Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
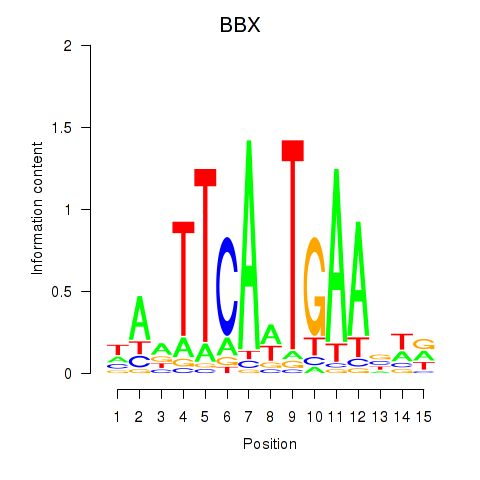
Results for BBX
Z-value: 0.71
Transcription factors associated with BBX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BBX
|
ENSG00000114439.14 | BBX high mobility group box domain containing |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| BBX | hg19_v2_chr3_+_107241783_107241850 | 0.26 | 1.1e-04 | Click! |
Activity profile of BBX motif
Sorted Z-values of BBX motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_28527152 | 14.36 |
ENST00000321830.5
|
AL353354.1
|
Uncharacterized protein |
| chr16_-_58768177 | 5.98 |
ENST00000434819.2
ENST00000245206.5 |
GOT2
|
glutamic-oxaloacetic transaminase 2, mitochondrial |
| chr15_+_65823092 | 5.47 |
ENST00000566074.1
|
PTPLAD1
|
protein tyrosine phosphatase-like A domain containing 1 |
| chr4_+_155484103 | 5.25 |
ENST00000302068.4
|
FGB
|
fibrinogen beta chain |
| chr6_-_150067696 | 4.75 |
ENST00000340413.2
ENST00000367403.3 |
NUP43
|
nucleoporin 43kDa |
| chr1_+_95616933 | 4.68 |
ENST00000604203.1
|
RP11-57H12.6
|
TMEM56-RWDD3 readthrough |
| chr5_+_74011328 | 4.03 |
ENST00000513336.1
|
HEXB
|
hexosaminidase B (beta polypeptide) |
| chr4_+_155484155 | 3.86 |
ENST00000509493.1
|
FGB
|
fibrinogen beta chain |
| chr11_-_14541872 | 3.76 |
ENST00000419365.2
ENST00000530457.1 ENST00000532256.1 ENST00000533068.1 |
PSMA1
|
proteasome (prosome, macropain) subunit, alpha type, 1 |
| chr7_-_16872932 | 3.20 |
ENST00000419572.2
ENST00000412973.1 |
AGR2
|
anterior gradient 2 |
| chr3_-_133380731 | 3.19 |
ENST00000260810.5
|
TOPBP1
|
topoisomerase (DNA) II binding protein 1 |
| chr5_-_148929848 | 3.14 |
ENST00000504676.1
ENST00000515435.1 |
CSNK1A1
|
casein kinase 1, alpha 1 |
| chrX_+_54834004 | 3.07 |
ENST00000375068.1
|
MAGED2
|
melanoma antigen family D, 2 |
| chr1_-_206785789 | 3.01 |
ENST00000437518.1
ENST00000367114.3 |
EIF2D
|
eukaryotic translation initiation factor 2D |
| chr9_-_116837249 | 2.99 |
ENST00000466610.2
|
AMBP
|
alpha-1-microglobulin/bikunin precursor |
| chr20_-_30310336 | 2.81 |
ENST00000434194.1
ENST00000376062.2 |
BCL2L1
|
BCL2-like 1 |
| chr1_+_52082751 | 2.76 |
ENST00000447887.1
ENST00000435686.2 ENST00000428468.1 ENST00000453295.1 |
OSBPL9
|
oxysterol binding protein-like 9 |
| chr3_-_168865522 | 2.74 |
ENST00000464456.1
|
MECOM
|
MDS1 and EVI1 complex locus |
| chr2_+_63816295 | 2.73 |
ENST00000539945.1
ENST00000544381.1 |
MDH1
|
malate dehydrogenase 1, NAD (soluble) |
| chr4_-_70080449 | 2.57 |
ENST00000446444.1
|
UGT2B11
|
UDP glucuronosyltransferase 2 family, polypeptide B11 |
| chr5_+_179921344 | 2.54 |
ENST00000261951.4
|
CNOT6
|
CCR4-NOT transcription complex, subunit 6 |
| chr3_+_130569429 | 2.32 |
ENST00000505330.1
ENST00000504381.1 ENST00000507488.2 ENST00000393221.4 |
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr10_+_115674530 | 2.31 |
ENST00000451472.1
|
AL162407.1
|
CDNA FLJ20147 fis, clone COL07954; HCG1781466; Uncharacterized protein |
| chr4_-_186733363 | 2.30 |
ENST00000393523.2
ENST00000393528.3 ENST00000449407.2 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr14_+_23235886 | 2.24 |
ENST00000604262.1
ENST00000431881.2 ENST00000412791.1 ENST00000358043.5 |
OXA1L
|
oxidase (cytochrome c) assembly 1-like |
| chr9_-_5833027 | 2.14 |
ENST00000339450.5
|
ERMP1
|
endoplasmic reticulum metallopeptidase 1 |
| chr22_+_32871224 | 2.10 |
ENST00000452138.1
ENST00000382058.3 ENST00000397426.1 |
FBXO7
|
F-box protein 7 |
| chr4_-_104021009 | 2.06 |
ENST00000509245.1
ENST00000296424.4 |
BDH2
|
3-hydroxybutyrate dehydrogenase, type 2 |
| chr20_-_30310693 | 2.02 |
ENST00000307677.4
ENST00000420653.1 |
BCL2L1
|
BCL2-like 1 |
| chr16_+_56672571 | 2.02 |
ENST00000290705.8
|
MT1A
|
metallothionein 1A |
| chr20_-_30310656 | 2.00 |
ENST00000376055.4
|
BCL2L1
|
BCL2-like 1 |
| chr7_+_5085452 | 1.96 |
ENST00000353796.3
ENST00000396912.1 ENST00000396904.2 |
RBAK
RBAK-RBAKDN
|
RB-associated KRAB zinc finger RBAK-RBAKDN readthrough |
| chr17_-_17184605 | 1.94 |
ENST00000268717.5
|
COPS3
|
COP9 signalosome subunit 3 |
| chr19_+_9361606 | 1.84 |
ENST00000456448.1
|
OR7E24
|
olfactory receptor, family 7, subfamily E, member 24 |
| chr4_-_186696425 | 1.75 |
ENST00000430503.1
ENST00000319454.6 ENST00000450341.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr2_-_55237484 | 1.73 |
ENST00000394609.2
|
RTN4
|
reticulon 4 |
| chrX_+_100663243 | 1.73 |
ENST00000316594.5
|
HNRNPH2
|
heterogeneous nuclear ribonucleoprotein H2 (H') |
| chr14_+_75230011 | 1.70 |
ENST00000552421.1
ENST00000325680.7 ENST00000238571.3 |
YLPM1
|
YLP motif containing 1 |
| chr12_-_10978957 | 1.68 |
ENST00000240619.2
|
TAS2R10
|
taste receptor, type 2, member 10 |
| chr4_-_159644507 | 1.40 |
ENST00000307720.3
|
PPID
|
peptidylprolyl isomerase D |
| chr4_+_108910870 | 1.39 |
ENST00000403312.1
ENST00000603302.1 ENST00000309522.3 |
HADH
|
hydroxyacyl-CoA dehydrogenase |
| chr5_+_147774275 | 1.39 |
ENST00000513826.1
|
FBXO38
|
F-box protein 38 |
| chr5_-_78809950 | 1.39 |
ENST00000334082.6
|
HOMER1
|
homer homolog 1 (Drosophila) |
| chr20_-_7921090 | 1.38 |
ENST00000378789.3
|
HAO1
|
hydroxyacid oxidase (glycolate oxidase) 1 |
| chr1_-_59043166 | 1.24 |
ENST00000371225.2
|
TACSTD2
|
tumor-associated calcium signal transducer 2 |
| chr11_+_118443098 | 1.21 |
ENST00000392859.3
ENST00000359415.4 ENST00000534182.2 ENST00000264028.4 |
ARCN1
|
archain 1 |
| chr2_+_102928009 | 1.08 |
ENST00000404917.2
ENST00000447231.1 |
IL1RL1
|
interleukin 1 receptor-like 1 |
| chr13_+_50570019 | 0.97 |
ENST00000442421.1
|
TRIM13
|
tripartite motif containing 13 |
| chr4_-_159080806 | 0.92 |
ENST00000590648.1
|
FAM198B
|
family with sequence similarity 198, member B |
| chr9_+_35161998 | 0.91 |
ENST00000396787.1
ENST00000378495.3 ENST00000378496.4 |
UNC13B
|
unc-13 homolog B (C. elegans) |
| chrX_+_54834159 | 0.90 |
ENST00000375053.2
ENST00000347546.4 ENST00000375062.4 |
MAGED2
|
melanoma antigen family D, 2 |
| chr5_+_140514782 | 0.81 |
ENST00000231134.5
|
PCDHB5
|
protocadherin beta 5 |
| chr3_-_47950745 | 0.68 |
ENST00000429422.1
|
MAP4
|
microtubule-associated protein 4 |
| chr5_+_125758813 | 0.61 |
ENST00000285689.3
ENST00000515200.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr5_+_140480083 | 0.53 |
ENST00000231130.2
|
PCDHB3
|
protocadherin beta 3 |
| chr11_+_22689648 | 0.52 |
ENST00000278187.3
|
GAS2
|
growth arrest-specific 2 |
| chr4_-_186697044 | 0.52 |
ENST00000437304.2
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr5_+_150040403 | 0.47 |
ENST00000517768.1
ENST00000297130.4 |
MYOZ3
|
myozenin 3 |
| chr6_-_161085291 | 0.34 |
ENST00000316300.5
|
LPA
|
lipoprotein, Lp(a) |
| chr6_+_31895254 | 0.34 |
ENST00000299367.5
ENST00000442278.2 |
C2
|
complement component 2 |
| chr18_-_13915530 | 0.31 |
ENST00000327606.3
|
MC2R
|
melanocortin 2 receptor (adrenocorticotropic hormone) |
| chr4_-_987217 | 0.29 |
ENST00000361661.2
ENST00000398516.2 |
SLC26A1
|
solute carrier family 26 (anion exchanger), member 1 |
| chr12_-_4754339 | 0.26 |
ENST00000228850.1
|
AKAP3
|
A kinase (PRKA) anchor protein 3 |
| chr15_+_49715293 | 0.25 |
ENST00000267843.4
ENST00000560270.1 |
FGF7
|
fibroblast growth factor 7 |
| chr9_-_13175823 | 0.23 |
ENST00000545857.1
|
MPDZ
|
multiple PDZ domain protein |
| chr6_-_25874440 | 0.23 |
ENST00000361703.6
ENST00000397060.4 |
SLC17A3
|
solute carrier family 17 (organic anion transporter), member 3 |
| chr7_+_117120017 | 0.17 |
ENST00000003084.6
ENST00000454343.1 |
CFTR
|
cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7) |
| chr13_+_33160553 | 0.16 |
ENST00000315596.10
|
PDS5B
|
PDS5, regulator of cohesion maintenance, homolog B (S. cerevisiae) |
| chr12_-_100378006 | 0.10 |
ENST00000547776.2
ENST00000329257.7 ENST00000547010.1 |
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr2_+_101437487 | 0.07 |
ENST00000427413.1
ENST00000542504.1 |
NPAS2
|
neuronal PAS domain protein 2 |
| chr6_-_49712123 | 0.06 |
ENST00000263045.4
|
CRISP3
|
cysteine-rich secretory protein 3 |
| chr12_-_8765446 | 0.06 |
ENST00000537228.1
ENST00000229335.6 |
AICDA
|
activation-induced cytidine deaminase |
| chr14_+_94492674 | 0.02 |
ENST00000203664.5
ENST00000553723.1 |
OTUB2
|
OTU domain, ubiquitin aldehyde binding 2 |
| chr2_+_232135245 | 0.02 |
ENST00000446447.1
|
ARMC9
|
armadillo repeat containing 9 |
Network of associatons between targets according to the STRING database.
First level regulatory network of BBX
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 6.0 | GO:0006106 | fumarate metabolic process(GO:0006106) aspartate catabolic process(GO:0006533) L-kynurenine metabolic process(GO:0097052) |
| 1.0 | 6.8 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.9 | 9.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.8 | 4.0 | GO:0007619 | courtship behavior(GO:0007619) |
| 0.7 | 2.2 | GO:0043461 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.6 | 3.2 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.6 | 4.0 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.5 | 3.2 | GO:1903899 | positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.5 | 1.4 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.4 | 3.1 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.4 | 3.0 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.4 | 2.5 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.4 | 2.3 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.4 | 1.4 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.3 | 1.2 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.2 | 2.7 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.2 | 0.9 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 0.2 | 3.0 | GO:0006787 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.2 | 1.7 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.2 | 1.4 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.2 | 2.1 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.2 | 1.2 | GO:0090191 | negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) |
| 0.2 | 4.6 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.2 | 2.1 | GO:1903599 | positive regulation of mitophagy(GO:1903599) |
| 0.1 | 1.9 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 0.7 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 4.7 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.1 | 0.3 | GO:0060437 | lung growth(GO:0060437) |
| 0.1 | 0.5 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.1 | 2.7 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.1 | 2.8 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.1 | 2.0 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.1 | 2.6 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.1 | 1.7 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 0.3 | GO:0050428 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.1 | 1.0 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.0 | 1.3 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 1.4 | GO:0010976 | positive regulation of neuron projection development(GO:0010976) |
| 0.0 | 3.8 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.0 | 1.4 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.0 | 0.2 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.1 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.3 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.3 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 9.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.5 | 6.8 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.4 | 2.1 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.3 | 3.8 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.3 | 0.9 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.3 | 4.7 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.3 | 3.1 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.2 | 3.2 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.1 | 1.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 1.8 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 1.7 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 1.0 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 4.3 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 3.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 1.9 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 6.0 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 2.2 | GO:0032592 | integral component of mitochondrial membrane(GO:0032592) |
| 0.0 | 4.0 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 6.4 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 1.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 2.7 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 1.4 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 2.8 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 3.0 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.2 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 2.7 | GO:0043209 | myelin sheath(GO:0043209) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 6.0 | GO:0080130 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.7 | 3.0 | GO:0019862 | IgA binding(GO:0019862) |
| 0.7 | 2.1 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.6 | 6.8 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.5 | 2.7 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.5 | 4.0 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.5 | 1.4 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.2 | 4.6 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.2 | 0.5 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.2 | 1.7 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 2.5 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 3.8 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 1.4 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 0.5 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.1 | 1.4 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.1 | 1.0 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 9.1 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.1 | 2.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 2.8 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 1.4 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.1 | 2.6 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 1.6 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 2.7 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.1 | 0.9 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 3.0 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.2 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 1.5 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.3 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.3 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 2.1 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 3.4 | GO:0008022 | protein C-terminus binding(GO:0008022) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 6.8 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.2 | 9.1 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 3.1 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.1 | 3.2 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 1.7 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 1.4 | PID CMYB PATHWAY | C-MYB transcription factor network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 9.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.2 | 6.8 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.2 | 6.0 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.2 | 4.0 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 3.1 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.1 | 4.7 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.1 | 1.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 2.6 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 1.4 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 2.5 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 3.8 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 2.7 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 2.3 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.3 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 1.7 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.3 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 1.7 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |