Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for CDC5L
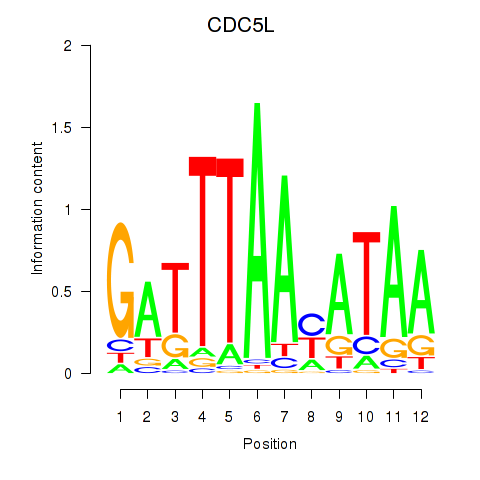
Z-value: 0.65
Transcription factors associated with CDC5L
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CDC5L
|
ENSG00000096401.7 | cell division cycle 5 like |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CDC5L | hg19_v2_chr6_+_44355257_44355315 | -0.34 | 4.0e-07 | Click! |
Activity profile of CDC5L motif
Sorted Z-values of CDC5L motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_69597810 | 18.70 |
ENST00000483798.2
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr6_-_32557610 | 10.71 |
ENST00000360004.5
|
HLA-DRB1
|
major histocompatibility complex, class II, DR beta 1 |
| chr16_-_71610985 | 10.48 |
ENST00000355962.4
|
TAT
|
tyrosine aminotransferase |
| chr12_-_10542617 | 10.06 |
ENST00000240618.6
|
KLRK1
|
killer cell lectin-like receptor subfamily K, member 1 |
| chr4_+_155484155 | 9.23 |
ENST00000509493.1
|
FGB
|
fibrinogen beta chain |
| chr12_-_53601055 | 7.99 |
ENST00000552972.1
ENST00000422257.3 ENST00000267082.5 |
ITGB7
|
integrin, beta 7 |
| chr10_-_69597915 | 7.96 |
ENST00000225171.2
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr11_-_118213331 | 7.55 |
ENST00000392884.2
|
CD3D
|
CD3d molecule, delta (CD3-TCR complex) |
| chr19_-_17185848 | 7.04 |
ENST00000593360.1
|
HAUS8
|
HAUS augmin-like complex, subunit 8 |
| chr12_-_91546926 | 7.04 |
ENST00000550758.1
|
DCN
|
decorin |
| chr6_-_33041378 | 6.99 |
ENST00000428995.1
|
HLA-DPA1
|
major histocompatibility complex, class II, DP alpha 1 |
| chr15_-_42076229 | 6.71 |
ENST00000597767.1
|
AC073657.1
|
Uncharacterized protein |
| chr6_-_32498046 | 6.70 |
ENST00000374975.3
|
HLA-DRB5
|
major histocompatibility complex, class II, DR beta 5 |
| chr12_-_53601000 | 6.48 |
ENST00000338737.4
ENST00000549086.2 |
ITGB7
|
integrin, beta 7 |
| chr12_-_91573249 | 6.32 |
ENST00000550099.1
ENST00000546391.1 ENST00000551354.1 |
DCN
|
decorin |
| chr11_-_118213455 | 6.21 |
ENST00000300692.4
|
CD3D
|
CD3d molecule, delta (CD3-TCR complex) |
| chr9_-_73029540 | 6.09 |
ENST00000377126.2
|
KLF9
|
Kruppel-like factor 9 |
| chr11_-_114466477 | 6.08 |
ENST00000375478.3
|
NXPE4
|
neurexophilin and PC-esterase domain family, member 4 |
| chr14_-_21516590 | 6.01 |
ENST00000555026.1
|
NDRG2
|
NDRG family member 2 |
| chr13_-_38172863 | 6.01 |
ENST00000541481.1
ENST00000379743.4 ENST00000379742.4 ENST00000379749.4 ENST00000541179.1 ENST00000379747.4 |
POSTN
|
periostin, osteoblast specific factor |
| chr4_+_155484103 | 5.92 |
ENST00000302068.4
|
FGB
|
fibrinogen beta chain |
| chr2_+_90248739 | 5.79 |
ENST00000468879.1
|
IGKV1D-43
|
immunoglobulin kappa variable 1D-43 |
| chr5_+_54398463 | 5.74 |
ENST00000274306.6
|
GZMA
|
granzyme A (granzyme 1, cytotoxic T-lymphocyte-associated serine esterase 3) |
| chr1_+_152635854 | 5.65 |
ENST00000368784.1
|
LCE2D
|
late cornified envelope 2D |
| chr2_+_127413481 | 5.65 |
ENST00000259254.4
|
GYPC
|
glycophorin C (Gerbich blood group) |
| chr17_+_56315936 | 5.65 |
ENST00000543544.1
|
LPO
|
lactoperoxidase |
| chr11_+_31391381 | 5.44 |
ENST00000465995.1
ENST00000536040.1 |
DNAJC24
|
DnaJ (Hsp40) homolog, subfamily C, member 24 |
| chr1_+_244998602 | 5.42 |
ENST00000411948.2
|
COX20
|
COX20 cytochrome C oxidase assembly factor |
| chr8_-_86253888 | 5.28 |
ENST00000522389.1
ENST00000432364.2 ENST00000517618.1 |
CA1
|
carbonic anhydrase I |
| chr2_-_100939195 | 5.26 |
ENST00000393437.3
|
LONRF2
|
LON peptidase N-terminal domain and ring finger 2 |
| chr1_+_25598989 | 5.17 |
ENST00000454452.2
|
RHD
|
Rh blood group, D antigen |
| chr9_-_93405352 | 4.95 |
ENST00000375765.3
|
DIRAS2
|
DIRAS family, GTP-binding RAS-like 2 |
| chr1_+_158985457 | 4.94 |
ENST00000567661.1
ENST00000474473.1 |
IFI16
|
interferon, gamma-inducible protein 16 |
| chr11_-_118213360 | 4.81 |
ENST00000529594.1
|
CD3D
|
CD3d molecule, delta (CD3-TCR complex) |
| chr12_-_91573316 | 4.79 |
ENST00000393155.1
|
DCN
|
decorin |
| chr5_+_54320078 | 4.76 |
ENST00000231009.2
|
GZMK
|
granzyme K (granzyme 3; tryptase II) |
| chr2_+_189839046 | 4.68 |
ENST00000304636.3
ENST00000317840.5 |
COL3A1
|
collagen, type III, alpha 1 |
| chr2_+_102721023 | 4.65 |
ENST00000409589.1
ENST00000409329.1 |
IL1R1
|
interleukin 1 receptor, type I |
| chr14_-_106725723 | 4.65 |
ENST00000390609.2
|
IGHV3-23
|
immunoglobulin heavy variable 3-23 |
| chr12_+_133757995 | 4.51 |
ENST00000536435.2
ENST00000228289.5 ENST00000541211.2 ENST00000500625.3 ENST00000539248.2 ENST00000542711.2 ENST00000536899.2 ENST00000542986.2 ENST00000537565.1 ENST00000541975.2 |
ZNF268
|
zinc finger protein 268 |
| chr6_-_52705641 | 4.45 |
ENST00000370989.2
|
GSTA5
|
glutathione S-transferase alpha 5 |
| chr17_-_2996290 | 4.45 |
ENST00000331459.1
|
OR1D2
|
olfactory receptor, family 1, subfamily D, member 2 |
| chr4_-_68749745 | 4.42 |
ENST00000283916.6
|
TMPRSS11D
|
transmembrane protease, serine 11D |
| chr1_+_152974218 | 4.13 |
ENST00000331860.3
ENST00000443178.1 ENST00000295367.4 |
SPRR3
|
small proline-rich protein 3 |
| chr11_-_114466471 | 4.06 |
ENST00000424261.2
|
NXPE4
|
neurexophilin and PC-esterase domain family, member 4 |
| chr3_+_101443476 | 4.05 |
ENST00000327230.4
ENST00000494050.1 |
CEP97
|
centrosomal protein 97kDa |
| chr14_-_99737822 | 4.02 |
ENST00000345514.2
ENST00000443726.2 |
BCL11B
|
B-cell CLL/lymphoma 11B (zinc finger protein) |
| chr6_+_161123270 | 3.88 |
ENST00000366924.2
ENST00000308192.9 ENST00000418964.1 |
PLG
|
plasminogen |
| chr5_+_156607829 | 3.86 |
ENST00000422843.3
|
ITK
|
IL2-inducible T-cell kinase |
| chr2_+_166095898 | 3.84 |
ENST00000424833.1
ENST00000375437.2 ENST00000357398.3 |
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit |
| chr1_-_226926864 | 3.81 |
ENST00000429204.1
ENST00000366784.1 |
ITPKB
|
inositol-trisphosphate 3-kinase B |
| chr12_-_371994 | 3.81 |
ENST00000343164.4
ENST00000436453.1 ENST00000445055.2 ENST00000546319.1 |
SLC6A13
|
solute carrier family 6 (neurotransmitter transporter), member 13 |
| chr15_+_32933866 | 3.80 |
ENST00000300175.4
ENST00000413748.2 ENST00000494364.1 ENST00000497208.1 |
SCG5
|
secretogranin V (7B2 protein) |
| chr14_-_99737565 | 3.78 |
ENST00000357195.3
|
BCL11B
|
B-cell CLL/lymphoma 11B (zinc finger protein) |
| chr11_+_55029628 | 3.76 |
ENST00000417545.2
|
TRIM48
|
tripartite motif containing 48 |
| chr1_+_89829610 | 3.75 |
ENST00000370456.4
ENST00000535065.1 |
GBP6
|
guanylate binding protein family, member 6 |
| chr21_+_10862622 | 3.74 |
ENST00000302092.5
ENST00000559480.1 |
IGHV1OR21-1
|
immunoglobulin heavy variable 1/OR21-1 (non-functional) |
| chr2_-_175462456 | 3.72 |
ENST00000409891.1
ENST00000410117.1 |
WIPF1
|
WAS/WASL interacting protein family, member 1 |
| chr17_+_73452695 | 3.65 |
ENST00000582186.1
ENST00000582455.1 ENST00000581252.1 ENST00000579208.1 |
KIAA0195
|
KIAA0195 |
| chr2_-_89459813 | 3.63 |
ENST00000390256.2
|
IGKV6-21
|
immunoglobulin kappa variable 6-21 (non-functional) |
| chr17_+_37856214 | 3.59 |
ENST00000445658.2
|
ERBB2
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 |
| chr1_+_196621002 | 3.56 |
ENST00000367429.4
ENST00000439155.2 |
CFH
|
complement factor H |
| chr17_+_56315787 | 3.56 |
ENST00000262290.4
ENST00000421678.2 |
LPO
|
lactoperoxidase |
| chr12_-_71182695 | 3.54 |
ENST00000342084.4
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr14_+_22458631 | 3.53 |
ENST00000390444.1
|
TRAV16
|
T cell receptor alpha variable 16 |
| chr1_-_89591749 | 3.51 |
ENST00000370466.3
|
GBP2
|
guanylate binding protein 2, interferon-inducible |
| chr1_+_198608146 | 3.48 |
ENST00000367376.2
ENST00000352140.3 ENST00000594404.1 ENST00000598951.1 ENST00000530727.1 ENST00000442510.2 ENST00000367367.4 ENST00000348564.6 ENST00000367364.1 ENST00000413409.2 |
PTPRC
|
protein tyrosine phosphatase, receptor type, C |
| chr4_+_75023816 | 3.47 |
ENST00000395759.2
ENST00000331145.6 ENST00000359107.5 ENST00000325278.6 |
MTHFD2L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2-like |
| chr5_-_177207634 | 3.42 |
ENST00000513554.1
ENST00000440605.3 |
FAM153A
|
family with sequence similarity 153, member A |
| chr4_-_68749699 | 3.42 |
ENST00000545541.1
|
TMPRSS11D
|
transmembrane protease, serine 11D |
| chr10_-_73848764 | 3.41 |
ENST00000317376.4
ENST00000412663.1 |
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr6_+_32709119 | 3.41 |
ENST00000374940.3
|
HLA-DQA2
|
major histocompatibility complex, class II, DQ alpha 2 |
| chr12_-_45307711 | 3.30 |
ENST00000333837.4
ENST00000551949.1 |
NELL2
|
NEL-like 2 (chicken) |
| chr6_+_26156551 | 3.29 |
ENST00000304218.3
|
HIST1H1E
|
histone cluster 1, H1e |
| chr3_-_194072019 | 3.24 |
ENST00000429275.1
ENST00000323830.3 |
CPN2
|
carboxypeptidase N, polypeptide 2 |
| chr8_+_21911054 | 3.22 |
ENST00000519850.1
ENST00000381470.3 |
DMTN
|
dematin actin binding protein |
| chr3_+_122044084 | 3.15 |
ENST00000264474.3
ENST00000479204.1 |
CSTA
|
cystatin A (stefin A) |
| chr3_+_73110810 | 3.09 |
ENST00000533473.1
|
EBLN2
|
endogenous Bornavirus-like nucleoprotein 2 |
| chr9_-_21368075 | 3.05 |
ENST00000449498.1
|
IFNA13
|
interferon, alpha 13 |
| chr6_-_39197226 | 3.05 |
ENST00000359534.3
|
KCNK5
|
potassium channel, subfamily K, member 5 |
| chr2_+_67624430 | 3.05 |
ENST00000272342.5
|
ETAA1
|
Ewing tumor-associated antigen 1 |
| chrX_-_48056199 | 3.03 |
ENST00000311798.1
ENST00000347757.1 |
SSX5
|
synovial sarcoma, X breakpoint 5 |
| chr15_-_22448819 | 3.02 |
ENST00000604066.1
|
IGHV1OR15-1
|
immunoglobulin heavy variable 1/OR15-1 (non-functional) |
| chr9_-_21351377 | 3.02 |
ENST00000380210.1
|
IFNA6
|
interferon, alpha 6 |
| chrX_+_77166172 | 3.00 |
ENST00000343533.5
ENST00000350425.4 ENST00000341514.6 |
ATP7A
|
ATPase, Cu++ transporting, alpha polypeptide |
| chr16_+_72088376 | 3.00 |
ENST00000570083.1
ENST00000355906.5 ENST00000398131.2 ENST00000569639.1 ENST00000564499.1 ENST00000357763.4 ENST00000562526.1 ENST00000565574.1 ENST00000568417.2 ENST00000356967.5 |
HP
HPR
|
haptoglobin haptoglobin-related protein |
| chr12_+_79439405 | 2.96 |
ENST00000552744.1
|
SYT1
|
synaptotagmin I |
| chr12_-_21927736 | 2.95 |
ENST00000240662.2
|
KCNJ8
|
potassium inwardly-rectifying channel, subfamily J, member 8 |
| chr2_+_85921522 | 2.94 |
ENST00000409696.3
|
GNLY
|
granulysin |
| chr10_-_73848531 | 2.93 |
ENST00000373109.2
|
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr2_+_85921414 | 2.90 |
ENST00000263863.4
ENST00000524600.1 |
GNLY
|
granulysin |
| chr21_+_43619796 | 2.89 |
ENST00000398457.2
|
ABCG1
|
ATP-binding cassette, sub-family G (WHITE), member 1 |
| chr3_+_48282587 | 2.88 |
ENST00000354698.3
ENST00000427617.2 ENST00000412564.1 ENST00000440261.2 |
ZNF589
|
zinc finger protein 589 |
| chr17_-_76713100 | 2.85 |
ENST00000585509.1
|
CYTH1
|
cytohesin 1 |
| chr2_-_74570520 | 2.83 |
ENST00000394019.2
ENST00000346834.4 ENST00000359484.4 ENST00000423644.1 ENST00000377634.4 ENST00000436454.1 |
SLC4A5
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 5 |
| chr3_+_46412345 | 2.82 |
ENST00000292303.4
|
CCR5
|
chemokine (C-C motif) receptor 5 (gene/pseudogene) |
| chr1_+_196621156 | 2.81 |
ENST00000359637.2
|
CFH
|
complement factor H |
| chr11_+_73661364 | 2.77 |
ENST00000339764.1
|
DNAJB13
|
DnaJ (Hsp40) homolog, subfamily B, member 13 |
| chr6_-_26250835 | 2.76 |
ENST00000446824.2
|
HIST1H3F
|
histone cluster 1, H3f |
| chr4_+_170541678 | 2.69 |
ENST00000360642.3
ENST00000512813.1 |
CLCN3
|
chloride channel, voltage-sensitive 3 |
| chr18_+_77160282 | 2.68 |
ENST00000318065.5
ENST00000545796.1 ENST00000592223.1 ENST00000329101.4 ENST00000586434.1 |
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 |
| chr17_+_41003166 | 2.66 |
ENST00000308423.2
|
AOC3
|
amine oxidase, copper containing 3 |
| chr1_+_22351977 | 2.65 |
ENST00000420503.1
ENST00000416769.1 ENST00000404210.2 |
LINC00339
|
long intergenic non-protein coding RNA 339 |
| chr16_-_1843720 | 2.63 |
ENST00000415638.3
ENST00000215539.3 |
IGFALS
|
insulin-like growth factor binding protein, acid labile subunit |
| chrX_+_103031758 | 2.62 |
ENST00000303958.2
ENST00000361621.2 |
PLP1
|
proteolipid protein 1 |
| chr11_+_119076745 | 2.61 |
ENST00000264033.4
|
CBL
|
Cbl proto-oncogene, E3 ubiquitin protein ligase |
| chr3_+_178276488 | 2.60 |
ENST00000432997.1
ENST00000455865.1 |
KCNMB2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
| chr21_+_37692481 | 2.60 |
ENST00000400485.1
|
MORC3
|
MORC family CW-type zinc finger 3 |
| chr5_+_140213815 | 2.59 |
ENST00000525929.1
ENST00000378125.3 |
PCDHA7
|
protocadherin alpha 7 |
| chr14_-_106733624 | 2.55 |
ENST00000390610.2
|
IGHV1-24
|
immunoglobulin heavy variable 1-24 |
| chr17_+_15602891 | 2.53 |
ENST00000421016.1
ENST00000593105.1 ENST00000580259.1 ENST00000583566.1 ENST00000472486.1 ENST00000395894.2 ENST00000581529.1 ENST00000579694.1 ENST00000580393.1 ENST00000585194.1 ENST00000583031.1 ENST00000464847.2 |
ZNF286A
|
Homo sapiens zinc finger protein 286A (ZNF286A), transcript variant 6, mRNA. |
| chr17_+_37856253 | 2.53 |
ENST00000540147.1
ENST00000584450.1 |
ERBB2
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 |
| chr12_-_21487829 | 2.53 |
ENST00000445053.1
ENST00000452078.1 ENST00000458504.1 ENST00000422327.1 ENST00000421294.1 |
SLCO1A2
|
solute carrier organic anion transporter family, member 1A2 |
| chr11_-_13517565 | 2.53 |
ENST00000282091.1
ENST00000529816.1 |
PTH
|
parathyroid hormone |
| chr4_+_128554081 | 2.51 |
ENST00000335251.6
ENST00000296461.5 |
INTU
|
inturned planar cell polarity protein |
| chr13_+_23755099 | 2.51 |
ENST00000537476.1
|
SGCG
|
sarcoglycan, gamma (35kDa dystrophin-associated glycoprotein) |
| chr15_-_20170354 | 2.51 |
ENST00000338912.5
|
IGHV1OR15-9
|
immunoglobulin heavy variable 1/OR15-9 (non-functional) |
| chr17_-_47755338 | 2.50 |
ENST00000508805.1
ENST00000515508.2 ENST00000451526.2 ENST00000507970.1 |
SPOP
|
speckle-type POZ protein |
| chr1_+_152956549 | 2.48 |
ENST00000307122.2
|
SPRR1A
|
small proline-rich protein 1A |
| chr2_+_210517895 | 2.47 |
ENST00000447185.1
|
MAP2
|
microtubule-associated protein 2 |
| chr11_-_236326 | 2.45 |
ENST00000525237.1
ENST00000532956.1 ENST00000525319.1 ENST00000524564.1 ENST00000382743.4 |
SIRT3
|
sirtuin 3 |
| chr9_-_6470375 | 2.44 |
ENST00000355513.4
|
C9orf38
|
chromosome 9 open reading frame 38 |
| chr11_-_130786400 | 2.38 |
ENST00000265909.4
|
SNX19
|
sorting nexin 19 |
| chr13_+_111855414 | 2.37 |
ENST00000375737.5
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr10_+_96698406 | 2.36 |
ENST00000260682.6
|
CYP2C9
|
cytochrome P450, family 2, subfamily C, polypeptide 9 |
| chr2_+_66662510 | 2.35 |
ENST00000272369.9
ENST00000407092.2 |
MEIS1
|
Meis homeobox 1 |
| chr5_+_147443534 | 2.34 |
ENST00000398454.1
ENST00000359874.3 ENST00000508733.1 ENST00000256084.7 |
SPINK5
|
serine peptidase inhibitor, Kazal type 5 |
| chr5_-_127418755 | 2.33 |
ENST00000501702.2
ENST00000501173.2 ENST00000514573.1 ENST00000499346.2 ENST00000606251.1 |
CTC-228N24.3
|
CTC-228N24.3 |
| chr5_-_138210977 | 2.32 |
ENST00000274711.6
ENST00000521094.2 |
LRRTM2
|
leucine rich repeat transmembrane neuronal 2 |
| chr6_+_27107053 | 2.31 |
ENST00000354348.2
|
HIST1H4I
|
histone cluster 1, H4i |
| chr2_-_110962544 | 2.30 |
ENST00000355301.4
ENST00000445609.2 ENST00000417665.1 ENST00000418527.1 ENST00000316534.4 ENST00000393272.3 |
NPHP1
|
nephronophthisis 1 (juvenile) |
| chr10_-_74114714 | 2.30 |
ENST00000338820.3
ENST00000394903.2 ENST00000444643.2 |
DNAJB12
|
DnaJ (Hsp40) homolog, subfamily B, member 12 |
| chr1_+_38022513 | 2.29 |
ENST00000296218.7
|
DNALI1
|
dynein, axonemal, light intermediate chain 1 |
| chr14_-_106668095 | 2.28 |
ENST00000390606.2
|
IGHV3-20
|
immunoglobulin heavy variable 3-20 |
| chr12_+_9980069 | 2.28 |
ENST00000354855.3
ENST00000324214.4 ENST00000279544.3 |
KLRF1
|
killer cell lectin-like receptor subfamily F, member 1 |
| chr1_+_156338993 | 2.26 |
ENST00000368249.1
ENST00000368246.2 ENST00000537040.1 ENST00000400992.2 ENST00000255013.3 ENST00000451864.2 |
RHBG
|
Rh family, B glycoprotein (gene/pseudogene) |
| chr6_+_35996859 | 2.25 |
ENST00000472333.1
|
MAPK14
|
mitogen-activated protein kinase 14 |
| chrX_-_15402498 | 2.25 |
ENST00000297904.3
|
FIGF
|
c-fos induced growth factor (vascular endothelial growth factor D) |
| chr15_+_42066632 | 2.25 |
ENST00000457542.2
ENST00000221214.6 ENST00000260357.7 ENST00000456763.2 |
MAPKBP1
|
mitogen-activated protein kinase binding protein 1 |
| chr15_-_99789736 | 2.23 |
ENST00000560235.1
ENST00000394132.2 ENST00000560860.1 ENST00000558078.1 ENST00000394136.1 ENST00000262074.4 ENST00000558613.1 ENST00000394130.1 ENST00000560772.1 |
TTC23
|
tetratricopeptide repeat domain 23 |
| chr22_-_40929812 | 2.23 |
ENST00000422851.1
|
MKL1
|
megakaryoblastic leukemia (translocation) 1 |
| chr11_+_64794875 | 2.21 |
ENST00000377244.3
ENST00000534637.1 ENST00000524831.1 |
SNX15
|
sorting nexin 15 |
| chr15_-_20193370 | 2.21 |
ENST00000558565.2
|
IGHV3OR15-7
|
immunoglobulin heavy variable 3/OR15-7 (pseudogene) |
| chr7_+_138943265 | 2.20 |
ENST00000483726.1
|
UBN2
|
ubinuclein 2 |
| chr1_+_241695670 | 2.20 |
ENST00000366557.4
|
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr4_+_118955500 | 2.17 |
ENST00000296499.5
|
NDST3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr14_-_71001708 | 2.17 |
ENST00000256389.3
|
ADAM20
|
ADAM metallopeptidase domain 20 |
| chr5_+_161495038 | 2.16 |
ENST00000393933.4
|
GABRG2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr4_-_76957214 | 2.16 |
ENST00000306621.3
|
CXCL11
|
chemokine (C-X-C motif) ligand 11 |
| chr11_-_83984231 | 2.14 |
ENST00000330014.6
ENST00000537455.1 ENST00000376106.3 ENST00000418306.2 ENST00000531015.1 |
DLG2
|
discs, large homolog 2 (Drosophila) |
| chr2_-_113542063 | 2.13 |
ENST00000263339.3
|
IL1A
|
interleukin 1, alpha |
| chr5_+_140800638 | 2.12 |
ENST00000398587.2
ENST00000518882.1 |
PCDHGA11
|
protocadherin gamma subfamily A, 11 |
| chr20_+_1246908 | 2.10 |
ENST00000381873.3
ENST00000381867.1 |
SNPH
|
syntaphilin |
| chr7_-_113559104 | 2.10 |
ENST00000284601.3
|
PPP1R3A
|
protein phosphatase 1, regulatory subunit 3A |
| chr12_+_18414446 | 2.09 |
ENST00000433979.1
|
PIK3C2G
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 gamma |
| chr14_-_80677815 | 2.09 |
ENST00000557125.1
ENST00000555750.1 |
DIO2
|
deiodinase, iodothyronine, type II |
| chr7_+_96634850 | 2.07 |
ENST00000518156.2
|
DLX6
|
distal-less homeobox 6 |
| chr17_-_7493390 | 2.06 |
ENST00000538513.2
ENST00000570788.1 ENST00000250055.2 |
SOX15
|
SRY (sex determining region Y)-box 15 |
| chr1_-_150738261 | 2.06 |
ENST00000448301.2
ENST00000368985.3 |
CTSS
|
cathepsin S |
| chr2_+_241807870 | 2.05 |
ENST00000307503.3
|
AGXT
|
alanine-glyoxylate aminotransferase |
| chr6_-_27840099 | 2.05 |
ENST00000328488.2
|
HIST1H3I
|
histone cluster 1, H3i |
| chr3_+_151531810 | 2.02 |
ENST00000232892.7
|
AADAC
|
arylacetamide deacetylase |
| chr17_+_37856299 | 2.02 |
ENST00000269571.5
|
ERBB2
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 |
| chr2_+_128848740 | 2.01 |
ENST00000375990.3
|
UGGT1
|
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr2_+_113931513 | 2.01 |
ENST00000245796.6
ENST00000441564.3 |
PSD4
|
pleckstrin and Sec7 domain containing 4 |
| chr1_+_66458072 | 2.01 |
ENST00000423207.2
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr8_+_24772455 | 2.00 |
ENST00000433454.2
|
NEFM
|
neurofilament, medium polypeptide |
| chr14_-_69261310 | 2.00 |
ENST00000336440.3
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr17_-_64225508 | 1.99 |
ENST00000205948.6
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr10_+_51549498 | 1.98 |
ENST00000358559.2
ENST00000298239.6 |
MSMB
|
microseminoprotein, beta- |
| chr1_+_9599540 | 1.98 |
ENST00000302692.6
|
SLC25A33
|
solute carrier family 25 (pyrimidine nucleotide carrier), member 33 |
| chr4_-_48082192 | 1.95 |
ENST00000507351.1
|
TXK
|
TXK tyrosine kinase |
| chr14_+_70918874 | 1.95 |
ENST00000603540.1
|
ADAM21
|
ADAM metallopeptidase domain 21 |
| chr4_+_80584903 | 1.94 |
ENST00000506460.1
|
RP11-452C8.1
|
RP11-452C8.1 |
| chr6_+_41604747 | 1.93 |
ENST00000419164.1
ENST00000373051.2 |
MDFI
|
MyoD family inhibitor |
| chr4_+_106631966 | 1.92 |
ENST00000360505.5
ENST00000510865.1 ENST00000509336.1 |
GSTCD
|
glutathione S-transferase, C-terminal domain containing |
| chr14_-_106453155 | 1.90 |
ENST00000390594.2
|
IGHV1-2
|
immunoglobulin heavy variable 1-2 |
| chr1_+_110254850 | 1.90 |
ENST00000369812.5
ENST00000256593.3 ENST00000369813.1 |
GSTM5
|
glutathione S-transferase mu 5 |
| chr20_+_43104508 | 1.88 |
ENST00000262605.4
ENST00000372904.3 |
TTPAL
|
tocopherol (alpha) transfer protein-like |
| chr20_-_18477862 | 1.88 |
ENST00000337227.4
|
RBBP9
|
retinoblastoma binding protein 9 |
| chr19_-_33360647 | 1.86 |
ENST00000590341.1
ENST00000587772.1 ENST00000023064.4 |
SLC7A9
|
solute carrier family 7 (amino acid transporter light chain, bo,+ system), member 9 |
| chr17_-_59668550 | 1.85 |
ENST00000521764.1
|
NACA2
|
nascent polypeptide-associated complex alpha subunit 2 |
| chrX_+_138612889 | 1.84 |
ENST00000218099.2
ENST00000394090.2 |
F9
|
coagulation factor IX |
| chr2_+_54350316 | 1.84 |
ENST00000606865.1
|
ACYP2
|
acylphosphatase 2, muscle type |
| chr2_-_163100045 | 1.84 |
ENST00000188790.4
|
FAP
|
fibroblast activation protein, alpha |
| chr4_+_164265035 | 1.81 |
ENST00000338566.3
|
NPY5R
|
neuropeptide Y receptor Y5 |
| chr4_-_70725856 | 1.80 |
ENST00000226444.3
|
SULT1E1
|
sulfotransferase family 1E, estrogen-preferring, member 1 |
| chr14_+_22963806 | 1.79 |
ENST00000390493.1
|
TRAJ44
|
T cell receptor alpha joining 44 |
| chr1_-_104238912 | 1.79 |
ENST00000330330.5
|
AMY1B
|
amylase, alpha 1B (salivary) |
| chr7_+_134430212 | 1.78 |
ENST00000436461.2
|
CALD1
|
caldesmon 1 |
| chr19_+_15852203 | 1.74 |
ENST00000305892.1
|
OR10H3
|
olfactory receptor, family 10, subfamily H, member 3 |
| chr2_-_222436988 | 1.72 |
ENST00000409854.1
ENST00000281821.2 ENST00000392071.4 ENST00000443796.1 |
EPHA4
|
EPH receptor A4 |
| chr6_+_29424958 | 1.71 |
ENST00000377136.1
ENST00000377133.1 |
OR2H1
|
olfactory receptor, family 2, subfamily H, member 1 |
| chr18_-_10701979 | 1.67 |
ENST00000538948.1
ENST00000285141.4 |
PIEZO2
|
piezo-type mechanosensitive ion channel component 2 |
| chr12_+_62860581 | 1.67 |
ENST00000393632.2
ENST00000393630.3 ENST00000280379.6 ENST00000546600.1 ENST00000552738.1 ENST00000393629.2 ENST00000552115.1 |
MON2
|
MON2 homolog (S. cerevisiae) |
| chr6_-_41254403 | 1.66 |
ENST00000589614.1
ENST00000334475.6 ENST00000591620.1 ENST00000244709.4 |
TREM1
|
triggering receptor expressed on myeloid cells 1 |
| chr13_+_23755054 | 1.66 |
ENST00000218867.3
|
SGCG
|
sarcoglycan, gamma (35kDa dystrophin-associated glycoprotein) |
| chrX_-_48216101 | 1.65 |
ENST00000298396.2
ENST00000376893.3 |
SSX3
|
synovial sarcoma, X breakpoint 3 |
| chr14_+_39703112 | 1.64 |
ENST00000555143.1
ENST00000280082.3 |
MIA2
|
melanoma inhibitory activity 2 |
| chr7_+_29519486 | 1.62 |
ENST00000409041.4
|
CHN2
|
chimerin 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of CDC5L
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 14.5 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 3.4 | 10.1 | GO:1901624 | negative regulation of lymphocyte chemotaxis(GO:1901624) |
| 2.1 | 10.7 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 2.0 | 6.0 | GO:1990523 | bone regeneration(GO:1990523) |
| 2.0 | 5.9 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 1.9 | 7.8 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 1.9 | 5.8 | GO:0002818 | intracellular defense response(GO:0002818) |
| 1.7 | 22.4 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 1.6 | 4.7 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 1.5 | 15.2 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 1.5 | 18.2 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 1.5 | 10.5 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 1.3 | 3.9 | GO:2000048 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 1.2 | 2.3 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 1.1 | 4.6 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 1.0 | 8.1 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 1.0 | 3.0 | GO:0071284 | cellular response to lead ion(GO:0071284) |
| 1.0 | 3.0 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 1.0 | 3.0 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 1.0 | 2.9 | GO:0009726 | detection of endogenous stimulus(GO:0009726) |
| 1.0 | 4.8 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.9 | 6.1 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.8 | 2.5 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.8 | 0.8 | GO:0035378 | carbon dioxide transmembrane transport(GO:0035378) |
| 0.8 | 3.3 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 0.8 | 2.4 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.8 | 2.3 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.7 | 3.6 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.7 | 3.6 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.7 | 2.1 | GO:2000391 | positive regulation of neutrophil extravasation(GO:2000391) |
| 0.7 | 2.1 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.7 | 0.7 | GO:0035990 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.7 | 3.5 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 0.7 | 2.8 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.7 | 2.1 | GO:0048627 | myoblast development(GO:0048627) positive regulation of G0 to G1 transition(GO:0070318) |
| 0.7 | 2.0 | GO:0006524 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) |
| 0.7 | 5.4 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.7 | 2.0 | GO:0097359 | UDP-glucosylation(GO:0097359) |
| 0.7 | 2.0 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.7 | 2.0 | GO:0048382 | mesendoderm development(GO:0048382) |
| 0.7 | 2.0 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.7 | 2.0 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.6 | 1.8 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.6 | 3.5 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.6 | 4.7 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.6 | 5.7 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.6 | 1.7 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.6 | 2.8 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.6 | 2.3 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.6 | 6.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.5 | 3.2 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.5 | 2.7 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.5 | 6.0 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.5 | 1.5 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.5 | 3.3 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.5 | 2.8 | GO:0044800 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.4 | 2.2 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.4 | 2.5 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.4 | 3.3 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.4 | 3.9 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.4 | 10.3 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.4 | 2.6 | GO:0090650 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.4 | 2.2 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.4 | 1.1 | GO:0042495 | detection of bacterial lipoprotein(GO:0042494) detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) |
| 0.4 | 1.1 | GO:0048073 | regulation of eye pigmentation(GO:0048073) positive regulation of eye pigmentation(GO:0048075) |
| 0.4 | 2.9 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.3 | 2.4 | GO:0045423 | regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045423) positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.3 | 2.7 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.3 | 8.3 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.3 | 1.7 | GO:0072672 | neutrophil extravasation(GO:0072672) |
| 0.3 | 3.8 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.3 | 4.0 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.3 | 1.9 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.3 | 2.4 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.3 | 1.8 | GO:0002865 | negative regulation of acute inflammatory response to antigenic stimulus(GO:0002865) |
| 0.3 | 3.0 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.3 | 10.0 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.3 | 7.4 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.3 | 1.5 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.2 | 2.0 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.2 | 1.5 | GO:0035672 | oligopeptide transmembrane transport(GO:0035672) |
| 0.2 | 0.7 | GO:0045643 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.2 | 1.0 | GO:0072658 | positive regulation of cell communication by electrical coupling(GO:0010650) maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.2 | 4.9 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 | 11.1 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.2 | 1.8 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.2 | 16.0 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.2 | 2.2 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.2 | 2.2 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.2 | 2.0 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.2 | 3.0 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.2 | 3.8 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.2 | 0.8 | GO:1900004 | negative regulation of serine-type endopeptidase activity(GO:1900004) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.2 | 2.3 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.2 | 2.5 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.2 | 2.1 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.2 | 9.3 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.2 | 2.3 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.2 | 3.9 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.2 | 2.0 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.2 | 2.3 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.2 | 1.5 | GO:0015705 | iodide transport(GO:0015705) |
| 0.2 | 0.3 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.2 | 2.6 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.2 | 2.6 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.2 | 0.8 | GO:0042746 | regulation of circadian sleep/wake cycle, wakefulness(GO:0010840) circadian sleep/wake cycle, wakefulness(GO:0042746) |
| 0.2 | 2.3 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) inositol lipid-mediated signaling(GO:0048017) |
| 0.2 | 2.1 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.1 | 0.9 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.1 | 0.9 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.1 | 3.3 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.1 | 1.0 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 | 0.4 | GO:0061365 | positive regulation of triglyceride lipase activity(GO:0061365) |
| 0.1 | 2.4 | GO:2000394 | positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.1 | 17.8 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.1 | 1.3 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.1 | 1.6 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.1 | 3.1 | GO:0050974 | detection of mechanical stimulus involved in sensory perception(GO:0050974) |
| 0.1 | 0.9 | GO:0086073 | cardiac muscle cell-cardiac muscle cell adhesion(GO:0086042) bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.1 | 1.6 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.1 | 1.4 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.1 | 1.9 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.1 | 0.1 | GO:0002018 | renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.1 | 2.3 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.1 | 0.3 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.1 | 7.9 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.1 | 2.2 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.1 | 4.9 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.1 | 16.5 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 0.5 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.1 | 2.6 | GO:0048147 | negative regulation of fibroblast proliferation(GO:0048147) |
| 0.1 | 0.8 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.1 | 0.4 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 4.0 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.1 | 1.3 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 1.6 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) |
| 0.1 | 1.4 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.1 | 3.6 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 3.8 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 0.9 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.1 | 0.6 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.1 | 0.3 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) negative regulation of skeletal muscle cell proliferation(GO:0014859) regulation of skeletal muscle tissue growth(GO:0048631) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.1 | 0.7 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.1 | 2.8 | GO:0018146 | keratan sulfate biosynthetic process(GO:0018146) |
| 0.1 | 2.1 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.1 | 1.0 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.1 | 0.3 | GO:0061743 | motor learning(GO:0061743) |
| 0.1 | 4.6 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.1 | 0.8 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 0.8 | GO:0008595 | tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.1 | 1.3 | GO:0030220 | platelet formation(GO:0030220) |
| 0.1 | 0.8 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.1 | 1.3 | GO:0033622 | integrin activation(GO:0033622) |
| 0.1 | 0.2 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.1 | 2.1 | GO:0039694 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.1 | 1.3 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.1 | 0.3 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.1 | 0.6 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) |
| 0.1 | 0.2 | GO:1902358 | sulfate transmembrane transport(GO:1902358) |
| 0.1 | 1.0 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.0 | 1.4 | GO:0007184 | SMAD protein import into nucleus(GO:0007184) |
| 0.0 | 0.8 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 6.9 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.4 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.4 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 1.5 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 1.8 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 0.8 | GO:0006271 | DNA strand elongation involved in DNA replication(GO:0006271) |
| 0.0 | 1.6 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.2 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.8 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 2.0 | GO:0071346 | cellular response to interferon-gamma(GO:0071346) |
| 0.0 | 0.9 | GO:0045725 | positive regulation of glycogen biosynthetic process(GO:0045725) |
| 0.0 | 1.1 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.0 | 0.3 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 1.0 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.0 | 1.0 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.0 | 0.3 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.0 | 0.2 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.6 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.5 | GO:0008105 | asymmetric protein localization(GO:0008105) |
| 0.0 | 0.3 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.0 | 0.1 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.2 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.4 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.2 | GO:0070375 | ERK5 cascade(GO:0070375) |
| 0.0 | 0.7 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.4 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.5 | GO:0044042 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) energy reserve metabolic process(GO:0006112) glucan metabolic process(GO:0044042) cellular polysaccharide metabolic process(GO:0044264) |
| 0.0 | 2.5 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 0.7 | GO:0007595 | lactation(GO:0007595) |
| 0.0 | 1.2 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 1.9 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.0 | 0.0 | GO:1901877 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 0.0 | 1.5 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.5 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.0 | 0.9 | GO:0060021 | palate development(GO:0060021) |
| 0.0 | 3.2 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 0.2 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.6 | GO:0071805 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 1.4 | GO:0050954 | sensory perception of mechanical stimulus(GO:0050954) |
| 0.0 | 0.7 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.3 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.0 | 0.8 | GO:0046834 | lipid phosphorylation(GO:0046834) |
| 0.0 | 1.1 | GO:2000257 | regulation of complement activation(GO:0030449) regulation of protein activation cascade(GO:2000257) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 14.5 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 1.7 | 18.6 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 1.4 | 4.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 1.3 | 27.8 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 1.3 | 18.2 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 1.2 | 15.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.8 | 5.8 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 0.8 | 5.0 | GO:0044218 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.8 | 7.0 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.8 | 3.0 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.6 | 3.0 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.5 | 4.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.4 | 2.4 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.4 | 4.7 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.4 | 1.1 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.4 | 2.9 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.3 | 17.2 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.3 | 2.3 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.3 | 6.4 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.3 | 3.3 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.3 | 2.4 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.3 | 2.1 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.3 | 5.0 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.3 | 2.6 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.3 | 1.1 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.3 | 1.9 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.3 | 1.3 | GO:0030430 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.2 | 1.4 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.2 | 3.3 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.2 | 2.6 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.2 | 0.9 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.2 | 10.0 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.2 | 0.6 | GO:0033150 | perinuclear theca(GO:0033011) cytoskeletal calyx(GO:0033150) |
| 0.2 | 1.5 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.1 | 2.0 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 2.9 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.1 | 1.6 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 2.0 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 1.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.9 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 3.2 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.1 | 2.0 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.1 | 3.3 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 3.0 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.1 | 27.7 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 3.2 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 1.5 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 2.1 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 6.9 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 2.3 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 0.7 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 0.6 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 2.0 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 0.4 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.1 | 0.9 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.1 | 0.6 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.1 | 2.0 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.1 | 4.6 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.1 | 2.3 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 0.4 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 2.2 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 0.2 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.1 | 5.1 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.1 | 5.7 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 1.5 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 2.8 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 4.4 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 5.9 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 10.8 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 5.9 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.5 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 3.7 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 4.0 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 1.9 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 2.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 2.3 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 1.4 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 1.5 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.2 | GO:0098576 | lumenal side of membrane(GO:0098576) |
| 0.0 | 3.4 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 4.8 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 7.5 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 1.0 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.1 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.5 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 2.5 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 2.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 2.3 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.6 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.8 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 26.7 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.8 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 3.1 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 3.6 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.0 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.0 | 4.8 | GO:0005759 | mitochondrial matrix(GO:0005759) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 10.5 | GO:0080130 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 1.2 | 4.7 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 1.2 | 3.5 | GO:0004487 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 1.1 | 6.7 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 1.0 | 3.0 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 1.0 | 2.9 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.9 | 2.7 | GO:0052594 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.8 | 2.3 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 0.7 | 10.4 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.7 | 8.0 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.7 | 2.0 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.7 | 2.0 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.7 | 5.3 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.6 | 19.3 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.6 | 3.8 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.6 | 3.8 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.6 | 1.8 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.6 | 3.6 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.6 | 3.0 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.5 | 3.3 | GO:0032564 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.5 | 3.1 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.5 | 1.5 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.5 | 3.0 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.5 | 1.5 | GO:0015440 | peptide-transporting ATPase activity(GO:0015440) |
| 0.5 | 1.5 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.5 | 3.9 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.5 | 3.4 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.5 | 2.4 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.5 | 2.7 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.4 | 2.2 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.4 | 1.2 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.4 | 2.0 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.4 | 1.1 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.4 | 1.5 | GO:0047718 | enone reductase activity(GO:0035671) androsterone dehydrogenase activity(GO:0047023) indanol dehydrogenase activity(GO:0047718) |
| 0.4 | 26.6 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.4 | 2.2 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.3 | 2.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.3 | 10.1 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.3 | 4.7 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.3 | 4.9 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.3 | 10.5 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.3 | 17.2 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.3 | 1.8 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.3 | 3.0 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.3 | 2.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.3 | 1.5 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.3 | 1.5 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.3 | 0.9 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.3 | 6.4 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.3 | 0.8 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.3 | 1.3 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.3 | 2.6 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.3 | 7.0 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.3 | 1.8 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) steroid sulfotransferase activity(GO:0050294) |
| 0.2 | 0.7 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.2 | 3.6 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.2 | 1.0 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.2 | 3.4 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.2 | 2.6 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.2 | 1.3 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.2 | 2.8 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.2 | 1.5 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.2 | 5.0 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.2 | 3.7 | GO:0005522 | profilin binding(GO:0005522) |
| 0.2 | 1.0 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.2 | 0.6 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.2 | 0.8 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.2 | 0.8 | GO:0016499 | orexin receptor activity(GO:0016499) |
| 0.2 | 4.9 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.2 | 9.3 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.2 | 0.9 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.2 | 9.2 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.2 | 1.7 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) PH domain binding(GO:0042731) |
| 0.2 | 2.2 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.2 | 2.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.2 | 1.5 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.2 | 2.8 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.2 | 2.6 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 3.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 3.7 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 1.4 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.1 | 2.0 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 2.5 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.1 | 0.6 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.1 | 0.8 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.1 | 2.2 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 13.6 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.1 | 2.4 | GO:0017136 | NAD-dependent histone deacetylase activity(GO:0017136) |
| 0.1 | 3.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 4.1 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 9.4 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 0.7 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.1 | 1.2 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.1 | 2.4 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 10.3 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 2.0 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 2.1 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 1.3 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 2.8 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.7 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.9 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.1 | 3.3 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 2.6 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 1.0 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.1 | 2.6 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 22.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 3.0 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 0.5 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.1 | 1.5 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 1.0 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 3.4 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 0.3 | GO:0008112 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 0.1 | 1.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 2.0 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 0.4 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.1 | 6.8 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.1 | 2.3 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 2.0 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 0.2 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.1 | 2.0 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.1 | 6.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.7 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 0.4 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 0.9 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 0.9 | GO:0008392 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.1 | 4.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 0.3 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.1 | 1.9 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.2 | GO:0052795 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.3 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 1.0 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.8 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 1.1 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.5 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.9 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.9 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 4.3 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.9 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 1.7 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 1.0 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.1 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 2.7 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 1.8 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 1.6 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 2.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.4 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.6 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.6 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 2.4 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 2.3 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.6 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 4.9 | GO:0060089 | receptor activity(GO:0004872) molecular transducer activity(GO:0060089) |
| 0.0 | 0.7 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.1 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.3 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 1.4 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 1.1 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 12.1 | GO:0004888 | transmembrane signaling receptor activity(GO:0004888) |
| 0.0 | 1.0 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 1.2 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.2 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 1.7 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 1.8 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.2 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 1.2 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 28.3 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.5 | 14.5 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.4 | 22.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.4 | 18.1 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.3 | 3.8 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.3 | 8.1 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.2 | 15.5 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.2 | 15.1 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.2 | 8.2 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.2 | 4.9 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 5.7 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.1 | 3.8 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 10.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 1.0 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 2.1 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 1.7 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 2.1 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.1 | 0.7 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 2.2 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 4.0 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 3.4 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 2.6 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.1 | 1.5 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 12.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 2.4 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.3 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 1.3 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 0.9 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 8.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 4.1 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.8 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.6 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 1.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.9 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 7.1 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.4 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.7 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 46.4 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.9 | 15.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.7 | 2.8 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.7 | 3.5 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.7 | 2.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.7 | 18.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.3 | 3.4 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.3 | 8.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.3 | 6.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.3 | 3.6 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.3 | 22.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.2 | 5.0 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.2 | 3.7 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.2 | 4.2 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.2 | 1.6 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.2 | 9.0 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.2 | 1.8 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.2 | 4.9 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.2 | 5.1 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.2 | 9.3 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.2 | 3.3 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.2 | 5.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 3.0 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.1 | 2.9 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 2.0 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.1 | 2.1 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 1.5 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 3.9 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 2.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 1.1 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.1 | 6.4 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 0.8 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 1.6 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 2.1 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 1.6 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 6.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 2.9 | REACTOME TCR SIGNALING | Genes involved in TCR signaling |
| 0.1 | 2.8 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.1 | 1.9 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 1.7 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 2.2 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 2.2 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 2.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 4.2 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.1 | 3.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 4.5 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 0.7 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 1.4 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.0 | 0.7 | REACTOME DAG AND IP3 SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.0 | 1.4 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 1.9 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 2.3 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 3.2 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.1 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.8 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 1.4 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.7 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 1.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.5 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.8 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 1.1 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.6 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.6 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 3.7 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.4 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.6 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.5 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.3 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.3 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 1.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |