Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
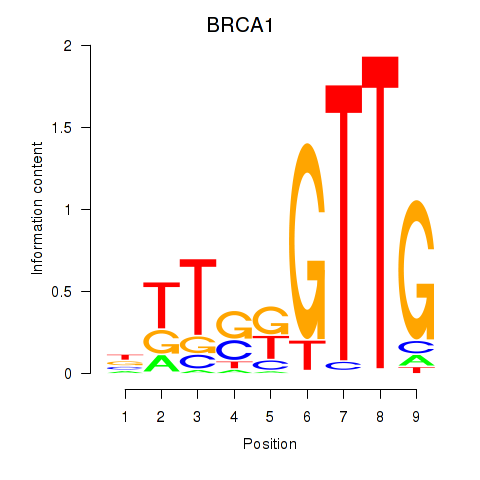
Results for BRCA1
Z-value: 0.41
Transcription factors associated with BRCA1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BRCA1
|
ENSG00000012048.15 | BRCA1 DNA repair associated |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| BRCA1 | hg19_v2_chr17_-_41277467_41277510 | 0.29 | 1.5e-05 | Click! |
Activity profile of BRCA1 motif
Sorted Z-values of BRCA1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_60097524 | 6.65 |
ENST00000342503.4
|
RTN1
|
reticulon 1 |
| chr14_-_60097297 | 6.07 |
ENST00000395090.1
|
RTN1
|
reticulon 1 |
| chr1_+_10271674 | 5.25 |
ENST00000377086.1
|
KIF1B
|
kinesin family member 1B |
| chr6_+_123100620 | 4.56 |
ENST00000368444.3
|
FABP7
|
fatty acid binding protein 7, brain |
| chr8_-_81083341 | 4.24 |
ENST00000519303.2
|
TPD52
|
tumor protein D52 |
| chr5_-_68665084 | 3.56 |
ENST00000509462.1
|
TAF9
|
TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 32kDa |
| chr2_+_223725652 | 3.54 |
ENST00000357430.3
ENST00000392066.3 |
ACSL3
|
acyl-CoA synthetase long-chain family member 3 |
| chr1_-_170043709 | 3.24 |
ENST00000367767.1
ENST00000361580.2 ENST00000538366.1 |
KIFAP3
|
kinesin-associated protein 3 |
| chr5_+_140186647 | 3.15 |
ENST00000512229.2
ENST00000356878.4 ENST00000530339.1 |
PCDHA4
|
protocadherin alpha 4 |
| chr1_+_159141397 | 2.39 |
ENST00000368124.4
ENST00000368125.4 ENST00000416746.1 |
CADM3
|
cell adhesion molecule 3 |
| chr5_-_68665469 | 2.28 |
ENST00000217893.5
|
TAF9
|
TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 32kDa |
| chr6_+_42981922 | 2.25 |
ENST00000326974.4
ENST00000244670.8 |
KLHDC3
|
kelch domain containing 3 |
| chr7_-_23510086 | 2.16 |
ENST00000258729.3
|
IGF2BP3
|
insulin-like growth factor 2 mRNA binding protein 3 |
| chr5_-_68665296 | 2.09 |
ENST00000512152.1
ENST00000503245.1 ENST00000512561.1 ENST00000380822.4 |
TAF9
|
TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 32kDa |
| chr11_+_117947724 | 2.08 |
ENST00000534111.1
|
TMPRSS4
|
transmembrane protease, serine 4 |
| chr11_+_117947782 | 2.03 |
ENST00000522307.1
ENST00000523251.1 ENST00000437212.3 ENST00000522824.1 ENST00000522151.1 |
TMPRSS4
|
transmembrane protease, serine 4 |
| chr3_-_127455200 | 1.88 |
ENST00000398101.3
|
MGLL
|
monoglyceride lipase |
| chr4_-_48782259 | 1.80 |
ENST00000507711.1
ENST00000358350.4 ENST00000537810.1 ENST00000264319.7 |
FRYL
|
FRY-like |
| chr18_-_49557 | 1.73 |
ENST00000308911.6
|
RP11-683L23.1
|
Tubulin beta-8 chain-like protein LOC260334 |
| chr3_-_176914238 | 1.72 |
ENST00000430069.1
ENST00000428970.1 |
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1 |
| chr11_-_71823266 | 1.71 |
ENST00000538919.1
ENST00000539395.1 ENST00000542531.1 |
ANAPC15
|
anaphase promoting complex subunit 15 |
| chr12_-_12491608 | 1.50 |
ENST00000545735.1
|
MANSC1
|
MANSC domain containing 1 |
| chr7_+_73868439 | 1.32 |
ENST00000424337.2
|
GTF2IRD1
|
GTF2I repeat domain containing 1 |
| chr1_-_173020056 | 1.28 |
ENST00000239468.2
ENST00000404377.3 |
TNFSF18
|
tumor necrosis factor (ligand) superfamily, member 18 |
| chr1_-_227505289 | 1.28 |
ENST00000366765.3
|
CDC42BPA
|
CDC42 binding protein kinase alpha (DMPK-like) |
| chr13_-_31038370 | 1.27 |
ENST00000399489.1
ENST00000339872.4 |
HMGB1
|
high mobility group box 1 |
| chr13_+_30002846 | 1.25 |
ENST00000542829.1
|
MTUS2
|
microtubule associated tumor suppressor candidate 2 |
| chr10_+_92631709 | 1.21 |
ENST00000413330.1
ENST00000277882.3 |
RPP30
|
ribonuclease P/MRP 30kDa subunit |
| chr19_+_45394477 | 1.20 |
ENST00000252487.5
ENST00000405636.2 ENST00000592434.1 ENST00000426677.2 ENST00000589649.1 |
TOMM40
|
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chrX_-_10851762 | 1.16 |
ENST00000380785.1
ENST00000380787.1 |
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr17_-_41738931 | 1.16 |
ENST00000329168.3
ENST00000549132.1 |
MEOX1
|
mesenchyme homeobox 1 |
| chrX_+_10124977 | 1.15 |
ENST00000380833.4
|
CLCN4
|
chloride channel, voltage-sensitive 4 |
| chr11_-_27722021 | 1.13 |
ENST00000356660.4
ENST00000418212.1 ENST00000533246.1 |
BDNF
|
brain-derived neurotrophic factor |
| chr8_-_116681221 | 1.07 |
ENST00000395715.3
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr20_-_48530230 | 1.05 |
ENST00000422556.1
|
SPATA2
|
spermatogenesis associated 2 |
| chr17_+_74372662 | 1.03 |
ENST00000591651.1
ENST00000545180.1 |
SPHK1
|
sphingosine kinase 1 |
| chr10_+_28822636 | 0.97 |
ENST00000442148.1
ENST00000448193.1 |
WAC
|
WW domain containing adaptor with coiled-coil |
| chr10_-_102289611 | 0.95 |
ENST00000299166.4
ENST00000370320.4 ENST00000531258.1 ENST00000370322.1 ENST00000535773.1 |
NDUFB8
SEC31B
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 8, 19kDa SEC31 homolog B (S. cerevisiae) |
| chr14_-_75179774 | 0.92 |
ENST00000555249.1
ENST00000556202.1 ENST00000356357.4 ENST00000338772.5 |
AREL1
AC007956.1
|
apoptosis resistant E3 ubiquitin protein ligase 1 Full-length cDNA 5-PRIME end of clone CS0CAP004YO05 of Thymus of Homo sapiens (human); Uncharacterized protein |
| chr10_-_69597810 | 0.90 |
ENST00000483798.2
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr20_+_9049682 | 0.85 |
ENST00000334005.3
ENST00000378473.3 |
PLCB4
|
phospholipase C, beta 4 |
| chr8_+_22462532 | 0.81 |
ENST00000389279.3
|
CCAR2
|
cell cycle and apoptosis regulator 2 |
| chr18_-_23671139 | 0.77 |
ENST00000579061.1
ENST00000542420.2 |
SS18
|
synovial sarcoma translocation, chromosome 18 |
| chr6_-_42981651 | 0.77 |
ENST00000244711.3
|
MEA1
|
male-enhanced antigen 1 |
| chr2_-_152118352 | 0.77 |
ENST00000331426.5
|
RBM43
|
RNA binding motif protein 43 |
| chr15_-_88799948 | 0.74 |
ENST00000394480.2
|
NTRK3
|
neurotrophic tyrosine kinase, receptor, type 3 |
| chr10_-_95209 | 0.74 |
ENST00000332708.5
ENST00000309812.4 |
TUBB8
|
tubulin, beta 8 class VIII |
| chr17_-_7082861 | 0.67 |
ENST00000269299.3
|
ASGR1
|
asialoglycoprotein receptor 1 |
| chr15_+_67430339 | 0.64 |
ENST00000439724.3
|
SMAD3
|
SMAD family member 3 |
| chr10_-_61900762 | 0.63 |
ENST00000355288.2
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr17_-_41739283 | 0.59 |
ENST00000393661.2
ENST00000318579.4 |
MEOX1
|
mesenchyme homeobox 1 |
| chr17_-_42019836 | 0.50 |
ENST00000225992.3
|
PPY
|
pancreatic polypeptide |
| chr1_-_160232312 | 0.50 |
ENST00000440682.1
|
DCAF8
|
DDB1 and CUL4 associated factor 8 |
| chr19_+_46850320 | 0.49 |
ENST00000391919.1
|
PPP5C
|
protein phosphatase 5, catalytic subunit |
| chr14_+_32030582 | 0.49 |
ENST00000550649.1
ENST00000281081.7 |
NUBPL
|
nucleotide binding protein-like |
| chr18_+_55888767 | 0.49 |
ENST00000431212.2
ENST00000586268.1 ENST00000587190.1 |
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr8_-_16859690 | 0.48 |
ENST00000180166.5
|
FGF20
|
fibroblast growth factor 20 |
| chr17_-_60142609 | 0.46 |
ENST00000397786.2
|
MED13
|
mediator complex subunit 13 |
| chr1_-_217250231 | 0.42 |
ENST00000493748.1
ENST00000463665.1 |
ESRRG
|
estrogen-related receptor gamma |
| chr19_+_46850251 | 0.41 |
ENST00000012443.4
|
PPP5C
|
protein phosphatase 5, catalytic subunit |
| chr4_-_140005443 | 0.40 |
ENST00000510408.1
ENST00000420916.2 ENST00000358635.3 |
ELF2
|
E74-like factor 2 (ets domain transcription factor) |
| chr10_-_95504 | 0.34 |
ENST00000447903.2
|
TUBB8
|
tubulin, beta 8 class VIII |
| chr1_+_63063152 | 0.29 |
ENST00000371129.3
|
ANGPTL3
|
angiopoietin-like 3 |
| chr3_+_185303962 | 0.29 |
ENST00000296257.5
|
SENP2
|
SUMO1/sentrin/SMT3 specific peptidase 2 |
| chr5_+_139493665 | 0.26 |
ENST00000331327.3
|
PURA
|
purine-rich element binding protein A |
| chr17_+_21730180 | 0.23 |
ENST00000584398.1
|
UBBP4
|
ubiquitin B pseudogene 4 |
| chr12_-_117318788 | 0.19 |
ENST00000550505.1
|
HRK
|
harakiri, BCL2 interacting protein (contains only BH3 domain) |
| chr12_-_117319236 | 0.17 |
ENST00000257572.5
|
HRK
|
harakiri, BCL2 interacting protein (contains only BH3 domain) |
| chr4_+_619347 | 0.16 |
ENST00000255622.6
|
PDE6B
|
phosphodiesterase 6B, cGMP-specific, rod, beta |
| chr11_+_47291645 | 0.16 |
ENST00000395336.3
ENST00000402192.2 |
MADD
|
MAP-kinase activating death domain |
| chrX_+_49644470 | 0.13 |
ENST00000508866.2
|
USP27X
|
ubiquitin specific peptidase 27, X-linked |
| chr9_+_35792151 | 0.11 |
ENST00000342694.2
|
NPR2
|
natriuretic peptide receptor B/guanylate cyclase B (atrionatriuretic peptide receptor B) |
| chr13_-_50018140 | 0.08 |
ENST00000410043.1
ENST00000347776.5 |
CAB39L
|
calcium binding protein 39-like |
| chr13_+_30002741 | 0.07 |
ENST00000380808.2
|
MTUS2
|
microtubule associated tumor suppressor candidate 2 |
| chr20_-_34025999 | 0.05 |
ENST00000374369.3
|
GDF5
|
growth differentiation factor 5 |
| chr13_-_50018241 | 0.05 |
ENST00000409308.1
|
CAB39L
|
calcium binding protein 39-like |
| chr17_-_56296580 | 0.05 |
ENST00000313863.6
ENST00000546108.1 ENST00000337050.7 ENST00000393119.2 |
MKS1
|
Meckel syndrome, type 1 |
| chr11_+_47291193 | 0.02 |
ENST00000428807.1
ENST00000402799.1 ENST00000406482.1 ENST00000349238.3 ENST00000311027.5 ENST00000407859.3 ENST00000395344.3 ENST00000444117.1 |
MADD
|
MAP-kinase activating death domain |
| chr7_-_99716952 | 0.02 |
ENST00000523306.1
ENST00000344095.4 ENST00000417349.1 ENST00000493322.1 ENST00000520135.1 ENST00000418432.2 ENST00000460673.2 ENST00000452041.1 ENST00000452438.2 ENST00000451699.1 ENST00000453269.2 |
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr1_-_160232291 | 0.01 |
ENST00000368074.1
ENST00000447377.1 |
DCAF8
|
DDB1 and CUL4 associated factor 8 |
Network of associatons between targets according to the STRING database.
First level regulatory network of BRCA1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.3 | GO:1904647 | response to rotenone(GO:1904647) |
| 1.2 | 3.5 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 1.1 | 3.2 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.6 | 7.9 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.4 | 1.7 | GO:0001757 | somite specification(GO:0001757) sclerotome development(GO:0061056) |
| 0.4 | 1.1 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.3 | 1.0 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.3 | 1.3 | GO:2000320 | negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.3 | 1.3 | GO:0002840 | plasmacytoid dendritic cell activation(GO:0002270) T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) regulation of restriction endodeoxyribonuclease activity(GO:0032072) negative regulation of apoptotic cell clearance(GO:2000426) |
| 0.3 | 1.7 | GO:0060613 | fat pad development(GO:0060613) |
| 0.3 | 1.9 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.3 | 1.3 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.2 | 0.7 | GO:0048687 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.2 | 1.0 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.2 | 1.1 | GO:0007056 | spindle assembly involved in female meiosis(GO:0007056) |
| 0.2 | 4.6 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.2 | 0.6 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.2 | 0.9 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.1 | 0.8 | GO:1901978 | positive regulation of cell cycle checkpoint(GO:1901978) |
| 0.1 | 0.6 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.1 | 1.8 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 0.5 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 1.2 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 0.5 | GO:1904338 | regulation of dopaminergic neuron differentiation(GO:1904338) |
| 0.1 | 1.7 | GO:1903504 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.1 | 1.2 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 1.2 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 2.3 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.0 | 0.3 | GO:0010519 | negative regulation of phospholipase activity(GO:0010519) |
| 0.0 | 2.4 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.3 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.2 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 1.1 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.3 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.1 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 4.0 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 3.2 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.2 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.0 | 2.2 | GO:0042035 | regulation of cytokine biosynthetic process(GO:0042035) |
| 0.0 | 0.5 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.0 | 1.4 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.4 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.1 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.0 | 0.4 | GO:0050855 | regulation of B cell receptor signaling pathway(GO:0050855) |
| 0.0 | 1.3 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.5 | GO:0055090 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.0 | 4.6 | GO:0006898 | receptor-mediated endocytosis(GO:0006898) |
| 0.0 | 0.9 | GO:0043647 | inositol phosphate metabolic process(GO:0043647) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 7.9 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.8 | 3.2 | GO:1990075 | kinesin II complex(GO:0016939) periciliary membrane compartment(GO:1990075) |
| 0.3 | 0.8 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.2 | 1.2 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.2 | 1.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 0.9 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 1.1 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.1 | 5.3 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.1 | 0.6 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 12.5 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 1.7 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 3.2 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 3.5 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 1.7 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.6 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.9 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 2.4 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.3 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 1.0 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.5 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.5 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 5.8 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 0.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 1.3 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 1.8 | GO:0030427 | site of polarized growth(GO:0030427) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 7.9 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.4 | 1.1 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.2 | 1.3 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.2 | 3.5 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.2 | 1.0 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 8.5 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 1.9 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 1.2 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 0.6 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.1 | 0.3 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 0.7 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.1 | 1.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 1.7 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 0.3 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.1 | 4.1 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 0.5 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 0.4 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.1 | 2.2 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 1.8 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 0.7 | GO:0005030 | GPI-linked ephrin receptor activity(GO:0005004) neurotrophin receptor activity(GO:0005030) |
| 0.1 | 0.9 | GO:0061659 | ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.1 | 1.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.9 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.5 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.9 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.5 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 1.5 | GO:0032813 | tumor necrosis factor receptor superfamily binding(GO:0032813) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 1.0 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 1.7 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 1.3 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.8 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.3 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.6 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:1990380 | Lys63-specific deubiquitinase activity(GO:0061578) Lys48-specific deubiquitinase activity(GO:1990380) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 7.9 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 1.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.0 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.6 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 2.4 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.9 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 1.3 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 1.7 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.3 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.5 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 8.0 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.1 | 3.2 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 1.9 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 1.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 1.7 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 4.2 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.5 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 1.0 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 2.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.6 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 1.2 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 2.0 | REACTOME DIABETES PATHWAYS | Genes involved in Diabetes pathways |
| 0.0 | 0.9 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.0 | 1.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |