | 0.1 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.0 | 1.5 | GO:0010977 | negative regulation of neuron projection development(GO:0010977) |
| 0.0 | 2.0 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.0 | 1.2 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 30.4 | 364.7 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 14.8 | 44.3 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 12.4 | 49.6 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 8.7 | 87.5 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 7.3 | 79.9 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 6.4 | 25.7 | GO:0070985 | TFIIK complex(GO:0070985) |
| 5.9 | 64.4 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 4.9 | 14.7 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 4.3 | 17.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 3.4 | 72.2 | GO:0000812 | Swr1 complex(GO:0000812) |
| 2.9 | 37.5 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 2.1 | 6.4 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 2.1 | 34.2 | GO:0031045 | dense core granule(GO:0031045) |
| 2.1 | 8.5 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 2.1 | 37.8 | GO:0031932 | TORC2 complex(GO:0031932) |
| 2.0 | 41.9 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 1.9 | 19.0 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 1.8 | 100.2 | GO:0000502 | proteasome complex(GO:0000502) |
| 1.7 | 7.0 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 1.7 | 37.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 1.6 | 137.7 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 1.2 | 5.9 | GO:0035061 | interchromatin granule(GO:0035061) |
| 1.1 | 7.9 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 1.1 | 12.9 | GO:0044754 | autolysosome(GO:0044754) |
| 1.0 | 7.3 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.8 | 9.1 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.8 | 28.8 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.6 | 4.5 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.5 | 28.4 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.4 | 7.6 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.4 | 21.4 | GO:0043034 | costamere(GO:0043034) |
| 0.4 | 74.5 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.4 | 9.0 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.4 | 10.9 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.3 | 22.9 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.3 | 2.7 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.3 | 6.6 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.3 | 12.1 | GO:0016592 | mediator complex(GO:0016592) |
| 0.3 | 47.5 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.3 | 40.4 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.3 | 15.0 | GO:0009986 | cell surface(GO:0009986) |
| 0.3 | 5.9 | GO:0031105 | septin complex(GO:0031105) |
| 0.2 | 2.7 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.2 | 2.9 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.2 | 14.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.2 | 87.1 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.2 | 12.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.2 | 64.3 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.2 | 13.6 | GO:0005903 | brush border(GO:0005903) |
| 0.2 | 23.7 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.2 | 0.8 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.1 | 4.0 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.4 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 13.3 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.1 | 32.6 | GO:0030425 | dendrite(GO:0030425) |
| 0.1 | 6.5 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.1 | 19.0 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 3.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 9.0 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 15.1 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.1 | 4.5 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.1 | 4.2 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 4.1 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 0.7 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 273.0 | GO:0005829 | cytosol(GO:0005829) |
| 0.0 | 2.9 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 1.0 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 5.5 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 0.3 | GO:0071565 | npBAF complex(GO:0071564) nBAF complex(GO:0071565) |
| 0.0 | 3.3 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 25.1 | 176.0 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 23.0 | 68.9 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 17.4 | 52.3 | GO:0050610 | oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor(GO:0016672) glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 16.5 | 49.6 | GO:0032551 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) sulfonylurea receptor binding(GO:0017098) pyrimidine ribonucleoside binding(GO:0032551) |
| 16.5 | 49.5 | GO:0098625 | methylselenol reductase activity(GO:0098625) methylseleninic acid reductase activity(GO:0098626) |
| 15.9 | 47.7 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 15.6 | 46.8 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 13.0 | 390.3 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 12.5 | 112.4 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 8.9 | 44.3 | GO:0070026 | nitric oxide binding(GO:0070026) |
| 8.4 | 67.5 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 7.4 | 80.9 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 7.3 | 79.9 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 7.2 | 28.8 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 5.6 | 16.8 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 4.9 | 29.2 | GO:0008948 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) oxaloacetate decarboxylase activity(GO:0008948) |
| 4.7 | 14.2 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 4.5 | 31.6 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 3.9 | 19.3 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 3.6 | 10.9 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 3.4 | 17.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 3.4 | 6.7 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 3.2 | 12.9 | GO:0047718 | indanol dehydrogenase activity(GO:0047718) |
| 2.7 | 37.5 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 2.6 | 18.3 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 2.5 | 37.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 2.1 | 6.4 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 1.9 | 49.0 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 1.8 | 7.2 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 1.5 | 35.4 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 1.5 | 79.7 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 1.4 | 8.5 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 1.1 | 6.8 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 1.1 | 15.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 1.1 | 23.1 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 1.1 | 25.7 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 1.0 | 14.7 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 1.0 | 7.9 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.9 | 5.6 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.9 | 5.4 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.8 | 7.0 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.7 | 10.2 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.7 | 12.9 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.7 | 7.0 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.7 | 44.4 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.6 | 9.4 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.5 | 2.2 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.5 | 6.0 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.5 | 2.2 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.5 | 89.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.5 | 2.9 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.5 | 22.9 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.5 | 22.0 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.4 | 4.4 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.4 | 9.2 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.4 | 9.1 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.4 | 6.0 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.4 | 6.6 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.4 | 17.1 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.4 | 1.2 | GO:0001181 | transcription factor activity, core RNA polymerase I binding(GO:0001181) |
| 0.4 | 9.0 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.4 | 2.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.4 | 14.9 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.3 | 4.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.3 | 10.4 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.3 | 7.3 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.3 | 2.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.3 | 2.8 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.2 | 20.1 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.2 | 12.9 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.2 | 0.9 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.2 | 1.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.2 | 3.3 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.2 | 0.8 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) aspartic-type endopeptidase inhibitor activity(GO:0019828) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.2 | 21.8 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.2 | 2.5 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.2 | 9.6 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.2 | 5.6 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.2 | 36.9 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.2 | 11.6 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.2 | 2.0 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 5.9 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 6.1 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 2.0 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 7.3 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 8.7 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 2.4 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.1 | 0.4 | GO:0004951 | cholecystokinin receptor activity(GO:0004951) |
| 0.1 | 1.3 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 0.5 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.1 | 1.9 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 7.0 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 3.9 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 1.2 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 8.4 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.1 | 2.7 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.1 | 8.8 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.1 | 6.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 6.0 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.1 | 2.6 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.1 | 12.7 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 9.5 | GO:0016758 | transferase activity, transferring hexosyl groups(GO:0016758) |
| 0.0 | 0.9 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 1.2 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 4.5 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 7.8 | GO:0003682 | chromatin binding(GO:0003682) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 145.8 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 1.8 | 25.7 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 1.8 | 119.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 1.2 | 34.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 1.1 | 97.0 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.9 | 104.0 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.9 | 59.3 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.9 | 12.3 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.7 | 36.9 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.6 | 17.1 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.6 | 2.5 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.6 | 40.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.6 | 55.9 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.6 | 39.0 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.5 | 49.1 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.4 | 7.0 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.4 | 7.9 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.4 | 28.8 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.4 | 12.4 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.3 | 7.2 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.3 | 13.5 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.2 | 14.4 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.2 | 14.8 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.2 | 2.8 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.2 | 2.4 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.2 | 2.7 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.2 | 9.8 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 2.9 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 5.0 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 4.4 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.1 | 2.0 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 3.9 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 1.9 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 14.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 4.9 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 5.3 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 2.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 2.9 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 1.6 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 3.7 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 1.1 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.5 | PID TNF PATHWAY | TNF receptor signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.6 | 200.7 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 9.0 | 699.1 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 4.1 | 111.5 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 4.0 | 140.4 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 3.6 | 116.1 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 3.3 | 88.7 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 3.1 | 37.5 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 1.8 | 37.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 1.6 | 81.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 1.5 | 46.8 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 1.1 | 12.4 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 1.0 | 10.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 1.0 | 25.7 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.9 | 14.7 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.9 | 31.6 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.8 | 15.1 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.6 | 7.5 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.5 | 9.1 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.5 | 30.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.5 | 14.2 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.5 | 5.6 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.4 | 7.6 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.4 | 7.9 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.4 | 12.3 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.4 | 9.0 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.4 | 49.9 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.3 | 5.5 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.3 | 2.7 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.3 | 9.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.3 | 22.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.3 | 7.0 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.3 | 6.6 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.2 | 7.3 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.2 | 1.5 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.2 | 5.6 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.2 | 4.9 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.2 | 12.7 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.2 | 3.9 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 5.9 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.1 | 2.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 19.3 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.1 | 14.0 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 7.0 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 6.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 13.6 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 2.9 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 3.1 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 5.5 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 8.7 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.2 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 1.1 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 2.7 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 1.7 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.2 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |