Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
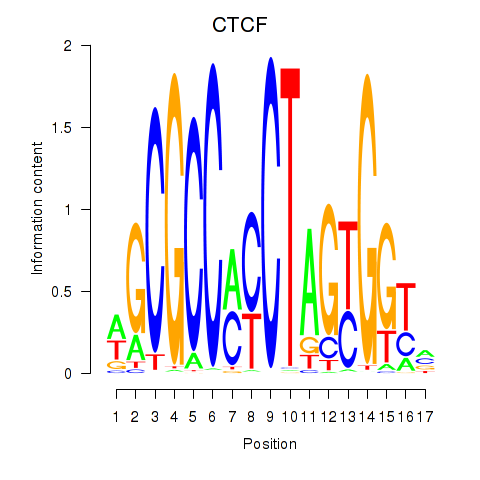
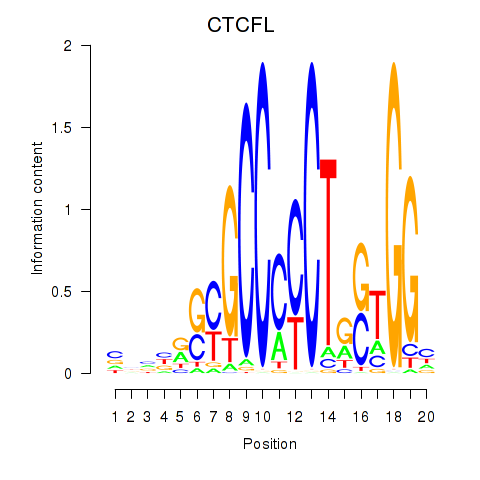
Results for CTCF_CTCFL
Z-value: 1.03
Transcription factors associated with CTCF_CTCFL
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CTCF
|
ENSG00000102974.10 | CCCTC-binding factor |
|
CTCFL
|
ENSG00000124092.8 | CCCTC-binding factor like |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CTCF | hg19_v2_chr16_+_67596310_67596353 | 0.50 | 4.3e-15 | Click! |
| CTCFL | hg19_v2_chr20_-_56100155_56100163 | -0.49 | 1.6e-14 | Click! |
Activity profile of CTCF_CTCFL motif
Sorted Z-values of CTCF_CTCFL motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_132249955 | 32.67 |
ENST00000309451.6
|
MZT2A
|
mitotic spindle organizing protein 2A |
| chr2_+_130939827 | 30.27 |
ENST00000409255.1
ENST00000455239.1 |
MZT2B
|
mitotic spindle organizing protein 2B |
| chr12_-_82752565 | 27.18 |
ENST00000256151.7
|
CCDC59
|
coiled-coil domain containing 59 |
| chr12_+_56618102 | 24.79 |
ENST00000267023.4
ENST00000380198.2 ENST00000341463.5 |
NABP2
|
nucleic acid binding protein 2 |
| chr1_+_203830703 | 23.46 |
ENST00000414487.2
|
SNRPE
|
small nuclear ribonucleoprotein polypeptide E |
| chr10_+_81107216 | 23.28 |
ENST00000394579.3
ENST00000225174.3 |
PPIF
|
peptidylprolyl isomerase F |
| chr19_+_41768401 | 21.74 |
ENST00000352456.3
ENST00000595018.1 ENST00000597725.1 |
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr17_-_48474828 | 20.95 |
ENST00000576448.1
ENST00000225972.7 |
LRRC59
|
leucine rich repeat containing 59 |
| chr4_-_106395135 | 20.35 |
ENST00000310267.7
|
PPA2
|
pyrophosphatase (inorganic) 2 |
| chr22_+_24204375 | 20.27 |
ENST00000433835.3
|
AP000350.10
|
Uncharacterized protein |
| chr19_+_49496705 | 18.90 |
ENST00000595090.1
|
RUVBL2
|
RuvB-like AAA ATPase 2 |
| chr16_-_66864806 | 18.76 |
ENST00000566336.1
ENST00000394074.2 ENST00000563185.2 ENST00000359087.4 ENST00000379463.2 ENST00000565535.1 ENST00000290810.3 |
NAE1
|
NEDD8 activating enzyme E1 subunit 1 |
| chr5_+_145826867 | 18.31 |
ENST00000296702.5
ENST00000394421.2 |
TCERG1
|
transcription elongation regulator 1 |
| chr1_-_94374946 | 17.84 |
ENST00000370238.3
|
GCLM
|
glutamate-cysteine ligase, modifier subunit |
| chr3_+_172468505 | 17.62 |
ENST00000427830.1
ENST00000417960.1 ENST00000428567.1 ENST00000366090.2 ENST00000426894.1 |
ECT2
|
epithelial cell transforming sequence 2 oncogene |
| chr11_-_6704513 | 16.95 |
ENST00000532203.1
ENST00000288937.6 |
MRPL17
|
mitochondrial ribosomal protein L17 |
| chr2_+_130939235 | 16.94 |
ENST00000425361.1
ENST00000457492.1 |
MZT2B
|
mitotic spindle organizing protein 2B |
| chr4_-_106395197 | 16.77 |
ENST00000508518.1
ENST00000354147.3 ENST00000432483.2 ENST00000510015.1 ENST00000504028.1 ENST00000348706.5 ENST00000357415.4 ENST00000380004.2 ENST00000341695.5 |
PPA2
|
pyrophosphatase (inorganic) 2 |
| chr3_+_172468472 | 16.61 |
ENST00000232458.5
ENST00000392692.3 |
ECT2
|
epithelial cell transforming sequence 2 oncogene |
| chr19_+_49496782 | 15.59 |
ENST00000601968.1
ENST00000596837.1 |
RUVBL2
|
RuvB-like AAA ATPase 2 |
| chr12_+_48357340 | 14.98 |
ENST00000256686.6
ENST00000549288.1 ENST00000552561.1 ENST00000546749.1 ENST00000552546.1 ENST00000550552.1 |
TMEM106C
|
transmembrane protein 106C |
| chr7_+_141438118 | 14.82 |
ENST00000265304.6
ENST00000498107.1 ENST00000467681.1 ENST00000465582.1 ENST00000463093.1 |
SSBP1
|
single-stranded DNA binding protein 1, mitochondrial |
| chr8_-_90996837 | 14.74 |
ENST00000519426.1
ENST00000265433.3 |
NBN
|
nibrin |
| chr1_+_186344883 | 14.67 |
ENST00000367470.3
|
C1orf27
|
chromosome 1 open reading frame 27 |
| chr1_+_186344945 | 14.58 |
ENST00000419367.3
ENST00000287859.6 |
C1orf27
|
chromosome 1 open reading frame 27 |
| chr17_-_5342380 | 14.53 |
ENST00000225698.4
|
C1QBP
|
complement component 1, q subcomponent binding protein |
| chr12_+_48357401 | 14.50 |
ENST00000429772.2
ENST00000449758.2 |
TMEM106C
|
transmembrane protein 106C |
| chr6_+_13615554 | 14.36 |
ENST00000451315.2
|
NOL7
|
nucleolar protein 7, 27kDa |
| chr12_-_48099754 | 14.35 |
ENST00000380650.4
|
RPAP3
|
RNA polymerase II associated protein 3 |
| chr7_-_102119342 | 14.28 |
ENST00000393794.3
ENST00000292614.5 |
POLR2J
|
polymerase (RNA) II (DNA directed) polypeptide J, 13.3kDa |
| chr21_-_26979786 | 13.62 |
ENST00000419219.1
ENST00000352957.4 ENST00000307301.7 |
MRPL39
|
mitochondrial ribosomal protein L39 |
| chr5_+_167913450 | 13.09 |
ENST00000231572.3
ENST00000538719.1 |
RARS
|
arginyl-tRNA synthetase |
| chr14_-_50319758 | 12.74 |
ENST00000298310.5
|
NEMF
|
nuclear export mediator factor |
| chr4_-_140216948 | 12.67 |
ENST00000265500.4
|
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr12_+_123237321 | 12.62 |
ENST00000280557.6
ENST00000455982.2 |
DENR
|
density-regulated protein |
| chr6_+_138725343 | 12.61 |
ENST00000607197.1
ENST00000367697.3 |
HEBP2
|
heme binding protein 2 |
| chr14_-_54908043 | 12.44 |
ENST00000556113.1
ENST00000553660.1 ENST00000395573.4 ENST00000557690.1 ENST00000216416.4 |
CNIH1
|
cornichon family AMPA receptor auxiliary protein 1 |
| chr1_+_212208919 | 12.41 |
ENST00000366991.4
ENST00000542077.1 |
DTL
|
denticleless E3 ubiquitin protein ligase homolog (Drosophila) |
| chr12_-_48099773 | 12.31 |
ENST00000432584.3
ENST00000005386.3 |
RPAP3
|
RNA polymerase II associated protein 3 |
| chr2_+_171785824 | 12.22 |
ENST00000452526.2
|
GORASP2
|
golgi reassembly stacking protein 2, 55kDa |
| chr12_+_104609550 | 12.14 |
ENST00000525566.1
ENST00000429002.2 |
TXNRD1
|
thioredoxin reductase 1 |
| chr14_-_50319482 | 11.88 |
ENST00000546046.1
ENST00000555970.1 ENST00000554626.1 ENST00000545773.1 ENST00000556672.1 |
NEMF
|
nuclear export mediator factor |
| chr9_+_71650645 | 11.42 |
ENST00000396366.2
|
FXN
|
frataxin |
| chr16_+_22308717 | 11.36 |
ENST00000299853.5
ENST00000564209.1 ENST00000565358.1 ENST00000418581.2 ENST00000564883.1 ENST00000359210.4 ENST00000563024.1 |
POLR3E
|
polymerase (RNA) III (DNA directed) polypeptide E (80kD) |
| chr1_+_29063119 | 11.19 |
ENST00000474884.1
ENST00000542507.1 |
YTHDF2
|
YTH domain family, member 2 |
| chr8_-_90996459 | 10.90 |
ENST00000517337.1
ENST00000409330.1 |
NBN
|
nibrin |
| chr16_+_4674814 | 10.87 |
ENST00000415496.1
ENST00000587747.1 ENST00000399577.5 ENST00000588994.1 ENST00000586183.1 |
MGRN1
|
mahogunin ring finger 1, E3 ubiquitin protein ligase |
| chr19_-_19051927 | 10.52 |
ENST00000600077.1
|
HOMER3
|
homer homolog 3 (Drosophila) |
| chr8_-_102217515 | 10.38 |
ENST00000520347.1
ENST00000523922.1 ENST00000520984.1 |
ZNF706
|
zinc finger protein 706 |
| chr19_+_5690297 | 10.34 |
ENST00000582463.1
ENST00000579446.1 ENST00000394580.2 |
RPL36
|
ribosomal protein L36 |
| chr15_-_66790146 | 10.29 |
ENST00000316634.5
|
SNAPC5
|
small nuclear RNA activating complex, polypeptide 5, 19kDa |
| chr6_+_155054459 | 10.29 |
ENST00000367178.3
ENST00000417268.1 ENST00000367186.4 |
SCAF8
|
SR-related CTD-associated factor 8 |
| chr7_-_5569588 | 10.22 |
ENST00000417101.1
|
ACTB
|
actin, beta |
| chr1_+_95699704 | 10.21 |
ENST00000370202.4
|
RWDD3
|
RWD domain containing 3 |
| chr9_-_139371533 | 10.16 |
ENST00000290037.6
ENST00000431893.2 ENST00000371706.3 |
SEC16A
|
SEC16 homolog A (S. cerevisiae) |
| chr2_+_171785012 | 10.11 |
ENST00000234160.4
|
GORASP2
|
golgi reassembly stacking protein 2, 55kDa |
| chr19_+_10982336 | 10.03 |
ENST00000344150.4
|
CARM1
|
coactivator-associated arginine methyltransferase 1 |
| chr20_+_44441271 | 10.03 |
ENST00000335046.3
ENST00000243893.6 |
UBE2C
|
ubiquitin-conjugating enzyme E2C |
| chr13_+_76123883 | 9.92 |
ENST00000377595.3
|
UCHL3
|
ubiquitin carboxyl-terminal esterase L3 (ubiquitin thiolesterase) |
| chr20_+_44441626 | 9.90 |
ENST00000372568.4
|
UBE2C
|
ubiquitin-conjugating enzyme E2C |
| chr16_-_47177874 | 9.75 |
ENST00000562435.1
|
NETO2
|
neuropilin (NRP) and tolloid (TLL)-like 2 |
| chr4_-_120988229 | 9.67 |
ENST00000296509.6
|
MAD2L1
|
MAD2 mitotic arrest deficient-like 1 (yeast) |
| chr8_-_102217796 | 9.62 |
ENST00000519744.1
ENST00000311212.4 ENST00000521272.1 ENST00000519882.1 |
ZNF706
|
zinc finger protein 706 |
| chr7_-_96339132 | 9.62 |
ENST00000413065.1
|
SHFM1
|
split hand/foot malformation (ectrodactyly) type 1 |
| chr7_-_16840820 | 9.60 |
ENST00000450569.1
|
AGR2
|
anterior gradient 2 |
| chr12_+_7033616 | 9.59 |
ENST00000356654.4
|
ATN1
|
atrophin 1 |
| chr19_+_17416457 | 9.52 |
ENST00000252602.1
|
MRPL34
|
mitochondrial ribosomal protein L34 |
| chr19_+_5690207 | 9.47 |
ENST00000347512.3
|
RPL36
|
ribosomal protein L36 |
| chr20_+_48807351 | 9.38 |
ENST00000303004.3
|
CEBPB
|
CCAAT/enhancer binding protein (C/EBP), beta |
| chr19_+_17416609 | 9.37 |
ENST00000602206.1
|
MRPL34
|
mitochondrial ribosomal protein L34 |
| chr1_-_37980344 | 9.31 |
ENST00000448519.2
ENST00000373075.2 ENST00000373073.4 ENST00000296214.5 |
MEAF6
|
MYST/Esa1-associated factor 6 |
| chr16_+_4674787 | 9.29 |
ENST00000262370.7
|
MGRN1
|
mahogunin ring finger 1, E3 ubiquitin protein ligase |
| chr1_+_193091080 | 9.23 |
ENST00000367435.3
|
CDC73
|
cell division cycle 73 |
| chr3_+_184080790 | 9.15 |
ENST00000430783.1
|
POLR2H
|
polymerase (RNA) II (DNA directed) polypeptide H |
| chr17_-_76183111 | 9.10 |
ENST00000405273.1
ENST00000590862.1 ENST00000590430.1 ENST00000586613.1 |
TK1
|
thymidine kinase 1, soluble |
| chr11_+_46368975 | 9.10 |
ENST00000527911.1
|
DGKZ
|
diacylglycerol kinase, zeta |
| chr11_+_8008867 | 8.92 |
ENST00000309828.4
ENST00000449102.2 |
EIF3F
|
eukaryotic translation initiation factor 3, subunit F |
| chr11_-_66206260 | 8.89 |
ENST00000329819.4
ENST00000310999.7 ENST00000430466.2 |
MRPL11
|
mitochondrial ribosomal protein L11 |
| chr19_-_2050852 | 8.88 |
ENST00000541165.1
ENST00000591601.1 |
MKNK2
|
MAP kinase interacting serine/threonine kinase 2 |
| chr1_+_10490779 | 8.79 |
ENST00000477755.1
|
APITD1
|
apoptosis-inducing, TAF9-like domain 1 |
| chr2_-_96874553 | 8.79 |
ENST00000337288.5
ENST00000443962.1 |
STARD7
|
StAR-related lipid transfer (START) domain containing 7 |
| chr10_+_81107271 | 8.77 |
ENST00000448165.1
|
PPIF
|
peptidylprolyl isomerase F |
| chr10_+_13628921 | 8.71 |
ENST00000378572.3
|
PRPF18
|
pre-mRNA processing factor 18 |
| chr1_+_29063439 | 8.65 |
ENST00000541996.1
ENST00000496288.1 |
YTHDF2
|
YTH domain family, member 2 |
| chr20_-_1447467 | 8.59 |
ENST00000353088.2
ENST00000350991.4 |
NSFL1C
|
NSFL1 (p97) cofactor (p47) |
| chr5_-_179050066 | 8.48 |
ENST00000329433.6
ENST00000510411.1 |
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 (H) |
| chr20_+_60878005 | 8.42 |
ENST00000253003.2
|
ADRM1
|
adhesion regulating molecule 1 |
| chr17_+_38137073 | 8.37 |
ENST00000541736.1
|
PSMD3
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 3 |
| chr2_+_231921574 | 8.36 |
ENST00000308696.6
ENST00000373635.4 ENST00000440838.1 ENST00000409643.1 |
PSMD1
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 1 |
| chr1_-_6614565 | 8.33 |
ENST00000377705.5
|
NOL9
|
nucleolar protein 9 |
| chr19_-_19051993 | 8.27 |
ENST00000594794.1
ENST00000355887.6 ENST00000392351.3 ENST00000596482.1 |
HOMER3
|
homer homolog 3 (Drosophila) |
| chr20_+_44441304 | 8.24 |
ENST00000352551.5
|
UBE2C
|
ubiquitin-conjugating enzyme E2C |
| chr1_+_29063271 | 8.21 |
ENST00000373812.3
|
YTHDF2
|
YTH domain family, member 2 |
| chr17_-_79827808 | 8.17 |
ENST00000580685.1
|
ARHGDIA
|
Rho GDP dissociation inhibitor (GDI) alpha |
| chr2_+_131100710 | 8.17 |
ENST00000452955.1
|
IMP4
|
IMP4, U3 small nucleolar ribonucleoprotein, homolog (yeast) |
| chr19_-_48894762 | 8.11 |
ENST00000600980.1
ENST00000330720.2 |
KDELR1
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 1 |
| chr6_+_32821924 | 8.10 |
ENST00000374859.2
ENST00000453265.2 |
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr17_-_79995553 | 8.06 |
ENST00000581584.1
ENST00000577712.1 ENST00000582900.1 ENST00000579155.1 ENST00000306869.2 |
DCXR
|
dicarbonyl/L-xylulose reductase |
| chr11_+_20409070 | 8.05 |
ENST00000331079.6
|
PRMT3
|
protein arginine methyltransferase 3 |
| chr20_+_44441215 | 7.98 |
ENST00000356455.4
ENST00000405520.1 |
UBE2C
|
ubiquitin-conjugating enzyme E2C |
| chr20_-_1447547 | 7.95 |
ENST00000476071.1
|
NSFL1C
|
NSFL1 (p97) cofactor (p47) |
| chr11_+_46368956 | 7.93 |
ENST00000543978.1
|
DGKZ
|
diacylglycerol kinase, zeta |
| chr21_-_44527613 | 7.90 |
ENST00000380276.2
ENST00000398137.1 ENST00000291552.4 |
U2AF1
|
U2 small nuclear RNA auxiliary factor 1 |
| chr3_-_64009658 | 7.74 |
ENST00000394431.2
|
PSMD6
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 6 |
| chr15_-_66679019 | 7.72 |
ENST00000568216.1
ENST00000562124.1 ENST00000570251.1 |
TIPIN
|
TIMELESS interacting protein |
| chr3_-_128369643 | 7.67 |
ENST00000296255.3
|
RPN1
|
ribophorin I |
| chr16_+_69345243 | 7.65 |
ENST00000254950.11
|
VPS4A
|
vacuolar protein sorting 4 homolog A (S. cerevisiae) |
| chr15_+_63340734 | 7.64 |
ENST00000560959.1
|
TPM1
|
tropomyosin 1 (alpha) |
| chr3_+_141457030 | 7.63 |
ENST00000273480.3
|
RNF7
|
ring finger protein 7 |
| chr10_-_96122682 | 7.62 |
ENST00000371361.3
|
NOC3L
|
nucleolar complex associated 3 homolog (S. cerevisiae) |
| chr19_-_55652290 | 7.58 |
ENST00000589745.1
|
TNNT1
|
troponin T type 1 (skeletal, slow) |
| chr3_+_184081175 | 7.57 |
ENST00000452961.1
ENST00000296223.3 |
POLR2H
|
polymerase (RNA) II (DNA directed) polypeptide H |
| chr3_+_184081213 | 7.57 |
ENST00000429568.1
|
POLR2H
|
polymerase (RNA) II (DNA directed) polypeptide H |
| chr17_+_49230897 | 7.55 |
ENST00000393196.3
ENST00000336097.3 ENST00000480143.1 ENST00000511355.1 ENST00000013034.3 ENST00000393198.3 ENST00000608447.1 ENST00000393193.2 ENST00000376392.6 ENST00000555572.1 |
NME1
NME1-NME2
NME2
|
NME/NM23 nucleoside diphosphate kinase 1 NME1-NME2 readthrough NME/NM23 nucleoside diphosphate kinase 2 |
| chr12_+_12966250 | 7.54 |
ENST00000352940.4
ENST00000358007.3 ENST00000544400.1 |
DDX47
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 47 |
| chr11_+_46369077 | 7.50 |
ENST00000456247.2
ENST00000421244.2 ENST00000318201.8 |
DGKZ
|
diacylglycerol kinase, zeta |
| chr6_+_119215308 | 7.50 |
ENST00000229595.5
|
ASF1A
|
anti-silencing function 1A histone chaperone |
| chr11_-_66103867 | 7.47 |
ENST00000424433.2
|
RIN1
|
Ras and Rab interactor 1 |
| chr3_+_58291965 | 7.41 |
ENST00000445193.3
ENST00000295959.5 ENST00000466547.1 |
RPP14
|
ribonuclease P/MRP 14kDa subunit |
| chr3_-_48723268 | 7.37 |
ENST00000439518.1
ENST00000416649.2 ENST00000341520.4 ENST00000294129.2 |
NCKIPSD
|
NCK interacting protein with SH3 domain |
| chr11_+_67374323 | 7.33 |
ENST00000322776.6
ENST00000532303.1 ENST00000532244.1 ENST00000528328.1 ENST00000529927.1 ENST00000532343.1 ENST00000415352.2 ENST00000533075.1 ENST00000529867.1 ENST00000530638.1 |
NDUFV1
|
NADH dehydrogenase (ubiquinone) flavoprotein 1, 51kDa |
| chr1_+_32687971 | 7.32 |
ENST00000373586.1
|
EIF3I
|
eukaryotic translation initiation factor 3, subunit I |
| chr2_+_190526111 | 7.32 |
ENST00000607062.1
ENST00000260952.4 ENST00000425590.1 ENST00000607535.1 ENST00000420250.1 ENST00000606910.1 ENST00000607690.1 ENST00000607829.1 |
ASNSD1
|
asparagine synthetase domain containing 1 |
| chr5_-_179233934 | 7.31 |
ENST00000292591.7
|
MGAT4B
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme B |
| chr17_+_28804380 | 7.24 |
ENST00000225724.5
ENST00000451249.2 ENST00000467337.2 ENST00000581721.1 ENST00000414833.2 |
GOSR1
|
golgi SNAP receptor complex member 1 |
| chr8_+_145149930 | 7.23 |
ENST00000318911.4
|
CYC1
|
cytochrome c-1 |
| chr1_-_156675535 | 7.22 |
ENST00000368221.1
|
CRABP2
|
cellular retinoic acid binding protein 2 |
| chr7_+_141438393 | 7.16 |
ENST00000484178.1
ENST00000473783.1 ENST00000481508.1 |
SSBP1
|
single-stranded DNA binding protein 1, mitochondrial |
| chr16_-_88851618 | 7.12 |
ENST00000301015.9
|
PIEZO1
|
piezo-type mechanosensitive ion channel component 1 |
| chr1_-_1284730 | 6.97 |
ENST00000378888.5
|
DVL1
|
dishevelled segment polarity protein 1 |
| chr19_+_23945768 | 6.96 |
ENST00000486528.1
ENST00000496398.1 |
RPSAP58
|
ribosomal protein SA pseudogene 58 |
| chr7_-_27219849 | 6.90 |
ENST00000396344.4
|
HOXA10
|
homeobox A10 |
| chr17_-_36891830 | 6.90 |
ENST00000578487.1
|
PCGF2
|
polycomb group ring finger 2 |
| chr2_+_177134134 | 6.86 |
ENST00000249442.6
ENST00000392529.2 ENST00000443241.1 |
MTX2
|
metaxin 2 |
| chr4_+_99916765 | 6.82 |
ENST00000296411.6
|
METAP1
|
methionyl aminopeptidase 1 |
| chr3_+_184081137 | 6.81 |
ENST00000443489.1
|
POLR2H
|
polymerase (RNA) II (DNA directed) polypeptide H |
| chr13_-_37573432 | 6.79 |
ENST00000413537.2
ENST00000443765.1 ENST00000239891.3 |
ALG5
|
ALG5, dolichyl-phosphate beta-glucosyltransferase |
| chr4_-_183838596 | 6.75 |
ENST00000508994.1
ENST00000512766.1 |
DCTD
|
dCMP deaminase |
| chr19_+_10982189 | 6.64 |
ENST00000327064.4
ENST00000588947.1 |
CARM1
|
coactivator-associated arginine methyltransferase 1 |
| chrX_+_47053208 | 6.58 |
ENST00000442035.1
ENST00000457753.1 ENST00000335972.6 |
UBA1
|
ubiquitin-like modifier activating enzyme 1 |
| chr12_-_56583243 | 6.57 |
ENST00000550164.1
|
SMARCC2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 2 |
| chr5_+_137688285 | 6.57 |
ENST00000314358.5
|
KDM3B
|
lysine (K)-specific demethylase 3B |
| chr11_-_64013663 | 6.51 |
ENST00000392210.2
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr1_-_36615051 | 6.49 |
ENST00000373163.1
|
TRAPPC3
|
trafficking protein particle complex 3 |
| chr3_-_48647470 | 6.48 |
ENST00000203407.5
|
UQCRC1
|
ubiquinol-cytochrome c reductase core protein I |
| chr3_-_64009102 | 6.46 |
ENST00000478185.1
ENST00000482510.1 ENST00000497323.1 ENST00000492933.1 ENST00000295901.4 |
PSMD6
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 6 |
| chr12_-_51663959 | 6.45 |
ENST00000604188.1
ENST00000398453.3 |
SMAGP
|
small cell adhesion glycoprotein |
| chr12_-_120638902 | 6.45 |
ENST00000551150.1
ENST00000313104.5 ENST00000547191.1 ENST00000546989.1 ENST00000392514.4 ENST00000228306.4 ENST00000550856.1 |
RPLP0
|
ribosomal protein, large, P0 |
| chr12_-_51664058 | 6.41 |
ENST00000605627.1
|
SMAGP
|
small cell adhesion glycoprotein |
| chr19_-_14682838 | 6.41 |
ENST00000215565.2
|
NDUFB7
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 7, 18kDa |
| chr17_-_3749515 | 6.39 |
ENST00000158149.3
ENST00000389005.4 |
C17orf85
|
chromosome 17 open reading frame 85 |
| chr12_-_120907374 | 6.34 |
ENST00000550458.1
|
SRSF9
|
serine/arginine-rich splicing factor 9 |
| chr4_-_183838372 | 6.28 |
ENST00000503820.1
ENST00000503988.1 |
DCTD
|
dCMP deaminase |
| chr3_-_183602515 | 6.25 |
ENST00000449306.1
ENST00000435888.1 ENST00000311101.5 ENST00000317096.4 |
PARL
|
presenilin associated, rhomboid-like |
| chr19_-_10530784 | 6.18 |
ENST00000593124.1
|
CDC37
|
cell division cycle 37 |
| chr2_+_90108504 | 6.18 |
ENST00000390271.2
|
IGKV6D-41
|
immunoglobulin kappa variable 6D-41 (non-functional) |
| chr22_-_22901477 | 6.16 |
ENST00000420709.1
ENST00000398741.1 ENST00000405655.3 |
PRAME
|
preferentially expressed antigen in melanoma |
| chrX_+_123094369 | 6.11 |
ENST00000455404.1
ENST00000218089.9 |
STAG2
|
stromal antigen 2 |
| chr2_-_151344172 | 6.07 |
ENST00000375734.2
ENST00000263895.4 ENST00000454202.1 |
RND3
|
Rho family GTPase 3 |
| chr17_+_38137050 | 6.00 |
ENST00000264639.4
|
PSMD3
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 3 |
| chr15_-_55489097 | 6.00 |
ENST00000260443.4
|
RSL24D1
|
ribosomal L24 domain containing 1 |
| chr12_-_102224704 | 5.98 |
ENST00000299314.7
|
GNPTAB
|
N-acetylglucosamine-1-phosphate transferase, alpha and beta subunits |
| chr8_-_71520513 | 5.97 |
ENST00000262213.2
ENST00000536748.1 ENST00000518678.1 |
TRAM1
|
translocation associated membrane protein 1 |
| chr17_-_1090599 | 5.94 |
ENST00000544583.2
|
ABR
|
active BCR-related |
| chr12_-_46384334 | 5.94 |
ENST00000369367.3
ENST00000266589.6 ENST00000395453.2 ENST00000395454.2 |
SCAF11
|
SR-related CTD-associated factor 11 |
| chr9_+_131451480 | 5.93 |
ENST00000322030.8
|
SET
|
SET nuclear oncogene |
| chr1_+_110527308 | 5.93 |
ENST00000369799.5
|
AHCYL1
|
adenosylhomocysteinase-like 1 |
| chr16_+_53088885 | 5.90 |
ENST00000566029.1
ENST00000447540.1 |
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chrX_-_153285395 | 5.85 |
ENST00000369980.3
|
IRAK1
|
interleukin-1 receptor-associated kinase 1 |
| chr2_+_118572226 | 5.83 |
ENST00000263239.2
|
DDX18
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 18 |
| chr16_-_30441293 | 5.79 |
ENST00000565758.1
ENST00000567983.1 ENST00000319285.4 |
DCTPP1
|
dCTP pyrophosphatase 1 |
| chr16_-_11891048 | 5.73 |
ENST00000571198.1
ENST00000572781.1 ENST00000355758.4 |
ZC3H7A
|
zinc finger CCCH-type containing 7A |
| chr7_-_96339167 | 5.69 |
ENST00000444799.1
ENST00000417009.1 ENST00000248566.2 |
SHFM1
|
split hand/foot malformation (ectrodactyly) type 1 |
| chr4_-_183838747 | 5.67 |
ENST00000438320.2
|
DCTD
|
dCMP deaminase |
| chr2_-_130939115 | 5.66 |
ENST00000441135.1
ENST00000339679.7 ENST00000426662.2 ENST00000443958.2 ENST00000351288.6 ENST00000453750.1 ENST00000452225.2 |
SMPD4
|
sphingomyelin phosphodiesterase 4, neutral membrane (neutral sphingomyelinase-3) |
| chrX_-_153285251 | 5.64 |
ENST00000444230.1
ENST00000393682.1 ENST00000393687.2 ENST00000429936.2 ENST00000369974.2 |
IRAK1
|
interleukin-1 receptor-associated kinase 1 |
| chr12_-_54070098 | 5.64 |
ENST00000394349.3
ENST00000549164.1 |
ATP5G2
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C2 (subunit 9) |
| chr8_+_22436248 | 5.63 |
ENST00000308354.7
|
PDLIM2
|
PDZ and LIM domain 2 (mystique) |
| chr15_+_63340647 | 5.58 |
ENST00000404484.4
|
TPM1
|
tropomyosin 1 (alpha) |
| chr3_+_141457105 | 5.58 |
ENST00000480908.1
ENST00000393000.3 |
RNF7
|
ring finger protein 7 |
| chr12_-_54069856 | 5.57 |
ENST00000602871.1
|
ATP5G2
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C2 (subunit 9) |
| chr1_-_75198940 | 5.52 |
ENST00000417775.1
|
CRYZ
|
crystallin, zeta (quinone reductase) |
| chr7_+_89783689 | 5.52 |
ENST00000297205.2
|
STEAP1
|
six transmembrane epithelial antigen of the prostate 1 |
| chr6_-_99797522 | 5.46 |
ENST00000389677.5
|
FAXC
|
failed axon connections homolog (Drosophila) |
| chr22_-_22901636 | 5.42 |
ENST00000406503.1
ENST00000439106.1 ENST00000402697.1 ENST00000543184.1 ENST00000398743.2 |
PRAME
|
preferentially expressed antigen in melanoma |
| chr10_+_13203543 | 5.40 |
ENST00000378714.3
ENST00000479669.1 ENST00000484800.2 |
MCM10
|
minichromosome maintenance complex component 10 |
| chr15_+_66679155 | 5.36 |
ENST00000307102.5
|
MAP2K1
|
mitogen-activated protein kinase kinase 1 |
| chr17_+_79670386 | 5.34 |
ENST00000333676.3
ENST00000571730.1 ENST00000541223.1 |
MRPL12
SLC25A10
SLC25A10
|
mitochondrial ribosomal protein L12 Mitochondrial dicarboxylate carrier; Uncharacterized protein; cDNA FLJ60124, highly similar to Mitochondrial dicarboxylate carrier solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 |
| chr11_+_66025938 | 5.31 |
ENST00000394066.2
|
KLC2
|
kinesin light chain 2 |
| chr13_-_48669232 | 5.30 |
ENST00000258648.2
ENST00000378586.1 |
MED4
|
mediator complex subunit 4 |
| chr4_-_100815525 | 5.26 |
ENST00000226522.8
ENST00000499666.2 |
LAMTOR3
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 3 |
| chr7_-_102213030 | 5.26 |
ENST00000511313.1
ENST00000513438.1 ENST00000513506.1 |
POLR2J3
|
polymerase (RNA) II (DNA directed) polypeptide J3 |
| chr11_+_60691924 | 5.25 |
ENST00000544065.1
ENST00000453848.2 ENST00000005286.4 |
TMEM132A
|
transmembrane protein 132A |
| chr12_-_51663728 | 5.25 |
ENST00000603864.1
ENST00000605426.1 |
SMAGP
|
small cell adhesion glycoprotein |
| chr2_+_177134201 | 5.22 |
ENST00000452865.1
|
MTX2
|
metaxin 2 |
| chr11_+_64781575 | 5.21 |
ENST00000246747.4
ENST00000529384.1 |
ARL2
|
ADP-ribosylation factor-like 2 |
| chr21_-_33975547 | 5.21 |
ENST00000431599.1
|
C21orf59
|
chromosome 21 open reading frame 59 |
| chr17_+_65713925 | 5.19 |
ENST00000253247.4
|
NOL11
|
nucleolar protein 11 |
| chr4_-_183838482 | 5.17 |
ENST00000357067.3
ENST00000510370.1 |
DCTD
|
dCMP deaminase |
| chr2_+_90060377 | 5.17 |
ENST00000436451.2
|
IGKV6D-21
|
immunoglobulin kappa variable 6D-21 (non-functional) |
Network of associatons between targets according to the STRING database.
First level regulatory network of CTCF_CTCFL
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.4 | 37.1 | GO:0071344 | diphosphate metabolic process(GO:0071344) |
| 10.7 | 32.1 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 8.6 | 34.5 | GO:0071733 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 5.9 | 23.8 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 5.2 | 20.6 | GO:0097068 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 5.0 | 39.9 | GO:0010994 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 4.8 | 33.6 | GO:1904690 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 4.4 | 13.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 4.2 | 12.7 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 4.2 | 12.6 | GO:0002188 | translation reinitiation(GO:0002188) |
| 3.8 | 11.4 | GO:0018282 | metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) |
| 3.8 | 18.8 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 3.6 | 14.5 | GO:1901165 | negative regulation of MDA-5 signaling pathway(GO:0039534) positive regulation of trophoblast cell migration(GO:1901165) |
| 3.4 | 23.9 | GO:0006226 | dUMP biosynthetic process(GO:0006226) |
| 3.3 | 16.7 | GO:0034971 | regulation of growth plate cartilage chondrocyte proliferation(GO:0003420) histone H3-R17 methylation(GO:0034971) |
| 2.9 | 5.8 | GO:0009213 | pyrimidine nucleoside triphosphate catabolic process(GO:0009149) pyrimidine deoxyribonucleoside triphosphate catabolic process(GO:0009213) |
| 2.7 | 21.8 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 2.5 | 7.6 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 2.5 | 4.9 | GO:0002352 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 2.4 | 7.1 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 2.3 | 9.3 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 2.3 | 7.0 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 2.3 | 34.2 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 2.3 | 9.1 | GO:0046104 | thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 2.3 | 2.3 | GO:0010868 | negative regulation of triglyceride biosynthetic process(GO:0010868) |
| 2.2 | 22.0 | GO:0051096 | positive regulation of helicase activity(GO:0051096) |
| 2.1 | 8.4 | GO:0043248 | proteasome assembly(GO:0043248) |
| 2.1 | 6.2 | GO:0014876 | response to injury involved in regulation of muscle adaptation(GO:0014876) |
| 2.0 | 20.0 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 2.0 | 5.9 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 1.9 | 5.7 | GO:0071030 | nuclear mRNA surveillance of spliceosomal pre-mRNA splicing(GO:0071030) nuclear retention of unspliced pre-mRNA at the site of transcription(GO:0071048) |
| 1.9 | 7.5 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 1.8 | 7.3 | GO:2000434 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 1.8 | 10.9 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 1.8 | 12.6 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 1.8 | 5.4 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 1.8 | 7.1 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 1.8 | 5.3 | GO:0015709 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) malate transmembrane transport(GO:0071423) oxaloacetate(2-) transmembrane transport(GO:1902356) |
| 1.7 | 15.3 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 1.7 | 20.2 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 1.6 | 4.8 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 1.5 | 7.7 | GO:0048478 | replication fork protection(GO:0048478) |
| 1.5 | 6.0 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 1.5 | 4.4 | GO:0018153 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 1.5 | 7.3 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 1.5 | 7.3 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 1.5 | 8.8 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 1.4 | 8.7 | GO:1903899 | positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 1.4 | 5.5 | GO:1903412 | response to bile acid(GO:1903412) |
| 1.4 | 4.1 | GO:0006258 | UDP-glucose catabolic process(GO:0006258) galactose catabolic process via UDP-galactose(GO:0033499) |
| 1.3 | 12.1 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 1.3 | 12.0 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 1.3 | 5.3 | GO:1904753 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) negative regulation of vascular associated smooth muscle cell migration(GO:1904753) |
| 1.2 | 2.5 | GO:0010725 | regulation of primitive erythrocyte differentiation(GO:0010725) eosinophil fate commitment(GO:0035854) |
| 1.2 | 9.9 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 1.2 | 4.8 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 1.2 | 13.2 | GO:0045116 | protein neddylation(GO:0045116) |
| 1.2 | 7.2 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 1.2 | 3.5 | GO:1901202 | negative regulation of extracellular matrix assembly(GO:1901202) |
| 1.2 | 3.5 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 1.1 | 21.8 | GO:0070200 | establishment of protein localization to telomere(GO:0070200) |
| 1.1 | 7.9 | GO:1901836 | regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901836) positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 1.1 | 3.3 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 1.1 | 3.3 | GO:1901535 | regulation of DNA demethylation(GO:1901535) negative regulation of DNA demethylation(GO:1901536) regulation of genetic imprinting(GO:2000653) |
| 1.1 | 8.6 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 1.0 | 3.1 | GO:0006535 | cysteine biosynthetic process from serine(GO:0006535) |
| 1.0 | 7.2 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 1.0 | 3.0 | GO:0072425 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 1.0 | 8.1 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 1.0 | 24.1 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 1.0 | 13.7 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 1.0 | 10.8 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 1.0 | 46.7 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.9 | 9.4 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.9 | 6.3 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.9 | 4.5 | GO:0098706 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.9 | 5.4 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.9 | 4.4 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.9 | 3.5 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.9 | 3.5 | GO:0043570 | maintenance of DNA repeat elements(GO:0043570) |
| 0.9 | 62.3 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.8 | 9.2 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.8 | 2.5 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.8 | 11.5 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.8 | 4.9 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.8 | 4.9 | GO:0016479 | negative regulation of transcription from RNA polymerase I promoter(GO:0016479) |
| 0.8 | 2.4 | GO:0018008 | N-terminal peptidyl-glycine N-myristoylation(GO:0018008) peptidyl-glycine modification(GO:0018201) |
| 0.8 | 3.2 | GO:0002545 | chronic inflammatory response to non-antigenic stimulus(GO:0002545) regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002880) |
| 0.8 | 2.3 | GO:0075528 | induction by symbiont of host defense response(GO:0044416) induction of host immune response by virus(GO:0046730) active induction of host immune response by virus(GO:0046732) modulation by symbiont of host defense response(GO:0052031) induction by organism of defense response of other organism involved in symbiotic interaction(GO:0052251) modulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052255) positive regulation by symbiont of host defense response(GO:0052509) positive regulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052510) modulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052552) modulation by symbiont of host immune response(GO:0052553) modulation by virus of host immune response(GO:0075528) |
| 0.8 | 7.6 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.8 | 7.5 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.8 | 8.3 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.8 | 2.3 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 0.7 | 7.5 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.7 | 13.4 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.7 | 8.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.7 | 6.6 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.7 | 7.2 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.7 | 2.8 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) positive regulation of potassium ion import(GO:1903288) |
| 0.7 | 8.3 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.7 | 6.9 | GO:0018202 | peptidyl-histidine modification(GO:0018202) |
| 0.7 | 17.7 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.7 | 2.7 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.7 | 2.0 | GO:0021827 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) regulation of negative chemotaxis(GO:0050923) |
| 0.6 | 10.4 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.6 | 5.2 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.6 | 4.5 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.6 | 3.2 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.6 | 15.4 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.6 | 10.1 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.6 | 4.4 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.6 | 11.4 | GO:0034162 | toll-like receptor 9 signaling pathway(GO:0034162) |
| 0.6 | 5.9 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.6 | 2.4 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 0.6 | 16.5 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.6 | 5.7 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.6 | 2.3 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.6 | 12.2 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.6 | 1.7 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.6 | 1.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.5 | 4.4 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.5 | 3.8 | GO:0048318 | axial mesoderm development(GO:0048318) |
| 0.5 | 4.3 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.5 | 1.6 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.5 | 2.7 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.5 | 2.1 | GO:0016334 | morphogenesis of follicular epithelium(GO:0016333) establishment or maintenance of polarity of follicular epithelium(GO:0016334) establishment of planar polarity of follicular epithelium(GO:0042247) |
| 0.5 | 51.4 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.5 | 8.8 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.5 | 2.0 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.5 | 12.6 | GO:0008334 | histone mRNA metabolic process(GO:0008334) |
| 0.5 | 9.8 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.5 | 2.0 | GO:0060708 | histone H3-K27 acetylation(GO:0043974) leukemia inhibitory factor signaling pathway(GO:0048861) spongiotrophoblast differentiation(GO:0060708) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.5 | 16.1 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.5 | 1.9 | GO:0061484 | inner cell mass cell differentiation(GO:0001826) hematopoietic stem cell homeostasis(GO:0061484) |
| 0.5 | 3.7 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.5 | 4.1 | GO:1900125 | regulation of hyaluronan biosynthetic process(GO:1900125) |
| 0.5 | 4.1 | GO:0010918 | positive regulation of mitochondrial membrane potential(GO:0010918) |
| 0.4 | 29.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.4 | 1.7 | GO:0060268 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) negative regulation of respiratory burst(GO:0060268) |
| 0.4 | 1.7 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.4 | 2.9 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.4 | 12.5 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.4 | 5.0 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.4 | 23.5 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.4 | 1.6 | GO:0046725 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of nephron tubule epithelial cell differentiation(GO:0072183) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) negative regulation of epithelial cell differentiation involved in kidney development(GO:2000697) |
| 0.4 | 1.2 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.4 | 3.4 | GO:0042407 | cristae formation(GO:0042407) |
| 0.4 | 15.6 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.4 | 4.3 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.4 | 1.1 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.4 | 2.1 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.3 | 27.8 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.3 | 2.7 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.3 | 1.6 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
| 0.3 | 14.3 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.3 | 1.3 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.3 | 4.1 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.3 | 0.9 | GO:0003220 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) |
| 0.3 | 6.2 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.3 | 0.6 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.3 | 5.5 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.3 | 1.7 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.3 | 2.0 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.3 | 1.1 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.3 | 9.0 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.3 | 7.8 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.3 | 1.0 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.3 | 1.0 | GO:0032933 | SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.3 | 3.3 | GO:0035058 | nonmotile primary cilium assembly(GO:0035058) regulation of nonmotile primary cilium assembly(GO:1902855) positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.3 | 3.0 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.2 | 6.5 | GO:0071174 | mitotic spindle checkpoint(GO:0071174) |
| 0.2 | 14.1 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.2 | 4.5 | GO:0098779 | mitophagy in response to mitochondrial depolarization(GO:0098779) |
| 0.2 | 30.0 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.2 | 7.0 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.2 | 22.8 | GO:0048199 | vesicle targeting, to, from or within Golgi(GO:0048199) |
| 0.2 | 10.1 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.2 | 2.6 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.2 | 4.7 | GO:0006206 | pyrimidine nucleobase metabolic process(GO:0006206) |
| 0.2 | 11.1 | GO:0018279 | peptidyl-asparagine modification(GO:0018196) protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.2 | 3.4 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.2 | 6.6 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.2 | 4.3 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.2 | 6.6 | GO:0060065 | uterus development(GO:0060065) |
| 0.2 | 7.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.2 | 1.1 | GO:0006581 | acetylcholine catabolic process in synaptic cleft(GO:0001507) acetylcholine catabolic process(GO:0006581) |
| 0.2 | 2.5 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.2 | 11.3 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.2 | 19.7 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.2 | 0.6 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.2 | 1.1 | GO:0042426 | choline catabolic process(GO:0042426) |
| 0.2 | 3.1 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.2 | 2.5 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.2 | 0.4 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.2 | 1.8 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.2 | 6.8 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.2 | 0.9 | GO:0090231 | regulation of spindle checkpoint(GO:0090231) |
| 0.2 | 2.9 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.2 | 5.2 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.2 | 6.4 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.2 | 6.0 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.2 | 2.2 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.2 | 0.8 | GO:2000812 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) regulation of barbed-end actin filament capping(GO:2000812) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.2 | 0.9 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.1 | 2.5 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 2.5 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.1 | 2.1 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.1 | 6.1 | GO:0099601 | regulation of neurotransmitter receptor activity(GO:0099601) |
| 0.1 | 3.7 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.1 | 1.8 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 1.6 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.1 | 5.7 | GO:0006739 | NADP metabolic process(GO:0006739) |
| 0.1 | 1.4 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.1 | 1.0 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.1 | 1.2 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.1 | 1.3 | GO:0015886 | heme transport(GO:0015886) |
| 0.1 | 0.5 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) |
| 0.1 | 0.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 1.6 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.1 | 1.6 | GO:0006295 | nucleotide-excision repair, preincision complex stabilization(GO:0006293) nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.1 | 0.7 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.1 | 2.6 | GO:0048169 | regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.1 | 0.3 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.1 | 0.7 | GO:0002911 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) |
| 0.1 | 1.0 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 | 0.4 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.1 | 3.3 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 3.8 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 1.9 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.1 | 0.4 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) positive regulation of blood vessel remodeling(GO:2000504) |
| 0.1 | 0.4 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.1 | 34.6 | GO:0008380 | RNA splicing(GO:0008380) |
| 0.1 | 8.8 | GO:0006413 | translational initiation(GO:0006413) |
| 0.1 | 1.8 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 1.8 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.1 | 4.4 | GO:0090280 | positive regulation of calcium ion import(GO:0090280) |
| 0.1 | 0.6 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 1.1 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.1 | 3.1 | GO:0006754 | ATP biosynthetic process(GO:0006754) |
| 0.1 | 0.5 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.1 | 2.0 | GO:0030042 | actin filament depolymerization(GO:0030042) |
| 0.1 | 1.2 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 3.1 | GO:0008637 | apoptotic mitochondrial changes(GO:0008637) |
| 0.1 | 3.8 | GO:0035722 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.1 | 1.1 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.1 | 0.4 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.1 | 0.4 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.1 | 0.5 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.1 | 2.9 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.1 | 0.5 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.1 | 1.1 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 2.5 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.1 | 0.9 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.1 | 1.8 | GO:0002260 | lymphocyte homeostasis(GO:0002260) |
| 0.1 | 1.1 | GO:0006511 | ubiquitin-dependent protein catabolic process(GO:0006511) |
| 0.1 | 3.5 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.1 | 8.0 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 5.1 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.1 | 5.7 | GO:0006364 | rRNA processing(GO:0006364) |
| 0.1 | 1.2 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 3.4 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.8 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.9 | GO:0048147 | negative regulation of fibroblast proliferation(GO:0048147) |
| 0.0 | 0.6 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.5 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.0 | 0.7 | GO:0032802 | low-density lipoprotein particle receptor catabolic process(GO:0032802) |
| 0.0 | 4.8 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.8 | GO:0030148 | sphingolipid biosynthetic process(GO:0030148) |
| 0.0 | 1.3 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 6.0 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.0 | 0.1 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.0 | 0.8 | GO:1900078 | positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.0 | 3.0 | GO:0051453 | regulation of intracellular pH(GO:0051453) |
| 0.0 | 0.9 | GO:0043304 | regulation of mast cell activation involved in immune response(GO:0033006) regulation of mast cell degranulation(GO:0043304) |
| 0.0 | 0.0 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.8 | GO:0045624 | positive regulation of T-helper cell differentiation(GO:0045624) |
| 0.0 | 1.3 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 0.3 | GO:0044839 | cell cycle G2/M phase transition(GO:0044839) |
| 0.0 | 0.2 | GO:0045955 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) |
| 0.0 | 0.7 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.9 | GO:0034383 | low-density lipoprotein particle clearance(GO:0034383) |
| 0.0 | 0.3 | GO:0032801 | receptor catabolic process(GO:0032801) |
| 0.0 | 3.2 | GO:0051168 | nuclear export(GO:0051168) |
| 0.0 | 0.7 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.2 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.8 | GO:0006897 | endocytosis(GO:0006897) |
| 0.0 | 1.5 | GO:0030819 | positive regulation of cAMP biosynthetic process(GO:0030819) |
| 0.0 | 0.2 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.1 | GO:0009099 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.0 | 0.1 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.1 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 1.7 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.9 | GO:0007052 | mitotic spindle organization(GO:0007052) |
| 0.0 | 0.8 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.4 | GO:0001755 | neural crest cell migration(GO:0001755) |
| 0.0 | 0.1 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.0 | 0.0 | GO:0032252 | secretory granule localization(GO:0032252) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.9 | 79.9 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 6.8 | 34.2 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 6.2 | 24.8 | GO:0070876 | SOSS complex(GO:0070876) |
| 6.1 | 61.2 | GO:0097255 | R2TP complex(GO:0097255) |
| 4.4 | 13.2 | GO:0043224 | nuclear SCF ubiquitin ligase complex(GO:0043224) |
| 4.1 | 16.5 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 3.7 | 25.6 | GO:0030870 | Mre11 complex(GO:0030870) |
| 2.9 | 23.5 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 2.6 | 7.9 | GO:0034455 | t-UTP complex(GO:0034455) |
| 2.3 | 11.5 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 2.3 | 22.8 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 1.9 | 5.7 | GO:0071020 | post-spliceosomal complex(GO:0071020) |
| 1.8 | 7.2 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 1.8 | 16.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 1.8 | 16.1 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 1.8 | 12.4 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 1.8 | 5.3 | GO:0016938 | kinesin I complex(GO:0016938) |
| 1.8 | 14.0 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 1.7 | 51.9 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 1.6 | 8.2 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 1.6 | 4.9 | GO:0031523 | Myb complex(GO:0031523) |
| 1.6 | 63.7 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 1.6 | 7.9 | GO:0089701 | U2AF(GO:0089701) |
| 1.6 | 4.7 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 1.4 | 44.6 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 1.4 | 25.9 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 1.3 | 3.8 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 1.2 | 13.7 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 1.2 | 9.7 | GO:0031931 | TORC1 complex(GO:0031931) |
| 1.2 | 36.2 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 1.2 | 10.7 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 1.1 | 4.4 | GO:0042025 | host cell nucleus(GO:0042025) host cell nuclear part(GO:0044094) |
| 1.1 | 5.3 | GO:0071986 | Ragulator complex(GO:0071986) |
| 1.0 | 22.6 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 1.0 | 7.2 | GO:1990635 | proximal dendrite(GO:1990635) |
| 1.0 | 10.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 1.0 | 9.8 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 1.0 | 11.6 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.9 | 9.3 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.9 | 5.2 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.9 | 3.5 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.8 | 11.7 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.8 | 8.8 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.8 | 10.1 | GO:0030681 | multimeric ribonuclease P complex(GO:0030681) |
| 0.8 | 39.8 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.7 | 10.3 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.7 | 5.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.7 | 9.5 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.6 | 9.7 | GO:0090543 | Flemming body(GO:0090543) |
| 0.6 | 1.9 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.6 | 1.9 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.6 | 7.7 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.6 | 15.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.6 | 13.1 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.6 | 2.4 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.6 | 2.9 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.6 | 3.5 | GO:0090661 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.6 | 7.0 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.6 | 8.1 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.5 | 1.6 | GO:0030689 | Noc complex(GO:0030689) |
| 0.5 | 1.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.5 | 55.4 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.5 | 17.8 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.5 | 4.8 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.5 | 3.4 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.5 | 3.7 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.5 | 4.1 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.4 | 29.3 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.4 | 5.6 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.4 | 2.1 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.4 | 6.3 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.4 | 1.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.4 | 2.5 | GO:0016589 | NURF complex(GO:0016589) |
| 0.3 | 2.7 | GO:0051286 | cell tip(GO:0051286) |
| 0.3 | 4.9 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.3 | 1.6 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.3 | 4.7 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.3 | 25.5 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.3 | 3.9 | GO:0097433 | dense body(GO:0097433) |
| 0.3 | 0.6 | GO:0031213 | RSF complex(GO:0031213) |
| 0.3 | 13.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.3 | 7.1 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.3 | 1.0 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.3 | 8.8 | GO:0032420 | stereocilium(GO:0032420) |
| 0.2 | 1.2 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.2 | 34.1 | GO:0005840 | ribosome(GO:0005840) |
| 0.2 | 0.7 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.2 | 16.5 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.2 | 1.5 | GO:0070522 | nucleotide-excision repair factor 1 complex(GO:0000110) ERCC4-ERCC1 complex(GO:0070522) |
| 0.2 | 2.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.2 | 1.8 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.2 | 2.3 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.2 | 1.1 | GO:1990130 | EGO complex(GO:0034448) Iml1 complex(GO:1990130) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.2 | 20.4 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.2 | 10.0 | GO:0015030 | Cajal body(GO:0015030) |
| 0.2 | 3.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.2 | 3.3 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.2 | 9.6 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 20.2 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 12.0 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 1.8 | GO:0044754 | autolysosome(GO:0044754) |
| 0.1 | 2.8 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 51.7 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 14.2 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.1 | 2.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 19.0 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 1.7 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 0.9 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.1 | 2.4 | GO:0018995 | host(GO:0018995) host cell(GO:0043657) |
| 0.1 | 12.5 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.1 | 0.9 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.1 | 3.1 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.1 | 3.5 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 3.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 14.9 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 4.8 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.1 | 16.9 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.1 | 5.2 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 4.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 6.3 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.1 | 0.4 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.1 | 0.3 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.1 | 4.2 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 1.1 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 1.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 4.6 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.1 | 1.5 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.1 | 0.4 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.1 | 6.4 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 3.2 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 1.6 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.1 | 26.4 | GO:0000785 | chromatin(GO:0000785) |
| 0.1 | 2.4 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 7.6 | GO:0101002 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.1 | 4.8 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.1 | 10.4 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 5.5 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 1.0 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.8 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 1.4 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 12.1 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.5 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 2.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 4.0 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.7 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 1.0 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.2 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.5 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.3 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 19.7 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 1.6 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.0 | 0.3 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 1.2 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 1.3 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 0.1 | GO:0033061 | DNA recombinase mediator complex(GO:0033061) Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.2 | 32.6 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 6.0 | 23.9 | GO:0004132 | dCMP deaminase activity(GO:0004132) |
| 5.7 | 34.5 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 4.8 | 14.5 | GO:0030984 | kininogen binding(GO:0030984) |
| 4.4 | 13.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 4.0 | 12.1 | GO:0098626 | methylselenol reductase activity(GO:0098625) methylseleninic acid reductase activity(GO:0098626) |
| 3.0 | 9.1 | GO:0004797 | thymidine kinase activity(GO:0004797) |
| 2.7 | 8.2 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 2.5 | 51.9 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 2.5 | 24.7 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 2.4 | 7.2 | GO:0045155 | electron transporter, transferring electrons from CoQH2-cytochrome c reductase complex and cytochrome c oxidase complex activity(GO:0045155) |
| 2.3 | 23.5 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 2.3 | 28.1 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 2.3 | 32.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 2.3 | 24.8 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 2.2 | 13.1 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 2.0 | 8.1 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 1.9 | 25.3 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 1.9 | 5.7 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 1.8 | 5.3 | GO:0015117 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) |
| 1.7 | 6.9 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 1.7 | 8.3 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 1.6 | 11.5 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 1.6 | 7.9 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 1.6 | 4.7 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 1.5 | 26.5 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 1.5 | 7.3 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 1.3 | 14.5 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) |
| 1.3 | 11.4 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 1.2 | 4.9 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 1.2 | 8.4 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 1.2 | 7.1 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 1.2 | 3.5 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 1.1 | 5.6 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 1.1 | 7.8 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 1.1 | 3.3 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 1.1 | 15.3 | GO:0031014 | troponin T binding(GO:0031014) |
| 1.1 | 4.3 | GO:0052795 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 1.0 | 3.1 | GO:0070025 | cystathionine beta-synthase activity(GO:0004122) oxidoreductase activity, acting on other nitrogenous compounds as donors, cytochrome as acceptor(GO:0016662) nitrite reductase (NO-forming) activity(GO:0050421) carbon monoxide binding(GO:0070025) nitrite reductase activity(GO:0098809) |
| 1.0 | 7.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 1.0 | 3.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 1.0 | 9.9 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 1.0 | 9.8 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 1.0 | 20.6 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 1.0 | 6.8 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.9 | 6.5 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.9 | 42.5 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.9 | 4.6 | GO:0048256 | flap endonuclease activity(GO:0048256) |
| 0.9 | 13.5 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.8 | 26.9 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.8 | 2.5 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.8 | 5.8 | GO:0032552 | deoxyribonucleotide binding(GO:0032552) |
| 0.8 | 8.2 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.8 | 2.4 | GO:0019107 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) myristoyltransferase activity(GO:0019107) |
| 0.8 | 2.3 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.8 | 4.5 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.8 | 2.3 | GO:0070404 | NADH binding(GO:0070404) |
| 0.7 | 28.9 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.7 | 7.3 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.7 | 10.2 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.7 | 13.4 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.7 | 1.4 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.7 | 6.2 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.7 | 7.5 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.7 | 2.7 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.7 | 42.6 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.6 | 10.1 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.6 | 1.8 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.6 | 3.5 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.6 | 37.3 | GO:0016655 | oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor(GO:0016655) |
| 0.6 | 2.8 | GO:0004905 | type I interferon receptor activity(GO:0004905) |
| 0.6 | 5.7 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.6 | 4.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.5 | 5.5 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.5 | 113.1 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.5 | 3.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.5 | 3.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.5 | 20.5 | GO:0043175 | RNA polymerase core enzyme binding(GO:0043175) |
| 0.5 | 12.1 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.5 | 3.0 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.5 | 2.9 | GO:0046979 | TAP2 binding(GO:0046979) |
| 0.5 | 10.7 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.5 | 5.3 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.5 | 9.5 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.5 | 7.2 | GO:0019841 | retinol binding(GO:0019841) |
| 0.4 | 4.9 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.4 | 3.1 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.4 | 4.7 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.4 | 3.3 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.4 | 11.2 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.4 | 4.4 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.4 | 9.6 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.4 | 11.5 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.4 | 5.4 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.4 | 8.5 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.4 | 7.0 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.4 | 11.0 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.4 | 8.8 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.3 | 13.3 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.3 | 11.7 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.3 | 18.5 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.3 | 4.0 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.3 | 31.4 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.3 | 2.9 | GO:0031386 | protein tag(GO:0031386) |
| 0.3 | 0.9 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.3 | 3.5 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.3 | 4.4 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.3 | 1.0 | GO:0022858 | L-alanine transmembrane transporter activity(GO:0015180) alanine transmembrane transporter activity(GO:0022858) |
| 0.3 | 4.6 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.3 | 2.5 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.2 | 4.7 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.2 | 1.0 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.2 | 3.7 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.2 | 4.4 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.2 | 9.9 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.2 | 3.8 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.2 | 2.6 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.2 | 0.9 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.2 | 7.2 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.2 | 2.7 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.2 | 2.0 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.2 | 1.6 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.2 | 1.1 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.2 | 2.6 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.2 | 1.5 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.2 | 6.3 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.2 | 24.0 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.2 | 2.9 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.2 | 9.7 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.2 | 0.8 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.2 | 8.5 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.2 | 4.4 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.2 | 0.6 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.2 | 5.0 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.2 | 2.0 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.2 | 0.9 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.2 | 0.7 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.2 | 2.8 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.2 | 0.9 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.2 | 5.0 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.2 | 0.9 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.2 | 6.6 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.2 | 14.6 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.2 | 2.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.2 | 1.6 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.2 | 4.1 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.2 | 2.5 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.2 | 1.7 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 4.2 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 3.8 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 1.6 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 2.9 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 1.3 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 5.3 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 1.1 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.1 | 2.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 2.4 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.1 | 4.5 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.1 | 3.1 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 10.1 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 1.8 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.1 | 0.5 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.1 | 1.4 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 1.1 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.1 | 2.4 | GO:0071949 | FAD binding(GO:0071949) |
| 0.1 | 1.7 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.3 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.1 | 5.4 | GO:0008186 | ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.1 | 5.9 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.1 | 3.2 | GO:0043531 | ADP binding(GO:0043531) |
| 0.1 | 2.9 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 2.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 0.5 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 2.0 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.1 | 33.4 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 0.4 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.1 | 0.6 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 1.8 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.1 | 2.0 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.1 | 0.2 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.1 | 1.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 1.9 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 0.5 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.1 | 9.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 1.1 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 0.4 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.1 | 1.9 | GO:0005262 | calcium channel activity(GO:0005262) calcium ion transmembrane transporter activity(GO:0015085) |
| 0.1 | 2.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 0.8 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.1 | 1.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 1.6 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.8 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 4.4 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 1.2 | GO:0008233 | peptidase activity(GO:0008233) |
| 0.0 | 0.7 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 2.8 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 1.5 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 0.8 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 1.3 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) |
| 0.0 | 6.2 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 1.5 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 1.0 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 1.1 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.2 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.1 | GO:0004084 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.1 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.0 | 0.6 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.7 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.4 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 1.8 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.0 | 0.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.6 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 16.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.7 | 34.5 | PID MYC PATHWAY | C-MYC pathway |
| 0.7 | 42.7 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.6 | 37.6 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.4 | 7.7 | PID ATR PATHWAY | ATR signaling pathway |
| 0.4 | 6.4 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.4 | 2.1 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.4 | 18.8 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.3 | 23.8 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.3 | 8.8 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.3 | 13.5 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.3 | 7.0 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.3 | 5.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.2 | 3.7 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.2 | 6.9 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.2 | 7.8 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.2 | 6.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.2 | 4.9 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.2 | 11.0 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.2 | 4.8 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.2 | 15.0 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.2 | 7.8 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.2 | 18.3 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.2 | 4.1 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.2 | 3.5 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.2 | 9.0 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.2 | 4.9 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.2 | 15.8 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.2 | 6.0 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.2 | 4.4 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 11.2 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 2.7 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 1.1 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.1 | 2.9 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.1 | 3.9 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 7.6 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.1 | 5.8 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 4.9 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 1.1 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 2.0 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 4.0 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 6.9 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 3.5 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 1.3 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 1.7 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 3.7 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 1.8 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.4 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.4 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 46.7 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 1.6 | 45.8 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 1.6 | 34.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 1.2 | 21.8 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 1.0 | 39.0 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.9 | 15.1 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.9 | 33.0 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.9 | 11.4 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.7 | 29.1 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.7 | 3.0 | REACTOME DNA STRAND ELONGATION | Genes involved in DNA strand elongation |
| 0.7 | 14.5 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.7 | 51.0 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.7 | 41.3 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.6 | 0.6 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.6 | 19.7 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.5 | 40.9 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.5 | 36.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.5 | 43.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.5 | 11.6 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.5 | 65.3 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.5 | 9.7 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.5 | 8.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.5 | 18.9 | REACTOME CLEAVAGE OF GROWING TRANSCRIPT IN THE TERMINATION REGION | Genes involved in Cleavage of Growing Transcript in the Termination Region |
| 0.4 | 2.7 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.4 | 5.4 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.4 | 1.7 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.4 | 6.8 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.4 | 3.7 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.4 | 13.1 | REACTOME TRNA AMINOACYLATION | Genes involved in tRNA Aminoacylation |
| 0.4 | 9.0 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.4 | 2.4 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.4 | 5.5 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.4 | 7.6 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.4 | 4.1 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.3 | 20.7 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.3 | 9.5 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.3 | 24.7 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.3 | 7.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.3 | 7.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.3 | 10.2 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.3 | 7.7 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.3 | 8.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.2 | 2.9 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.2 | 7.5 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.2 | 5.6 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.2 | 0.2 | REACTOME GLUCAGON SIGNALING IN METABOLIC REGULATION | Genes involved in Glucagon signaling in metabolic regulation |
| 0.2 | 9.4 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.2 | 1.9 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.2 | 4.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.2 | 7.0 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.2 | 4.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.2 | 4.4 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.1 | 8.5 | REACTOME HOST INTERACTIONS OF HIV FACTORS | Genes involved in Host Interactions of HIV factors |
| 0.1 | 3.6 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.1 | 2.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 3.2 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.1 | 10.8 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 3.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 2.7 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 2.7 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 0.7 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 2.8 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.1 | 6.0 | REACTOME INFLUENZA LIFE CYCLE | Genes involved in Influenza Life Cycle |
| 0.1 | 3.5 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 4.2 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.1 | 14.0 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 2.4 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.1 | 0.8 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.1 | 1.3 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 1.3 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 2.9 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.1 | 3.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 2.0 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 9.5 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 1.5 | REACTOME TRANSCRIPTION COUPLED NER TC NER | Genes involved in Transcription-coupled NER (TC-NER) |
| 0.1 | 1.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 2.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 2.5 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.0 | 1.1 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.8 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.4 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.8 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.0 | 0.3 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.4 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 2.3 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |