Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
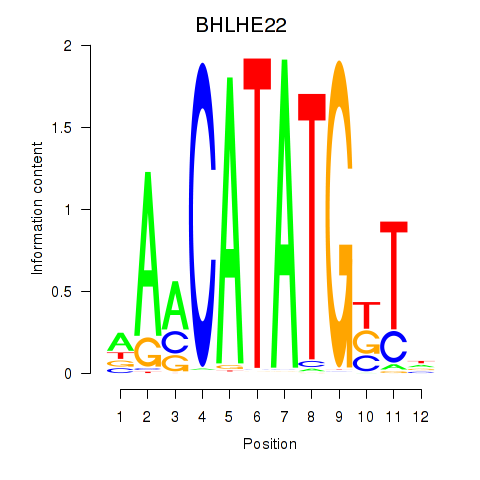
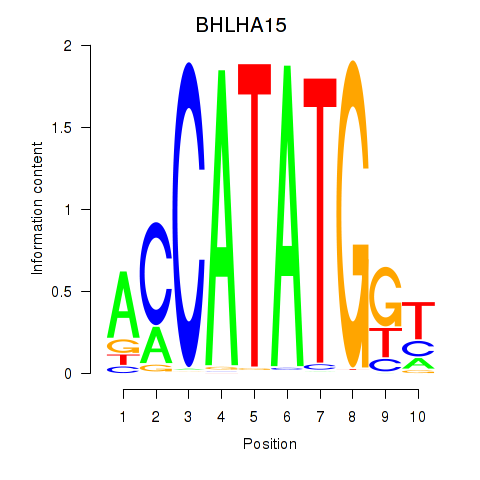
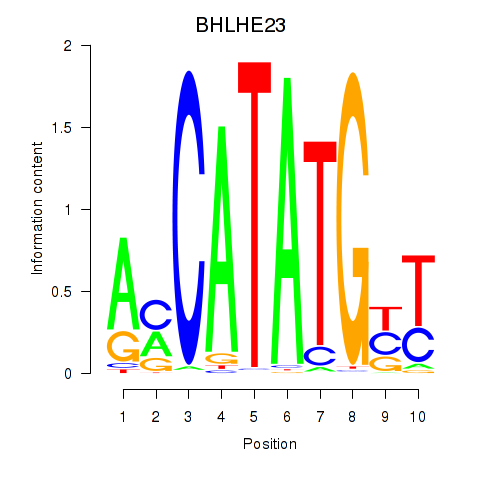
Results for BHLHE22_BHLHA15_BHLHE23
Z-value: 0.21
Transcription factors associated with BHLHE22_BHLHA15_BHLHE23
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BHLHE22
|
ENSG00000180828.1 | basic helix-loop-helix family member e22 |
|
BHLHA15
|
ENSG00000180535.3 | basic helix-loop-helix family member a15 |
|
BHLHE23
|
ENSG00000125533.4 | basic helix-loop-helix family member e23 |
Activity profile of BHLHE22_BHLHA15_BHLHE23 motif
Sorted Z-values of BHLHE22_BHLHA15_BHLHE23 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_121379739 | 18.86 |
ENST00000428394.2
ENST00000314583.3 |
HCLS1
|
hematopoietic cell-specific Lyn substrate 1 |
| chrX_+_128913906 | 12.09 |
ENST00000356892.3
|
SASH3
|
SAM and SH3 domain containing 3 |
| chr10_+_5135981 | 9.66 |
ENST00000380554.3
|
AKR1C3
|
aldo-keto reductase family 1, member C3 |
| chr1_+_244214577 | 8.78 |
ENST00000358704.4
|
ZBTB18
|
zinc finger and BTB domain containing 18 |
| chr5_-_111312622 | 7.71 |
ENST00000395634.3
|
NREP
|
neuronal regeneration related protein |
| chr19_-_54876558 | 7.62 |
ENST00000391742.2
ENST00000434277.2 |
LAIR1
|
leukocyte-associated immunoglobulin-like receptor 1 |
| chr3_-_18480260 | 5.90 |
ENST00000454909.2
|
SATB1
|
SATB homeobox 1 |
| chr11_+_65405556 | 5.55 |
ENST00000534313.1
ENST00000533361.1 ENST00000526137.1 |
SIPA1
|
signal-induced proliferation-associated 1 |
| chr6_-_112115074 | 5.35 |
ENST00000368667.2
|
FYN
|
FYN oncogene related to SRC, FGR, YES |
| chr6_+_135502501 | 5.07 |
ENST00000527615.1
ENST00000420123.2 ENST00000525369.1 ENST00000528774.1 ENST00000534121.1 ENST00000534044.1 ENST00000533624.1 |
MYB
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr3_-_114477787 | 4.65 |
ENST00000464560.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr19_+_18208603 | 4.60 |
ENST00000262811.6
|
MAST3
|
microtubule associated serine/threonine kinase 3 |
| chr3_-_114477962 | 4.53 |
ENST00000471418.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr7_-_55930443 | 4.31 |
ENST00000388975.3
|
SEPT14
|
septin 14 |
| chr20_+_57226841 | 4.18 |
ENST00000358029.4
ENST00000361830.3 |
STX16
|
syntaxin 16 |
| chrX_+_107288197 | 3.58 |
ENST00000415430.3
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chrX_+_107288239 | 3.42 |
ENST00000217957.5
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr7_-_27205136 | 2.86 |
ENST00000396345.1
ENST00000343483.6 |
HOXA9
|
homeobox A9 |
| chr14_-_107283278 | 2.83 |
ENST00000390639.2
|
IGHV7-81
|
immunoglobulin heavy variable 7-81 (non-functional) |
| chr4_+_160188889 | 2.77 |
ENST00000264431.4
|
RAPGEF2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr1_+_50569575 | 2.27 |
ENST00000371827.1
|
ELAVL4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr8_-_23315190 | 2.23 |
ENST00000356206.6
ENST00000358689.4 ENST00000417069.2 |
ENTPD4
|
ectonucleoside triphosphate diphosphohydrolase 4 |
| chr5_-_169725231 | 1.87 |
ENST00000046794.5
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) |
| chr5_+_156607829 | 1.79 |
ENST00000422843.3
|
ITK
|
IL2-inducible T-cell kinase |
| chr10_+_51565188 | 1.45 |
ENST00000430396.2
ENST00000374087.4 ENST00000414907.2 |
NCOA4
|
nuclear receptor coactivator 4 |
| chr10_+_51565108 | 1.39 |
ENST00000438493.1
ENST00000452682.1 |
NCOA4
|
nuclear receptor coactivator 4 |
| chr1_+_104293028 | 1.39 |
ENST00000370079.3
|
AMY1C
|
amylase, alpha 1C (salivary) |
| chr6_+_69942298 | 1.33 |
ENST00000238918.8
|
BAI3
|
brain-specific angiogenesis inhibitor 3 |
| chr3_+_113251143 | 1.29 |
ENST00000264852.4
ENST00000393830.3 |
SIDT1
|
SID1 transmembrane family, member 1 |
| chr4_-_153456153 | 1.22 |
ENST00000603548.1
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr13_+_49551020 | 1.16 |
ENST00000541916.1
|
FNDC3A
|
fibronectin type III domain containing 3A |
| chrX_+_105855160 | 1.16 |
ENST00000372544.2
ENST00000372548.4 |
CXorf57
|
chromosome X open reading frame 57 |
| chr9_+_2158485 | 1.12 |
ENST00000417599.1
ENST00000382185.1 ENST00000382183.1 |
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr2_+_219110149 | 1.11 |
ENST00000456575.1
|
ARPC2
|
actin related protein 2/3 complex, subunit 2, 34kDa |
| chr4_-_74088800 | 1.11 |
ENST00000509867.2
|
ANKRD17
|
ankyrin repeat domain 17 |
| chr9_+_2158443 | 1.04 |
ENST00000302401.3
ENST00000324954.5 ENST00000423555.1 ENST00000382186.1 |
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr1_+_159272111 | 1.00 |
ENST00000368114.1
|
FCER1A
|
Fc fragment of IgE, high affinity I, receptor for; alpha polypeptide |
| chr19_-_37019562 | 0.98 |
ENST00000523638.1
|
ZNF260
|
zinc finger protein 260 |
| chr1_+_110881945 | 0.96 |
ENST00000602849.1
ENST00000487146.2 |
RBM15
|
RNA binding motif protein 15 |
| chr18_-_52989525 | 0.94 |
ENST00000457482.3
|
TCF4
|
transcription factor 4 |
| chr1_-_205091115 | 0.92 |
ENST00000264515.6
ENST00000367164.1 |
RBBP5
|
retinoblastoma binding protein 5 |
| chr19_-_11450249 | 0.91 |
ENST00000222120.3
|
RAB3D
|
RAB3D, member RAS oncogene family |
| chr15_-_55700522 | 0.84 |
ENST00000564092.1
ENST00000310958.6 |
CCPG1
|
cell cycle progression 1 |
| chr1_+_12524965 | 0.81 |
ENST00000471923.1
|
VPS13D
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr3_-_150264272 | 0.79 |
ENST00000491660.1
ENST00000487153.1 ENST00000239944.2 |
SERP1
|
stress-associated endoplasmic reticulum protein 1 |
| chr2_-_163099885 | 0.73 |
ENST00000443424.1
|
FAP
|
fibroblast activation protein, alpha |
| chr22_+_36113919 | 0.72 |
ENST00000249044.2
|
APOL5
|
apolipoprotein L, 5 |
| chr6_+_37400974 | 0.65 |
ENST00000455891.1
ENST00000373451.4 |
CMTR1
|
cap methyltransferase 1 |
| chr7_+_116502527 | 0.59 |
ENST00000361183.3
|
CAPZA2
|
capping protein (actin filament) muscle Z-line, alpha 2 |
| chr16_-_4896205 | 0.55 |
ENST00000589389.1
|
GLYR1
|
glyoxylate reductase 1 homolog (Arabidopsis) |
| chr4_+_114214125 | 0.54 |
ENST00000509550.1
|
ANK2
|
ankyrin 2, neuronal |
| chr1_+_170633047 | 0.51 |
ENST00000239461.6
ENST00000497230.2 |
PRRX1
|
paired related homeobox 1 |
| chr4_+_144354644 | 0.50 |
ENST00000512843.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr8_+_21823726 | 0.47 |
ENST00000433566.4
|
XPO7
|
exportin 7 |
| chr7_-_38403077 | 0.42 |
ENST00000426402.2
|
TRGV2
|
T cell receptor gamma variable 2 |
| chr16_+_72142195 | 0.41 |
ENST00000563819.1
ENST00000567142.2 |
DHX38
|
DEAH (Asp-Glu-Ala-His) box polypeptide 38 |
| chr4_+_2814011 | 0.41 |
ENST00000502260.1
ENST00000435136.2 |
SH3BP2
|
SH3-domain binding protein 2 |
| chr17_+_7211280 | 0.38 |
ENST00000419711.2
ENST00000571955.1 ENST00000573714.1 |
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr2_-_163100045 | 0.38 |
ENST00000188790.4
|
FAP
|
fibroblast activation protein, alpha |
| chr3_+_137728842 | 0.36 |
ENST00000183605.5
|
CLDN18
|
claudin 18 |
| chr12_-_30887948 | 0.36 |
ENST00000433722.2
|
CAPRIN2
|
caprin family member 2 |
| chr2_-_183387283 | 0.35 |
ENST00000435564.1
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chrX_+_107334895 | 0.35 |
ENST00000372232.3
ENST00000345734.3 ENST00000372254.3 |
ATG4A
|
autophagy related 4A, cysteine peptidase |
| chr15_-_55700457 | 0.34 |
ENST00000442196.3
ENST00000563171.1 ENST00000425574.3 |
CCPG1
|
cell cycle progression 1 |
| chr7_+_116502605 | 0.33 |
ENST00000458284.2
ENST00000490693.1 |
CAPZA2
|
capping protein (actin filament) muscle Z-line, alpha 2 |
| chr2_-_182545603 | 0.32 |
ENST00000295108.3
|
NEUROD1
|
neuronal differentiation 1 |
| chr15_-_65503801 | 0.29 |
ENST00000261883.4
|
CILP
|
cartilage intermediate layer protein, nucleotide pyrophosphohydrolase |
| chr4_-_87028478 | 0.28 |
ENST00000515400.1
ENST00000395157.3 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr7_-_41742697 | 0.28 |
ENST00000242208.4
|
INHBA
|
inhibin, beta A |
| chr8_-_67976509 | 0.27 |
ENST00000518747.1
|
COPS5
|
COP9 signalosome subunit 5 |
| chr17_+_60758814 | 0.25 |
ENST00000579432.1
ENST00000446119.2 |
MRC2
|
mannose receptor, C type 2 |
| chr12_-_40013553 | 0.24 |
ENST00000308666.3
|
ABCD2
|
ATP-binding cassette, sub-family D (ALD), member 2 |
| chr1_+_180897269 | 0.24 |
ENST00000367587.1
|
KIAA1614
|
KIAA1614 |
| chr11_-_44972476 | 0.23 |
ENST00000527685.1
ENST00000308212.5 |
TP53I11
|
tumor protein p53 inducible protein 11 |
| chr3_-_33700589 | 0.21 |
ENST00000461133.3
ENST00000496954.2 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr7_-_97881429 | 0.20 |
ENST00000420697.1
ENST00000379795.3 ENST00000415086.1 ENST00000542604.1 ENST00000447648.2 |
TECPR1
|
tectonin beta-propeller repeat containing 1 |
| chr15_-_55700216 | 0.19 |
ENST00000569205.1
|
CCPG1
|
cell cycle progression 1 |
| chr4_-_70505358 | 0.19 |
ENST00000457664.2
ENST00000604629.1 ENST00000604021.1 |
UGT2A2
|
UDP glucuronosyltransferase 2 family, polypeptide A2 |
| chr19_-_52227221 | 0.19 |
ENST00000222115.1
ENST00000540069.2 |
HAS1
|
hyaluronan synthase 1 |
| chr10_-_48416849 | 0.18 |
ENST00000249598.1
|
GDF2
|
growth differentiation factor 2 |
| chr12_-_108714412 | 0.18 |
ENST00000412676.1
ENST00000550573.1 |
CMKLR1
|
chemokine-like receptor 1 |
| chr2_-_183387064 | 0.16 |
ENST00000536095.1
ENST00000331935.6 ENST00000358139.2 ENST00000456212.1 |
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chrX_-_15402498 | 0.16 |
ENST00000297904.3
|
FIGF
|
c-fos induced growth factor (vascular endothelial growth factor D) |
| chr2_-_165424973 | 0.13 |
ENST00000543549.1
|
GRB14
|
growth factor receptor-bound protein 14 |
| chr4_+_2813946 | 0.13 |
ENST00000442312.2
|
SH3BP2
|
SH3-domain binding protein 2 |
| chr8_-_38239732 | 0.10 |
ENST00000534155.1
ENST00000433384.2 ENST00000317025.8 ENST00000316985.3 |
WHSC1L1
|
Wolf-Hirschhorn syndrome candidate 1-like 1 |
| chr4_-_159094194 | 0.10 |
ENST00000592057.1
ENST00000585682.1 ENST00000393807.5 |
FAM198B
|
family with sequence similarity 198, member B |
| chr2_+_181845843 | 0.10 |
ENST00000602710.1
|
UBE2E3
|
ubiquitin-conjugating enzyme E2E 3 |
| chr4_+_74301880 | 0.09 |
ENST00000395792.2
ENST00000226359.2 |
AFP
|
alpha-fetoprotein |
| chr9_+_125796806 | 0.09 |
ENST00000373642.1
|
GPR21
|
G protein-coupled receptor 21 |
| chr10_+_18629628 | 0.09 |
ENST00000377329.4
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr3_-_33700933 | 0.08 |
ENST00000480013.1
|
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr8_+_67976593 | 0.07 |
ENST00000262210.5
ENST00000412460.1 |
CSPP1
|
centrosome and spindle pole associated protein 1 |
| chr10_-_94257512 | 0.07 |
ENST00000371581.5
|
IDE
|
insulin-degrading enzyme |
| chr4_+_55095264 | 0.06 |
ENST00000257290.5
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr1_+_100810575 | 0.05 |
ENST00000542213.1
|
CDC14A
|
cell division cycle 14A |
| chr11_-_44972418 | 0.04 |
ENST00000525680.1
ENST00000528290.1 ENST00000530035.1 |
TP53I11
|
tumor protein p53 inducible protein 11 |
| chr14_+_24540046 | 0.04 |
ENST00000397016.2
ENST00000537691.1 ENST00000560356.1 ENST00000558450.1 |
CPNE6
|
copine VI (neuronal) |
| chr15_-_63448973 | 0.04 |
ENST00000462430.1
|
RPS27L
|
ribosomal protein S27-like |
| chr5_+_175299743 | 0.02 |
ENST00000502265.1
|
CPLX2
|
complexin 2 |
| chr18_+_32290218 | 0.01 |
ENST00000348997.5
ENST00000588949.1 ENST00000597599.1 |
DTNA
|
dystrobrevin, alpha |
Network of associatons between targets according to the STRING database.
First level regulatory network of BHLHE22_BHLHA15_BHLHE23
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 9.7 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 1.7 | 5.1 | GO:1990922 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 1.1 | 5.6 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 1.1 | 18.9 | GO:0045651 | positive regulation of macrophage differentiation(GO:0045651) |
| 0.9 | 2.8 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.6 | 1.1 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 0.5 | 12.1 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.5 | 5.3 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.4 | 2.2 | GO:0009191 | ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.4 | 5.9 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.4 | 7.3 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.3 | 1.0 | GO:0001812 | positive regulation of type I hypersensitivity(GO:0001812) |
| 0.3 | 1.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.3 | 1.4 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.3 | 2.9 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.2 | 4.2 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.2 | 1.2 | GO:2000639 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.2 | 1.3 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.2 | 9.2 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.2 | 0.9 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.2 | 0.5 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.2 | 1.8 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.1 | 1.0 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.7 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.1 | 0.3 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.1 | 0.3 | GO:0060278 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.1 | 2.8 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 0.5 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.1 | 1.1 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.1 | 0.8 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.1 | 1.2 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 2.8 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.3 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 7.3 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.0 | 0.3 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.2 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.0 | 0.9 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.3 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.0 | 0.1 | GO:0072276 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.0 | 0.3 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.2 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.1 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.0 | 1.9 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 0.3 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.9 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.0 | 0.5 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.0 | 0.2 | GO:0010759 | positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.0 | 0.1 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.2 | GO:0030213 | negative regulation of fibroblast migration(GO:0010764) hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 5.0 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 7.0 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.0 | 2.2 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 0.2 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.2 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) |
| 0.4 | 1.9 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.4 | 1.1 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.2 | 2.8 | GO:0044754 | autolysosome(GO:0044754) |
| 0.2 | 1.2 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.1 | 5.9 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.1 | 7.9 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 1.1 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 0.3 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.1 | 2.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 0.9 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 0.9 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 4.2 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 7.6 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.1 | 0.9 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 2.8 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 19.9 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 7.1 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 5.1 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 2.5 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.2 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.8 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.5 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.5 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.3 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.9 | GO:0001669 | acrosomal vesicle(GO:0001669) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 9.7 | GO:0047718 | enone reductase activity(GO:0035671) androsterone dehydrogenase activity(GO:0047023) indanol dehydrogenase activity(GO:0047718) |
| 0.9 | 5.3 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.6 | 2.8 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.5 | 5.1 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.4 | 2.2 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.2 | 1.3 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.2 | 0.5 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.2 | 1.2 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.1 | 0.3 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 0.7 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.1 | 1.0 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) IgE binding(GO:0019863) |
| 0.1 | 19.4 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 4.2 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 0.9 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 2.3 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 0.2 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 9.7 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 0.9 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 2.8 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 0.7 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.1 | 1.1 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 5.9 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.3 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 2.5 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.5 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 2.8 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.3 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.9 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.2 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.3 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 1.8 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.1 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 5.0 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 4.3 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.5 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.2 | GO:0017070 | U6 snRNA binding(GO:0017070) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 28.9 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 5.1 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 5.9 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 7.4 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 3.8 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.2 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 2.2 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.3 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.3 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 4.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 1.4 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 5.6 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 5.9 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 3.7 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 1.2 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 5.1 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.3 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.5 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |