Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
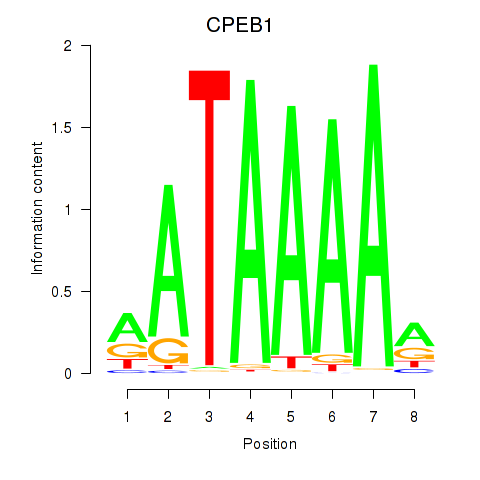
Results for CPEB1
Z-value: 0.65
Transcription factors associated with CPEB1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CPEB1
|
ENSG00000214575.5 | cytoplasmic polyadenylation element binding protein 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CPEB1 | hg19_v2_chr15_-_83240507_83240539 | -0.33 | 5.6e-07 | Click! |
Activity profile of CPEB1 motif
Sorted Z-values of CPEB1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_56078695 | 35.35 |
ENST00000416613.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr12_-_54071181 | 27.34 |
ENST00000338662.5
|
ATP5G2
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C2 (subunit 9) |
| chr2_-_151344172 | 25.35 |
ENST00000375734.2
ENST00000263895.4 ENST00000454202.1 |
RND3
|
Rho family GTPase 3 |
| chr11_-_11374904 | 23.37 |
ENST00000528848.2
|
CSNK2A3
|
casein kinase 2, alpha 3 polypeptide |
| chr1_-_32384693 | 20.74 |
ENST00000602683.1
ENST00000470404.1 |
PTP4A2
|
protein tyrosine phosphatase type IVA, member 2 |
| chr10_+_114709999 | 17.17 |
ENST00000355995.4
ENST00000545257.1 ENST00000543371.1 ENST00000536810.1 ENST00000355717.4 ENST00000538897.1 ENST00000534894.1 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr6_+_111195973 | 16.63 |
ENST00000368885.3
ENST00000368882.3 ENST00000451850.2 ENST00000368877.5 |
AMD1
|
adenosylmethionine decarboxylase 1 |
| chr17_-_38574169 | 15.86 |
ENST00000423485.1
|
TOP2A
|
topoisomerase (DNA) II alpha 170kDa |
| chr22_+_41258250 | 15.59 |
ENST00000544094.1
|
XPNPEP3
|
X-prolyl aminopeptidase (aminopeptidase P) 3, putative |
| chr3_-_141747439 | 15.58 |
ENST00000467667.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr16_-_2314222 | 15.55 |
ENST00000566397.1
|
RNPS1
|
RNA binding protein S1, serine-rich domain |
| chr4_+_174089904 | 14.66 |
ENST00000265000.4
|
GALNT7
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7) |
| chr12_-_52967600 | 14.33 |
ENST00000549343.1
ENST00000305620.2 |
KRT74
|
keratin 74 |
| chr14_+_56127989 | 14.23 |
ENST00000555573.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr19_+_36036477 | 13.79 |
ENST00000222284.5
ENST00000392204.2 |
TMEM147
|
transmembrane protein 147 |
| chr2_-_190044480 | 13.09 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr19_+_36036583 | 12.51 |
ENST00000392205.1
|
TMEM147
|
transmembrane protein 147 |
| chr3_+_142342228 | 12.36 |
ENST00000337777.3
|
PLS1
|
plastin 1 |
| chr4_-_102268628 | 12.23 |
ENST00000323055.6
ENST00000512215.1 ENST00000394854.3 |
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr11_-_27723158 | 12.15 |
ENST00000395980.2
|
BDNF
|
brain-derived neurotrophic factor |
| chr14_-_102552659 | 11.89 |
ENST00000441629.2
|
HSP90AA1
|
heat shock protein 90kDa alpha (cytosolic), class A member 1 |
| chr11_+_57529234 | 11.87 |
ENST00000360682.6
ENST00000361796.4 ENST00000529526.1 ENST00000426142.2 ENST00000399050.4 ENST00000361391.6 ENST00000361332.4 ENST00000532463.1 ENST00000529986.1 ENST00000358694.6 ENST00000532787.1 ENST00000533667.1 ENST00000532649.1 ENST00000528621.1 ENST00000530748.1 ENST00000428599.2 ENST00000527467.1 ENST00000528232.1 ENST00000531014.1 ENST00000526772.1 ENST00000529873.1 ENST00000525902.1 ENST00000532844.1 ENST00000526357.1 ENST00000530094.1 ENST00000415361.2 ENST00000532245.1 ENST00000534579.1 ENST00000526938.1 |
CTNND1
|
catenin (cadherin-associated protein), delta 1 |
| chr10_-_13390021 | 11.82 |
ENST00000537130.1
|
SEPHS1
|
selenophosphate synthetase 1 |
| chr20_+_30193083 | 11.69 |
ENST00000376112.3
ENST00000376105.3 |
ID1
|
inhibitor of DNA binding 1, dominant negative helix-loop-helix protein |
| chr3_+_159570722 | 11.62 |
ENST00000482804.1
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr2_+_181845843 | 11.48 |
ENST00000602710.1
|
UBE2E3
|
ubiquitin-conjugating enzyme E2E 3 |
| chr17_-_4852332 | 11.40 |
ENST00000572383.1
|
PFN1
|
profilin 1 |
| chrX_+_106871713 | 11.39 |
ENST00000372435.4
ENST00000372428.4 ENST00000372419.3 ENST00000543248.1 |
PRPS1
|
phosphoribosyl pyrophosphate synthetase 1 |
| chr18_-_33702078 | 11.29 |
ENST00000586829.1
|
SLC39A6
|
solute carrier family 39 (zinc transporter), member 6 |
| chr4_-_102267953 | 11.29 |
ENST00000523694.2
ENST00000507176.1 |
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chrX_-_20236970 | 10.79 |
ENST00000379548.4
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
| chr1_-_89357179 | 10.78 |
ENST00000448623.1
ENST00000418217.1 ENST00000370500.5 |
GTF2B
|
general transcription factor IIB |
| chr4_-_102268484 | 10.64 |
ENST00000394853.4
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr9_+_42671887 | 10.52 |
ENST00000456520.1
ENST00000377391.3 |
CBWD7
|
COBW domain containing 7 |
| chr10_-_126847276 | 10.41 |
ENST00000531469.1
|
CTBP2
|
C-terminal binding protein 2 |
| chr3_+_158991025 | 10.31 |
ENST00000337808.6
|
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr15_+_80364901 | 10.27 |
ENST00000560228.1
ENST00000559835.1 ENST00000559775.1 ENST00000558688.1 ENST00000560392.1 ENST00000560976.1 ENST00000558272.1 ENST00000558390.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr2_-_148779106 | 9.86 |
ENST00000416719.1
ENST00000264169.2 |
ORC4
|
origin recognition complex, subunit 4 |
| chr8_+_71485681 | 9.84 |
ENST00000391684.1
|
AC120194.1
|
AC120194.1 |
| chr12_+_28410128 | 9.83 |
ENST00000381259.1
ENST00000381256.1 |
CCDC91
|
coiled-coil domain containing 91 |
| chr18_+_3252265 | 9.74 |
ENST00000580887.1
ENST00000536605.1 |
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr17_-_39684550 | 9.68 |
ENST00000455635.1
ENST00000361566.3 |
KRT19
|
keratin 19 |
| chr15_-_60690163 | 9.55 |
ENST00000558998.1
ENST00000560165.1 ENST00000557986.1 ENST00000559780.1 ENST00000559467.1 ENST00000559956.1 ENST00000332680.4 ENST00000396024.3 ENST00000421017.2 ENST00000560466.1 ENST00000558132.1 ENST00000559113.1 ENST00000557906.1 ENST00000558558.1 ENST00000560468.1 ENST00000559370.1 ENST00000558169.1 ENST00000559725.1 ENST00000558985.1 ENST00000451270.2 |
ANXA2
|
annexin A2 |
| chr14_+_73563735 | 9.52 |
ENST00000532192.1
|
RBM25
|
RNA binding motif protein 25 |
| chr9_-_69229650 | 9.47 |
ENST00000416428.1
|
CBWD6
|
COBW domain containing 6 |
| chr6_+_44194762 | 9.40 |
ENST00000371708.1
|
SLC29A1
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1 |
| chr7_+_100464760 | 9.40 |
ENST00000200457.4
|
TRIP6
|
thyroid hormone receptor interactor 6 |
| chr6_+_119215308 | 9.36 |
ENST00000229595.5
|
ASF1A
|
anti-silencing function 1A histone chaperone |
| chr17_-_36358166 | 8.97 |
ENST00000537432.1
|
TBC1D3
|
TBC1 domain family, member 3 |
| chr1_+_84630645 | 8.95 |
ENST00000394839.2
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr5_-_140070897 | 8.85 |
ENST00000448240.1
ENST00000438307.2 ENST00000415192.2 ENST00000457527.2 ENST00000307633.3 ENST00000507746.1 ENST00000431330.2 |
HARS
|
histidyl-tRNA synthetase |
| chr11_+_35160709 | 8.84 |
ENST00000415148.2
ENST00000433354.2 ENST00000449691.2 ENST00000437706.2 ENST00000360158.4 ENST00000428726.2 ENST00000526669.2 ENST00000433892.2 ENST00000278386.6 ENST00000434472.2 ENST00000352818.4 ENST00000442151.2 |
CD44
|
CD44 molecule (Indian blood group) |
| chr8_+_104831554 | 8.80 |
ENST00000408894.2
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr9_-_86593238 | 8.79 |
ENST00000351839.3
|
HNRNPK
|
heterogeneous nuclear ribonucleoprotein K |
| chr5_-_78808617 | 8.57 |
ENST00000282260.6
ENST00000508576.1 ENST00000535690.1 |
HOMER1
|
homer homolog 1 (Drosophila) |
| chr1_+_169077172 | 8.52 |
ENST00000499679.3
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr5_-_111091948 | 8.50 |
ENST00000447165.2
|
NREP
|
neuronal regeneration related protein |
| chr10_+_114710211 | 8.40 |
ENST00000349937.2
ENST00000369397.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr3_-_69129501 | 8.26 |
ENST00000540295.1
ENST00000415609.2 ENST00000361055.4 ENST00000349511.4 |
UBA3
|
ubiquitin-like modifier activating enzyme 3 |
| chr17_-_4852243 | 8.16 |
ENST00000225655.5
|
PFN1
|
profilin 1 |
| chrX_-_100662881 | 8.05 |
ENST00000218516.3
|
GLA
|
galactosidase, alpha |
| chr19_+_36142147 | 8.03 |
ENST00000590618.1
|
COX6B1
|
cytochrome c oxidase subunit VIb polypeptide 1 (ubiquitous) |
| chr10_+_47894023 | 7.92 |
ENST00000358474.5
|
FAM21B
|
family with sequence similarity 21, member B |
| chr9_+_131452239 | 7.83 |
ENST00000372688.4
ENST00000372686.5 |
SET
|
SET nuclear oncogene |
| chr6_+_12290586 | 7.81 |
ENST00000379375.5
|
EDN1
|
endothelin 1 |
| chr9_-_70465758 | 7.80 |
ENST00000489273.1
|
CBWD5
|
COBW domain containing 5 |
| chr14_-_35344093 | 7.74 |
ENST00000382422.2
|
BAZ1A
|
bromodomain adjacent to zinc finger domain, 1A |
| chr13_-_31040060 | 7.71 |
ENST00000326004.4
ENST00000341423.5 |
HMGB1
|
high mobility group box 1 |
| chr18_-_33709268 | 7.68 |
ENST00000269187.5
ENST00000590986.1 ENST00000440549.2 |
SLC39A6
|
solute carrier family 39 (zinc transporter), member 6 |
| chr8_+_9953061 | 7.59 |
ENST00000522907.1
ENST00000528246.1 |
MSRA
|
methionine sulfoxide reductase A |
| chr7_-_27205136 | 7.41 |
ENST00000396345.1
ENST00000343483.6 |
HOXA9
|
homeobox A9 |
| chr1_+_26798955 | 7.40 |
ENST00000361427.5
|
HMGN2
|
high mobility group nucleosomal binding domain 2 |
| chr12_-_50616382 | 7.29 |
ENST00000552783.1
|
LIMA1
|
LIM domain and actin binding 1 |
| chr1_+_95616933 | 7.23 |
ENST00000604203.1
|
RP11-57H12.6
|
TMEM56-RWDD3 readthrough |
| chr2_-_175711133 | 7.21 |
ENST00000409597.1
ENST00000413882.1 |
CHN1
|
chimerin 1 |
| chr7_+_115862858 | 7.19 |
ENST00000393481.2
|
TES
|
testis derived transcript (3 LIM domains) |
| chr6_+_26104104 | 7.06 |
ENST00000377803.2
|
HIST1H4C
|
histone cluster 1, H4c |
| chr16_+_2820912 | 6.99 |
ENST00000570539.1
|
SRRM2
|
serine/arginine repetitive matrix 2 |
| chr3_+_158787041 | 6.95 |
ENST00000471575.1
ENST00000476809.1 ENST00000485419.1 |
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr4_-_111119804 | 6.93 |
ENST00000394607.3
ENST00000302274.3 |
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr2_-_207023918 | 6.87 |
ENST00000455934.2
ENST00000449699.1 ENST00000454195.1 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr1_+_44870866 | 6.86 |
ENST00000355387.2
ENST00000361799.2 |
RNF220
|
ring finger protein 220 |
| chr19_+_36119929 | 6.81 |
ENST00000588161.1
ENST00000262633.4 ENST00000592202.1 ENST00000586618.1 |
RBM42
|
RNA binding motif protein 42 |
| chr6_+_114178512 | 6.72 |
ENST00000368635.4
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate |
| chr1_+_244214577 | 6.69 |
ENST00000358704.4
|
ZBTB18
|
zinc finger and BTB domain containing 18 |
| chr15_-_56209306 | 6.69 |
ENST00000506154.1
ENST00000338963.2 ENST00000508342.1 |
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4, E3 ubiquitin protein ligase |
| chr16_+_1728257 | 6.63 |
ENST00000248098.3
ENST00000562684.1 ENST00000561516.1 ENST00000382711.5 ENST00000566742.1 |
HN1L
|
hematological and neurological expressed 1-like |
| chr6_+_21593972 | 6.59 |
ENST00000244745.1
ENST00000543472.1 |
SOX4
|
SRY (sex determining region Y)-box 4 |
| chr17_-_17480779 | 6.47 |
ENST00000395782.1
|
PEMT
|
phosphatidylethanolamine N-methyltransferase |
| chr8_+_9953214 | 6.32 |
ENST00000382490.5
|
MSRA
|
methionine sulfoxide reductase A |
| chr5_+_162932554 | 6.28 |
ENST00000321757.6
ENST00000421814.2 ENST00000518095.1 |
MAT2B
|
methionine adenosyltransferase II, beta |
| chr3_+_107241783 | 6.15 |
ENST00000415149.2
ENST00000402543.1 ENST00000325805.8 ENST00000427402.1 |
BBX
|
bobby sox homolog (Drosophila) |
| chr7_-_27219849 | 6.12 |
ENST00000396344.4
|
HOXA10
|
homeobox A10 |
| chr15_-_37390482 | 5.96 |
ENST00000559085.1
ENST00000397624.3 |
MEIS2
|
Meis homeobox 2 |
| chr4_-_48782259 | 5.89 |
ENST00000507711.1
ENST00000358350.4 ENST00000537810.1 ENST00000264319.7 |
FRYL
|
FRY-like |
| chr20_-_30310336 | 5.88 |
ENST00000434194.1
ENST00000376062.2 |
BCL2L1
|
BCL2-like 1 |
| chr5_+_135496675 | 5.72 |
ENST00000507637.1
|
SMAD5
|
SMAD family member 5 |
| chr1_-_156675535 | 5.67 |
ENST00000368221.1
|
CRABP2
|
cellular retinoic acid binding protein 2 |
| chr12_-_49412541 | 5.61 |
ENST00000547306.1
ENST00000548857.1 ENST00000551696.1 ENST00000316299.5 |
PRKAG1
|
protein kinase, AMP-activated, gamma 1 non-catalytic subunit |
| chr10_-_118764862 | 5.60 |
ENST00000260777.10
|
KIAA1598
|
KIAA1598 |
| chr10_-_118765081 | 5.56 |
ENST00000392903.2
ENST00000355371.4 |
KIAA1598
|
KIAA1598 |
| chr6_-_27114577 | 5.56 |
ENST00000356950.1
ENST00000396891.4 |
HIST1H2BK
|
histone cluster 1, H2bk |
| chr17_+_45286706 | 5.55 |
ENST00000393450.1
ENST00000572303.1 |
MYL4
|
myosin, light chain 4, alkali; atrial, embryonic |
| chr6_+_27114861 | 5.51 |
ENST00000377459.1
|
HIST1H2AH
|
histone cluster 1, H2ah |
| chr6_+_15246501 | 5.46 |
ENST00000341776.2
|
JARID2
|
jumonji, AT rich interactive domain 2 |
| chr17_+_45286387 | 5.42 |
ENST00000572316.1
ENST00000354968.1 ENST00000576874.1 ENST00000536623.2 |
MYL4
|
myosin, light chain 4, alkali; atrial, embryonic |
| chr12_-_49412588 | 5.36 |
ENST00000547082.1
ENST00000395170.3 |
PRKAG1
|
protein kinase, AMP-activated, gamma 1 non-catalytic subunit |
| chr2_-_85645545 | 5.29 |
ENST00000409275.1
|
CAPG
|
capping protein (actin filament), gelsolin-like |
| chr19_+_36120009 | 5.26 |
ENST00000589871.1
|
RBM42
|
RNA binding motif protein 42 |
| chr1_+_84630053 | 5.22 |
ENST00000394838.2
ENST00000370682.3 ENST00000432111.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr3_-_37216055 | 5.19 |
ENST00000336686.4
|
LRRFIP2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr1_+_164528866 | 5.18 |
ENST00000420696.2
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr13_+_28519343 | 5.13 |
ENST00000381026.3
|
ATP5EP2
|
ATP synthase, H+ transporting, mitochondrial F1 complex, epsilon subunit pseudogene 2 |
| chr1_-_115259337 | 5.08 |
ENST00000369535.4
|
NRAS
|
neuroblastoma RAS viral (v-ras) oncogene homolog |
| chr10_-_13390270 | 5.00 |
ENST00000378614.4
ENST00000545675.1 ENST00000327347.5 |
SEPHS1
|
selenophosphate synthetase 1 |
| chr17_+_7462103 | 4.98 |
ENST00000396545.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr3_-_33700589 | 4.97 |
ENST00000461133.3
ENST00000496954.2 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr10_-_104262460 | 4.95 |
ENST00000446605.2
ENST00000369905.4 ENST00000545684.1 |
ACTR1A
|
ARP1 actin-related protein 1 homolog A, centractin alpha (yeast) |
| chr1_-_32110467 | 4.82 |
ENST00000440872.2
ENST00000373703.4 |
PEF1
|
penta-EF-hand domain containing 1 |
| chr7_+_116654935 | 4.81 |
ENST00000432298.1
ENST00000422922.1 |
ST7
|
suppression of tumorigenicity 7 |
| chr8_+_77593474 | 4.80 |
ENST00000455469.2
ENST00000050961.6 |
ZFHX4
|
zinc finger homeobox 4 |
| chr6_+_30585486 | 4.74 |
ENST00000259873.4
ENST00000506373.2 |
MRPS18B
|
mitochondrial ribosomal protein S18B |
| chr2_+_8822113 | 4.66 |
ENST00000396290.1
ENST00000331129.3 |
ID2
|
inhibitor of DNA binding 2, dominant negative helix-loop-helix protein |
| chr6_+_108977520 | 4.64 |
ENST00000540898.1
|
FOXO3
|
forkhead box O3 |
| chr5_-_73937244 | 4.64 |
ENST00000302351.4
ENST00000510316.1 ENST00000508331.1 |
ENC1
|
ectodermal-neural cortex 1 (with BTB domain) |
| chr10_+_47894572 | 4.60 |
ENST00000355876.5
|
FAM21B
|
family with sequence similarity 21, member B |
| chr1_+_145883868 | 4.57 |
ENST00000447947.2
|
GPR89C
|
G protein-coupled receptor 89C |
| chr4_+_74735102 | 4.56 |
ENST00000395761.3
|
CXCL1
|
chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) |
| chr1_-_9129895 | 4.52 |
ENST00000473209.1
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr2_+_163175394 | 4.47 |
ENST00000446271.1
ENST00000429691.2 |
GCA
|
grancalcin, EF-hand calcium binding protein |
| chr6_-_31697977 | 4.47 |
ENST00000375787.2
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr5_-_98262240 | 4.44 |
ENST00000284049.3
|
CHD1
|
chromodomain helicase DNA binding protein 1 |
| chr17_-_27418537 | 4.42 |
ENST00000408971.2
|
TIAF1
|
TGFB1-induced anti-apoptotic factor 1 |
| chr2_-_2334888 | 4.42 |
ENST00000428368.2
ENST00000399161.2 |
MYT1L
|
myelin transcription factor 1-like |
| chr20_+_31407692 | 4.41 |
ENST00000375571.5
|
MAPRE1
|
microtubule-associated protein, RP/EB family, member 1 |
| chrX_+_123095546 | 4.40 |
ENST00000371157.3
ENST00000371145.3 ENST00000371144.3 |
STAG2
|
stromal antigen 2 |
| chr4_+_160188889 | 4.30 |
ENST00000264431.4
|
RAPGEF2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr2_-_55277654 | 4.26 |
ENST00000337526.6
ENST00000317610.7 ENST00000357732.4 |
RTN4
|
reticulon 4 |
| chr10_+_111967345 | 4.24 |
ENST00000332674.5
ENST00000453116.1 |
MXI1
|
MAX interactor 1, dimerization protein |
| chr17_+_7462031 | 4.23 |
ENST00000380535.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr11_-_62473706 | 4.21 |
ENST00000403550.1
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr10_+_102106829 | 4.20 |
ENST00000370355.2
|
SCD
|
stearoyl-CoA desaturase (delta-9-desaturase) |
| chr18_+_3252206 | 4.15 |
ENST00000578562.2
|
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr11_-_62432641 | 4.15 |
ENST00000528405.1
ENST00000524958.1 ENST00000525675.1 |
RP11-831H9.11
C11orf48
|
Uncharacterized protein chromosome 11 open reading frame 48 |
| chr4_-_40631859 | 4.12 |
ENST00000295971.7
ENST00000319592.4 |
RBM47
|
RNA binding motif protein 47 |
| chr17_+_39846114 | 4.10 |
ENST00000586699.1
|
EIF1
|
eukaryotic translation initiation factor 1 |
| chr3_-_196911002 | 4.04 |
ENST00000452595.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr19_+_36706024 | 4.01 |
ENST00000443387.2
|
ZNF146
|
zinc finger protein 146 |
| chr12_-_49412525 | 3.99 |
ENST00000551121.1
ENST00000552212.1 ENST00000548605.1 ENST00000548950.1 ENST00000547125.1 |
PRKAG1
|
protein kinase, AMP-activated, gamma 1 non-catalytic subunit |
| chr10_-_74856608 | 3.99 |
ENST00000307116.2
ENST00000373008.2 ENST00000412021.2 ENST00000394890.2 ENST00000263556.3 ENST00000440381.1 |
P4HA1
|
prolyl 4-hydroxylase, alpha polypeptide I |
| chr1_-_149814478 | 3.91 |
ENST00000369161.3
|
HIST2H2AA3
|
histone cluster 2, H2aa3 |
| chr8_+_77593448 | 3.88 |
ENST00000521891.2
|
ZFHX4
|
zinc finger homeobox 4 |
| chr13_-_46543805 | 3.85 |
ENST00000378921.2
|
ZC3H13
|
zinc finger CCCH-type containing 13 |
| chr4_-_105416039 | 3.79 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chr19_+_36119975 | 3.78 |
ENST00000589559.1
ENST00000360475.4 |
RBM42
|
RNA binding motif protein 42 |
| chr15_-_55562479 | 3.73 |
ENST00000564609.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr14_+_103058948 | 3.72 |
ENST00000262241.6
|
RCOR1
|
REST corepressor 1 |
| chr15_-_55541227 | 3.71 |
ENST00000566877.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr10_-_104262426 | 3.67 |
ENST00000487599.1
|
ACTR1A
|
ARP1 actin-related protein 1 homolog A, centractin alpha (yeast) |
| chr17_+_7461613 | 3.65 |
ENST00000438470.1
ENST00000436057.1 |
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr3_-_33700933 | 3.63 |
ENST00000480013.1
|
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr3_-_196910721 | 3.53 |
ENST00000443183.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr1_+_23345930 | 3.51 |
ENST00000356634.3
|
KDM1A
|
lysine (K)-specific demethylase 1A |
| chr6_-_41909191 | 3.50 |
ENST00000512426.1
ENST00000372987.4 |
CCND3
|
cyclin D3 |
| chr12_-_118797475 | 3.50 |
ENST00000541786.1
ENST00000419821.2 ENST00000541878.1 |
TAOK3
|
TAO kinase 3 |
| chrX_-_77395186 | 3.44 |
ENST00000341864.5
|
TAF9B
|
TAF9B RNA polymerase II, TATA box binding protein (TBP)-associated factor, 31kDa |
| chr5_+_92919043 | 3.38 |
ENST00000327111.3
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr11_+_34642656 | 3.37 |
ENST00000257831.3
ENST00000450654.2 |
EHF
|
ets homologous factor |
| chr11_-_62473776 | 3.35 |
ENST00000278893.7
ENST00000407022.3 ENST00000421906.1 |
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr1_+_167298281 | 3.34 |
ENST00000367862.5
|
POU2F1
|
POU class 2 homeobox 1 |
| chr5_+_112074029 | 3.34 |
ENST00000512211.2
|
APC
|
adenomatous polyposis coli |
| chr18_+_19749386 | 3.32 |
ENST00000269216.3
|
GATA6
|
GATA binding protein 6 |
| chr1_-_156675368 | 3.22 |
ENST00000368222.3
|
CRABP2
|
cellular retinoic acid binding protein 2 |
| chr1_+_161736072 | 3.16 |
ENST00000367942.3
|
ATF6
|
activating transcription factor 6 |
| chr1_+_23345943 | 3.14 |
ENST00000400181.4
ENST00000542151.1 |
KDM1A
|
lysine (K)-specific demethylase 1A |
| chr1_-_167059830 | 3.13 |
ENST00000367868.3
|
GPA33
|
glycoprotein A33 (transmembrane) |
| chr2_-_145277569 | 3.12 |
ENST00000303660.4
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr1_+_82266053 | 3.06 |
ENST00000370715.1
ENST00000370713.1 ENST00000319517.6 ENST00000370717.2 ENST00000394879.1 ENST00000271029.4 ENST00000335786.5 |
LPHN2
|
latrophilin 2 |
| chr13_+_22245522 | 3.03 |
ENST00000382353.5
|
FGF9
|
fibroblast growth factor 9 |
| chr6_+_31683117 | 2.89 |
ENST00000375825.3
ENST00000375824.1 |
LY6G6D
|
lymphocyte antigen 6 complex, locus G6D |
| chr4_-_74864386 | 2.88 |
ENST00000296027.4
|
CXCL5
|
chemokine (C-X-C motif) ligand 5 |
| chr17_+_7461580 | 2.74 |
ENST00000483039.1
ENST00000396542.1 |
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr12_-_81763127 | 2.70 |
ENST00000541017.1
|
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr3_-_141747459 | 2.70 |
ENST00000477292.1
ENST00000478006.1 ENST00000495310.1 ENST00000486111.1 |
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr3_+_69928256 | 2.69 |
ENST00000394355.2
|
MITF
|
microphthalmia-associated transcription factor |
| chr17_+_39411636 | 2.64 |
ENST00000394008.1
|
KRTAP9-9
|
keratin associated protein 9-9 |
| chr5_-_142782862 | 2.58 |
ENST00000415690.2
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr18_-_25616519 | 2.57 |
ENST00000399380.3
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal) |
| chrX_-_122756660 | 2.54 |
ENST00000441692.1
|
THOC2
|
THO complex 2 |
| chr18_-_3219847 | 2.45 |
ENST00000261606.7
|
MYOM1
|
myomesin 1 |
| chr8_+_104831472 | 2.44 |
ENST00000262231.10
ENST00000507740.1 |
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr19_-_57967854 | 2.42 |
ENST00000321039.3
|
VN1R1
|
vomeronasal 1 receptor 1 |
| chr9_+_2159850 | 2.41 |
ENST00000416751.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chrX_-_114252193 | 2.40 |
ENST00000243213.1
|
IL13RA2
|
interleukin 13 receptor, alpha 2 |
| chr17_-_34417479 | 2.38 |
ENST00000225245.5
|
CCL3
|
chemokine (C-C motif) ligand 3 |
| chr5_-_88119580 | 2.35 |
ENST00000539796.1
|
MEF2C
|
myocyte enhancer factor 2C |
| chr8_-_95274536 | 2.32 |
ENST00000297596.2
ENST00000396194.2 |
GEM
|
GTP binding protein overexpressed in skeletal muscle |
| chr3_-_197300194 | 2.32 |
ENST00000358186.2
ENST00000431056.1 |
BDH1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr3_+_51575596 | 2.22 |
ENST00000409535.2
|
RAD54L2
|
RAD54-like 2 (S. cerevisiae) |
| chr5_-_82969405 | 2.16 |
ENST00000510978.1
|
HAPLN1
|
hyaluronan and proteoglycan link protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of CPEB1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.4 | 34.2 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 5.6 | 16.8 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 5.5 | 16.6 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 4.1 | 16.2 | GO:0044111 | development involved in symbiotic interaction(GO:0044111) |
| 4.0 | 12.1 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 3.9 | 11.7 | GO:0010621 | negative regulation of transcription by transcription factor localization(GO:0010621) |
| 3.4 | 10.3 | GO:0006427 | histidyl-tRNA aminoacylation(GO:0006427) |
| 3.3 | 6.6 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 3.3 | 13.1 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) |
| 3.0 | 11.9 | GO:0043335 | protein unfolding(GO:0043335) |
| 2.9 | 8.6 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 2.8 | 11.4 | GO:0046101 | hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 2.8 | 14.2 | GO:0097338 | response to clozapine(GO:0097338) |
| 2.8 | 8.3 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 2.7 | 10.8 | GO:1904798 | positive regulation of core promoter binding(GO:1904798) |
| 2.6 | 7.8 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 2.5 | 12.4 | GO:1902896 | terminal web assembly(GO:1902896) |
| 2.4 | 14.1 | GO:1902730 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 2.2 | 8.8 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 2.2 | 6.6 | GO:0035905 | ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 2.1 | 8.5 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) positive regulation of potassium ion import(GO:1903288) |
| 2.0 | 18.3 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 2.0 | 19.6 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 1.9 | 7.7 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) regulation of restriction endodeoxyribonuclease activity(GO:0032072) negative regulation of apoptotic cell clearance(GO:2000426) |
| 1.9 | 19.0 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 1.9 | 9.4 | GO:0015862 | uridine transport(GO:0015862) |
| 1.9 | 9.4 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 1.6 | 15.9 | GO:0030263 | resolution of meiotic recombination intermediates(GO:0000712) apoptotic chromosome condensation(GO:0030263) |
| 1.6 | 6.3 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 1.5 | 4.6 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 1.5 | 13.9 | GO:0030091 | protein repair(GO:0030091) |
| 1.5 | 4.4 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 1.4 | 4.3 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 1.4 | 8.5 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 1.3 | 11.7 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 1.3 | 7.6 | GO:1903286 | regulation of potassium ion import(GO:1903286) |
| 1.2 | 4.8 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 1.2 | 4.8 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 1.2 | 4.7 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 1.2 | 10.4 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 1.2 | 6.9 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 1.1 | 8.9 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 1.1 | 3.3 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 1.1 | 8.8 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 1.1 | 6.5 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 1.1 | 8.6 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 1.0 | 31.4 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 1.0 | 8.1 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 1.0 | 5.7 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.9 | 9.4 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.9 | 2.7 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.9 | 2.6 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.9 | 27.3 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.8 | 5.9 | GO:0060154 | response to cycloheximide(GO:0046898) cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.8 | 4.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.8 | 9.8 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.8 | 2.3 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.7 | 4.8 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.7 | 2.7 | GO:0060025 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) regulation of synaptic activity(GO:0060025) |
| 0.6 | 3.2 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.6 | 7.4 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.6 | 4.2 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.6 | 4.1 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.6 | 3.4 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.6 | 1.7 | GO:0071676 | negative regulation of mononuclear cell migration(GO:0071676) |
| 0.5 | 1.6 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.5 | 4.6 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.5 | 5.1 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.5 | 4.0 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.5 | 4.0 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.5 | 3.5 | GO:0015755 | fructose transport(GO:0015755) fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.5 | 26.3 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.5 | 1.9 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.5 | 0.5 | GO:1900133 | regulation of renin secretion into blood stream(GO:1900133) |
| 0.5 | 1.9 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.5 | 4.3 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.5 | 2.3 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.5 | 5.9 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.4 | 4.5 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.4 | 1.7 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.4 | 11.9 | GO:0035635 | entry of bacterium into host cell(GO:0035635) |
| 0.4 | 5.5 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.4 | 18.3 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.4 | 2.1 | GO:0035989 | tendon development(GO:0035989) |
| 0.4 | 10.3 | GO:0072663 | protein targeting to peroxisome(GO:0006625) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.4 | 8.0 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.4 | 4.4 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.4 | 1.9 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.4 | 3.0 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.4 | 7.8 | GO:0035067 | negative regulation of histone acetylation(GO:0035067) |
| 0.4 | 11.4 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.4 | 6.7 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.3 | 11.3 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.3 | 3.7 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.3 | 11.3 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.3 | 5.3 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.3 | 7.6 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.3 | 2.6 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.3 | 1.6 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.3 | 2.4 | GO:0043305 | negative regulation of mast cell degranulation(GO:0043305) |
| 0.3 | 2.4 | GO:0043308 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil activation(GO:0043307) eosinophil degranulation(GO:0043308) |
| 0.3 | 2.0 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.3 | 3.4 | GO:1902166 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902166) |
| 0.3 | 2.0 | GO:0006537 | glutamate biosynthetic process(GO:0006537) gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.3 | 10.2 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.3 | 14.7 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.3 | 11.5 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.3 | 2.5 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.3 | 11.1 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.3 | 2.3 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.2 | 0.7 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.2 | 8.0 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.2 | 1.6 | GO:1904783 | positive regulation of NMDA glutamate receptor activity(GO:1904783) |
| 0.2 | 1.1 | GO:0045577 | regulation of B cell differentiation(GO:0045577) |
| 0.2 | 1.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.2 | 0.7 | GO:1901624 | negative regulation of lymphocyte chemotaxis(GO:1901624) |
| 0.2 | 4.4 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.2 | 1.8 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.2 | 3.6 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.2 | 7.3 | GO:0030835 | negative regulation of actin filament depolymerization(GO:0030835) |
| 0.2 | 0.6 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 0.2 | 5.1 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.2 | 5.5 | GO:0043555 | regulation of translation in response to stress(GO:0043555) |
| 0.2 | 4.4 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.2 | 4.2 | GO:0042994 | cytoplasmic sequestering of transcription factor(GO:0042994) |
| 0.2 | 0.7 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.2 | 0.5 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.2 | 0.5 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.2 | 3.5 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.2 | 2.3 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.2 | 0.2 | GO:1900157 | regulation of bone mineralization involved in bone maturation(GO:1900157) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.2 | 1.1 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.2 | 5.1 | GO:0072600 | establishment of protein localization to Golgi(GO:0072600) |
| 0.2 | 18.7 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 15.0 | GO:0006110 | regulation of glycolytic process(GO:0006110) |
| 0.1 | 0.9 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.1 | 0.8 | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.1 | 0.6 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 2.6 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 1.2 | GO:0032364 | oxygen homeostasis(GO:0032364) hemoglobin biosynthetic process(GO:0042541) |
| 0.1 | 4.3 | GO:0046949 | fatty-acyl-CoA biosynthetic process(GO:0046949) |
| 0.1 | 0.7 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.1 | 31.6 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.1 | 8.1 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.1 | 2.9 | GO:1905145 | acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.1 | 8.6 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.1 | 4.1 | GO:0060706 | cell differentiation involved in embryonic placenta development(GO:0060706) |
| 0.1 | 6.9 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 4.4 | GO:0044364 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) |
| 0.1 | 6.0 | GO:0031016 | pancreas development(GO:0031016) |
| 0.1 | 0.5 | GO:2000503 | regulation of natural killer cell chemotaxis(GO:2000501) positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.1 | 4.6 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 10.5 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.1 | 0.7 | GO:2000111 | positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.1 | 0.6 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.1 | 1.2 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.1 | 4.0 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.1 | 0.6 | GO:0060048 | cardiac muscle contraction(GO:0060048) |
| 0.1 | 1.2 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.1 | 1.1 | GO:0061436 | regulation of water loss via skin(GO:0033561) establishment of skin barrier(GO:0061436) |
| 0.1 | 2.8 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.1 | 12.7 | GO:0030307 | positive regulation of cell growth(GO:0030307) |
| 0.1 | 5.5 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.1 | 0.5 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.1 | 0.3 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) arterial endothelial cell differentiation(GO:0060842) |
| 0.1 | 1.2 | GO:0035815 | positive regulation of renal sodium excretion(GO:0035815) |
| 0.1 | 1.3 | GO:0010458 | exit from mitosis(GO:0010458) |
| 0.1 | 0.3 | GO:0048023 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.1 | 4.7 | GO:0070126 | mitochondrial translational elongation(GO:0070125) mitochondrial translational termination(GO:0070126) |
| 0.1 | 0.8 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.1 | 1.5 | GO:0030516 | regulation of axon extension(GO:0030516) |
| 0.0 | 1.4 | GO:0006260 | DNA replication(GO:0006260) |
| 0.0 | 0.2 | GO:1990481 | tRNA pseudouridine synthesis(GO:0031119) mRNA pseudouridine synthesis(GO:1990481) |
| 0.0 | 1.9 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.4 | GO:0071600 | otic vesicle formation(GO:0030916) otic vesicle morphogenesis(GO:0071600) |
| 0.0 | 0.4 | GO:0046325 | negative regulation of glucose import(GO:0046325) |
| 0.0 | 5.7 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 3.4 | GO:0010977 | negative regulation of neuron projection development(GO:0010977) |
| 0.0 | 3.3 | GO:0009301 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 2.0 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.0 | 0.9 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.7 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 1.1 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 14.0 | GO:0007264 | small GTPase mediated signal transduction(GO:0007264) |
| 0.0 | 0.7 | GO:0007565 | female pregnancy(GO:0007565) |
| 0.0 | 2.8 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 0.4 | GO:0021983 | pituitary gland development(GO:0021983) |
| 0.0 | 0.4 | GO:0030193 | regulation of blood coagulation(GO:0030193) regulation of hemostasis(GO:1900046) |
| 0.0 | 0.2 | GO:1903209 | positive regulation of oxidative stress-induced cell death(GO:1903209) |
| 0.0 | 0.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.6 | GO:0060348 | bone development(GO:0060348) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.3 | 15.9 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 4.4 | 13.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 3.8 | 34.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 3.7 | 22.0 | GO:1990357 | terminal web(GO:1990357) |
| 2.8 | 11.4 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 2.4 | 14.1 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 2.1 | 6.3 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 2.0 | 7.8 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 1.9 | 9.6 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 1.9 | 9.4 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 1.7 | 15.5 | GO:0061574 | ASAP complex(GO:0061574) |
| 1.5 | 27.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 1.4 | 8.6 | GO:0002177 | manchette(GO:0002177) |
| 1.4 | 6.9 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 1.3 | 4.0 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 1.3 | 8.8 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 1.1 | 4.4 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 1.0 | 14.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 1.0 | 10.8 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 1.0 | 6.8 | GO:0016342 | catenin complex(GO:0016342) |
| 0.9 | 3.7 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.9 | 10.7 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.9 | 8.6 | GO:0045180 | basal cortex(GO:0045180) |
| 0.9 | 11.1 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.8 | 10.4 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.8 | 2.3 | GO:0034676 | integrin alpha6-beta4 complex(GO:0034676) |
| 0.8 | 8.5 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.7 | 15.0 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.7 | 5.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.7 | 7.7 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.6 | 7.6 | GO:0097025 | lateral loop(GO:0043219) MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.6 | 19.0 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.5 | 2.7 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.5 | 5.9 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.4 | 8.5 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.4 | 6.7 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.4 | 2.9 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.4 | 2.5 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.4 | 5.3 | GO:0008290 | F-actin capping protein complex(GO:0008290) Flemming body(GO:0090543) |
| 0.3 | 2.3 | GO:0098576 | lumenal side of membrane(GO:0098576) |
| 0.3 | 15.7 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.3 | 1.7 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.3 | 23.5 | GO:0016459 | myosin complex(GO:0016459) |
| 0.2 | 5.7 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.2 | 2.1 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.2 | 3.4 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.2 | 5.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.2 | 4.3 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.2 | 4.8 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.2 | 10.4 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.2 | 4.7 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.2 | 0.8 | GO:0030425 | dendrite(GO:0030425) |
| 0.1 | 4.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 6.7 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 1.3 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 44.5 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 24.5 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.1 | 7.0 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 12.3 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 7.1 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 10.4 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.1 | 2.3 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.1 | 7.0 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 2.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 0.7 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.1 | 6.5 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 3.6 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 12.4 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 4.4 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 1.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 6.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 3.5 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.1 | 22.3 | GO:0005938 | cell cortex(GO:0005938) |
| 0.1 | 26.2 | GO:0005769 | early endosome(GO:0005769) |
| 0.1 | 3.5 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 0.6 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 1.2 | GO:0043234 | protein complex(GO:0043234) |
| 0.1 | 0.8 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 8.4 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 21.1 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 3.8 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 2.8 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 2.4 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 7.1 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 3.8 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 0.3 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 10.3 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 1.1 | GO:0031975 | organelle envelope(GO:0031967) envelope(GO:0031975) |
| 0.0 | 23.2 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 5.9 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 2.9 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 15.7 | GO:0005815 | microtubule organizing center(GO:0005815) |
| 0.0 | 6.3 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 3.8 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 1.7 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 1.2 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 4.4 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 0.7 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 3.0 | GO:0000785 | chromatin(GO:0000785) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.6 | 16.8 | GO:0004756 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 5.3 | 15.9 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 5.2 | 20.7 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 4.0 | 12.1 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 3.4 | 10.3 | GO:0004821 | histidine-tRNA ligase activity(GO:0004821) |
| 3.4 | 34.2 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 2.8 | 13.9 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 2.7 | 11.0 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 2.2 | 6.5 | GO:0000773 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 2.0 | 8.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 1.9 | 11.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 1.8 | 29.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 1.7 | 6.6 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 1.6 | 19.6 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 1.6 | 9.6 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 1.6 | 7.8 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 1.5 | 10.4 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 1.4 | 4.2 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 1.3 | 7.7 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 1.2 | 32.5 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 1.2 | 7.2 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 1.2 | 11.9 | GO:0030911 | TPR domain binding(GO:0030911) |
| 1.2 | 6.9 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 1.1 | 21.3 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 1.0 | 8.3 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 1.0 | 19.0 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 1.0 | 14.7 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 1.0 | 6.7 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.9 | 4.5 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.9 | 4.3 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.8 | 4.0 | GO:0019798 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.8 | 2.3 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.8 | 43.6 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.7 | 7.6 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.7 | 11.1 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.6 | 8.9 | GO:0016918 | retinal binding(GO:0016918) |
| 0.6 | 8.6 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.5 | 13.0 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.5 | 2.7 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.5 | 8.5 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.5 | 2.6 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.5 | 3.5 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.5 | 1.5 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.5 | 2.0 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.5 | 2.9 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.5 | 2.4 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.5 | 20.2 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.4 | 9.4 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.4 | 5.7 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.4 | 6.5 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.4 | 1.2 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.4 | 18.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.3 | 10.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.3 | 7.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.3 | 3.4 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.3 | 8.5 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.3 | 6.9 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.3 | 10.8 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.3 | 16.6 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.3 | 15.0 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.3 | 2.0 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.3 | 8.0 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.3 | 11.5 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.3 | 7.0 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.3 | 2.0 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.2 | 9.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.2 | 5.5 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.2 | 1.7 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.2 | 8.7 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.2 | 7.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 9.4 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.2 | 7.4 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.2 | 7.2 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.2 | 2.3 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.2 | 3.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.2 | 1.9 | GO:0050544 | icosanoid binding(GO:0050542) arachidonic acid binding(GO:0050544) fatty acid derivative binding(GO:1901567) |
| 0.2 | 1.6 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.2 | 1.8 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.2 | 3.7 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 6.7 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 7.7 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 2.7 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 4.3 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.1 | 1.7 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 2.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 4.8 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 20.9 | GO:0042393 | histone binding(GO:0042393) |
| 0.1 | 0.9 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.1 | 3.3 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 1.3 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 0.4 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.1 | 9.1 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 8.5 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.1 | 0.6 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 3.0 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 16.9 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 32.8 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 4.3 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.1 | 1.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 1.6 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 2.7 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 0.5 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.1 | 5.9 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 32.5 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 1.6 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 1.9 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.8 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 0.5 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) steroid sulfotransferase activity(GO:0050294) |
| 0.1 | 0.3 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.1 | 1.1 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 2.7 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 0.7 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 4.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.1 | GO:0070404 | NADH binding(GO:0070404) |
| 0.0 | 0.5 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.7 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 1.3 | GO:0001071 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 6.6 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.2 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 7.6 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.7 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.8 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 14.0 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 1.6 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 2.3 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 4.7 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 6.7 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.5 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.3 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.2 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 1.3 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 1.4 | GO:0004386 | helicase activity(GO:0004386) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 31.1 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.6 | 20.7 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.5 | 16.5 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.5 | 2.4 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.5 | 11.5 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.5 | 11.9 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.4 | 27.5 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.4 | 8.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.4 | 5.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.3 | 15.8 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.3 | 18.2 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.3 | 5.7 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.3 | 16.6 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.3 | 8.8 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.3 | 7.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.2 | 24.2 | PID E2F PATHWAY | E2F transcription factor network |
| 0.2 | 14.7 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.2 | 9.8 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.2 | 10.0 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.2 | 16.0 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.2 | 18.5 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.2 | 7.6 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.2 | 14.6 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.2 | 3.9 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.2 | 2.0 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.2 | 1.7 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 2.4 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.1 | 11.3 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 3.5 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 7.2 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 1.9 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 3.9 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 2.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 2.7 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.1 | 5.5 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 4.4 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 5.0 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 1.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 4.8 | PID P73PATHWAY | p73 transcription factor network |
| 0.1 | 1.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 2.6 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 4.7 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.1 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 1.7 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 8.8 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.7 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.4 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.2 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 19.0 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.9 | 11.1 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.7 | 15.0 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.7 | 11.9 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.7 | 40.7 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.7 | 16.6 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.6 | 6.7 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.5 | 23.4 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.5 | 8.8 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.4 | 5.1 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.4 | 7.6 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.4 | 13.1 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.4 | 6.0 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.4 | 18.9 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.3 | 15.5 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.3 | 14.7 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.3 | 8.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.3 | 16.5 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.3 | 6.9 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.3 | 7.7 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.2 | 8.8 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.2 | 15.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.2 | 6.5 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.2 | 14.9 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.2 | 3.0 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.2 | 6.3 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.2 | 8.5 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.2 | 2.7 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.2 | 9.1 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.2 | 8.1 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.2 | 10.8 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.2 | 14.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.2 | 3.5 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.2 | 13.0 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.2 | 4.6 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.2 | 2.7 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 1.6 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 3.3 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.1 | 5.9 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 31.4 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 4.3 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.1 | 1.5 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 4.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 5.2 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.1 | 1.6 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 4.5 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 2.6 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 1.3 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 1.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.8 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.5 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.7 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 2.4 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 6.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.1 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.8 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.4 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.5 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.8 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |